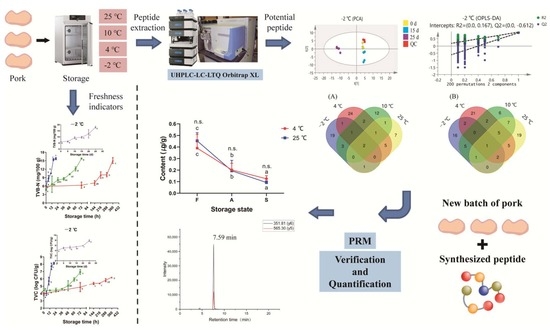
Identification of Potential Peptide Marker(s) for Evaluating Pork Meat Freshness via Mass Spectrometry-Based Peptidomics during Storage under Different Temperatures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Sample Preparation
2.2. Color Measurement
2.3. Microbiological Analysis
2.4. pH
2.5. Total Volatile Basic Nitrogen
2.6. Sample Preparation for Peptidomics
2.7. Peptide Identification and Data Analysis
2.8. Quantification of Potential Peptide
2.9. Statistical Analysis
3. Results and Discussion
3.1. Changes in Color
3.2. Microbiological Analysis
3.3. Changes in pH
3.4. Total Volatile Basic Nitrogen Analysis
3.5. Identification of Peptides
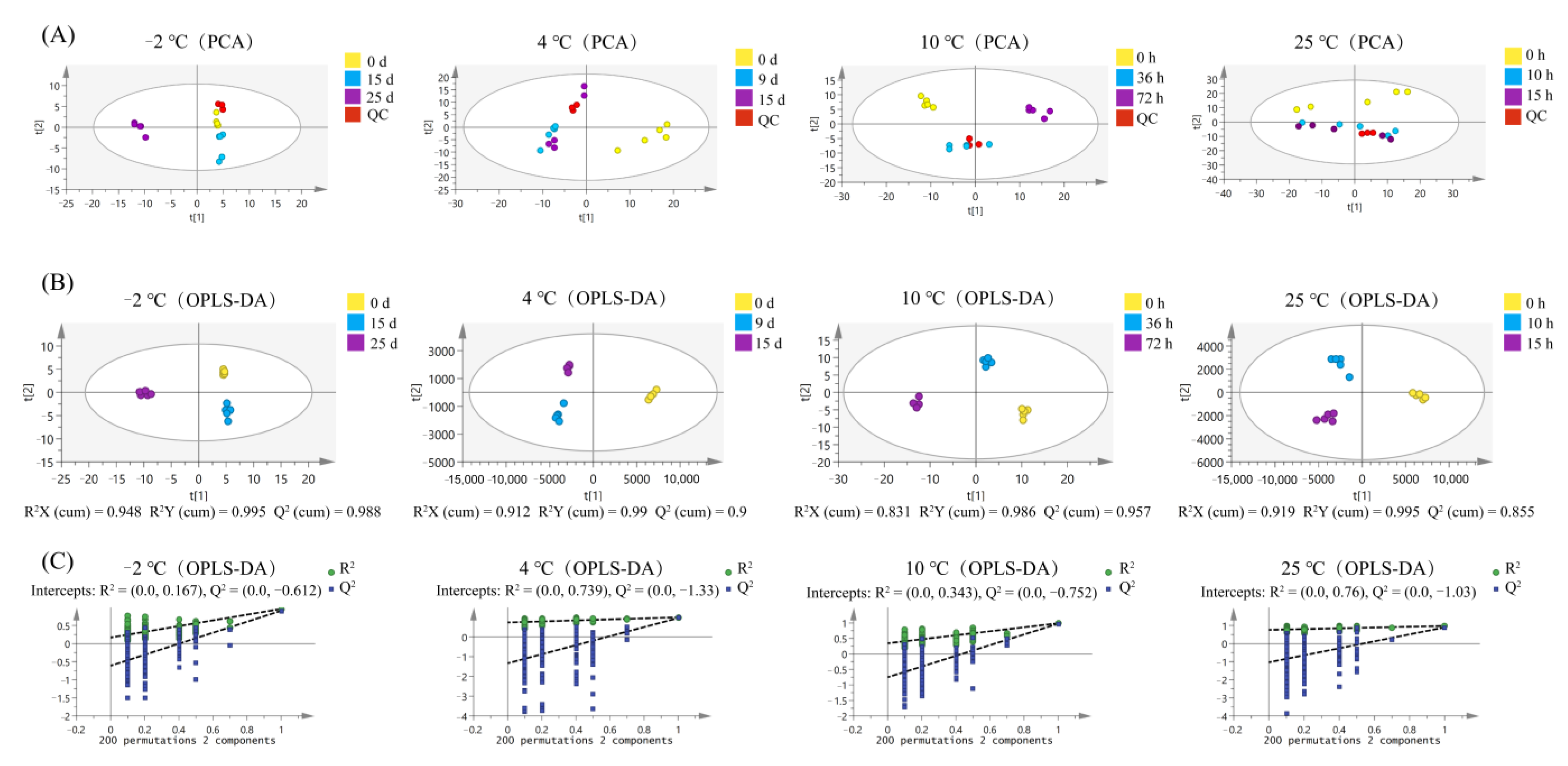
3.6. Multivariate Analyses of Pork Peptidome
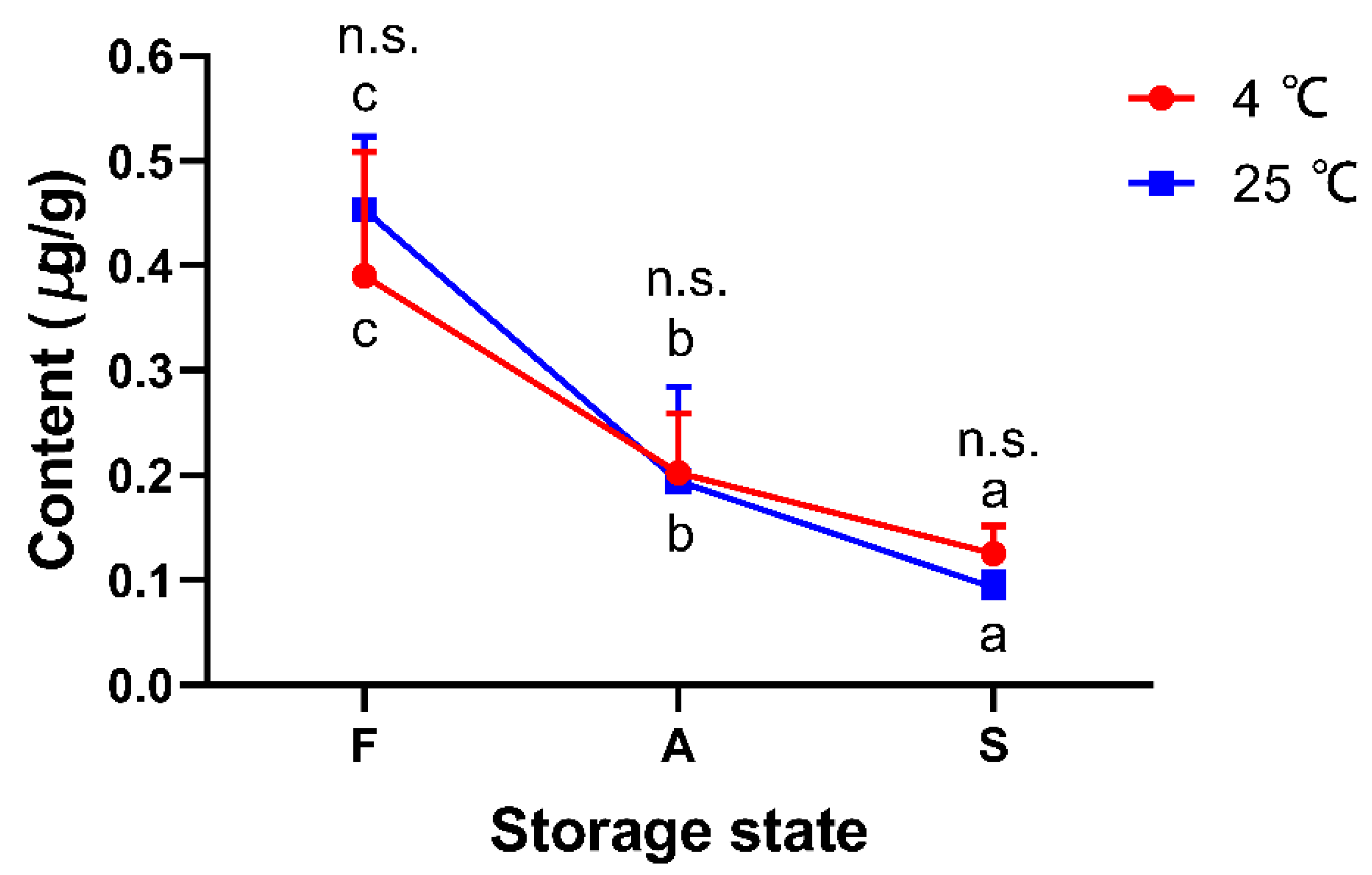
3.7. Selection of Potential Peptide Biomarkers
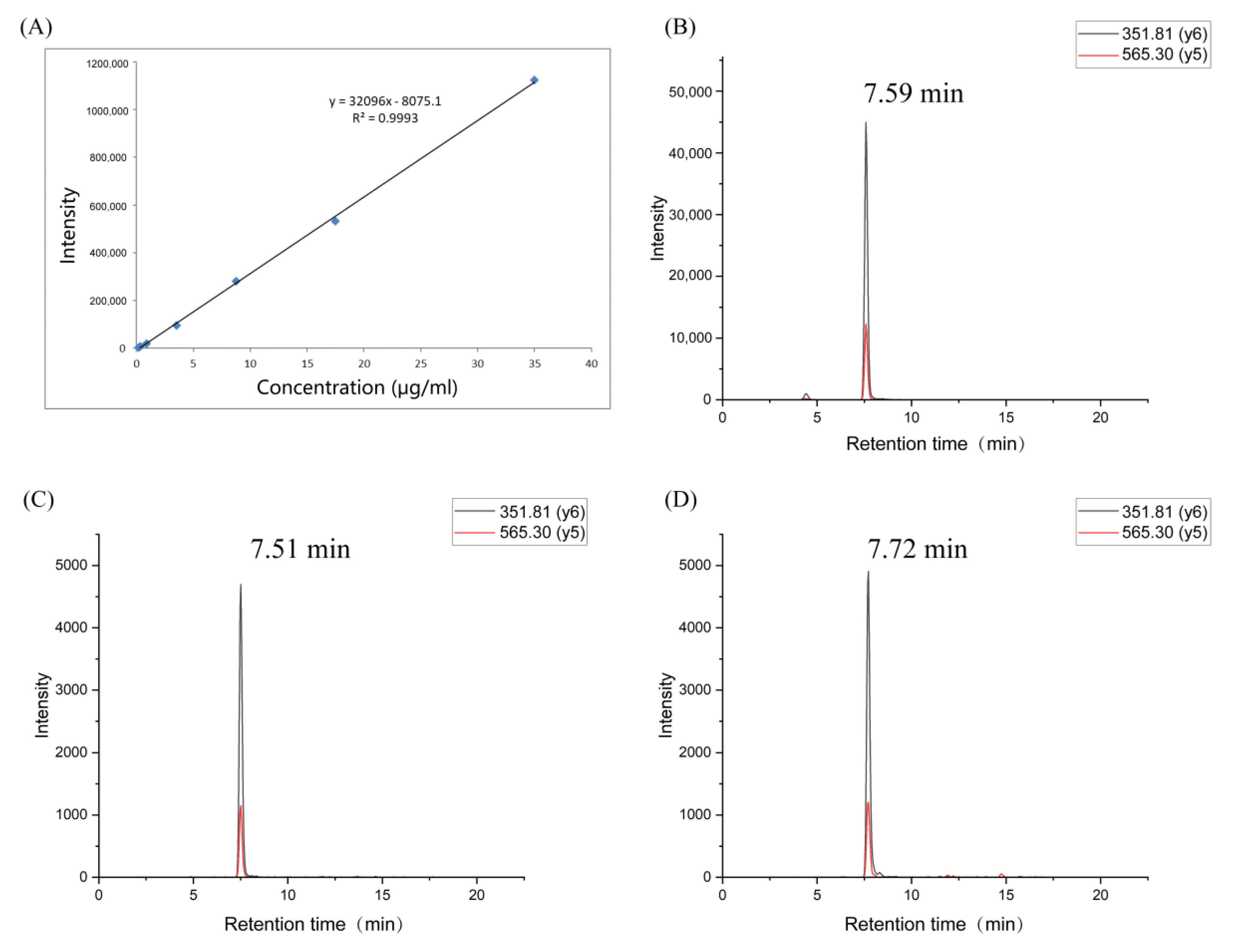
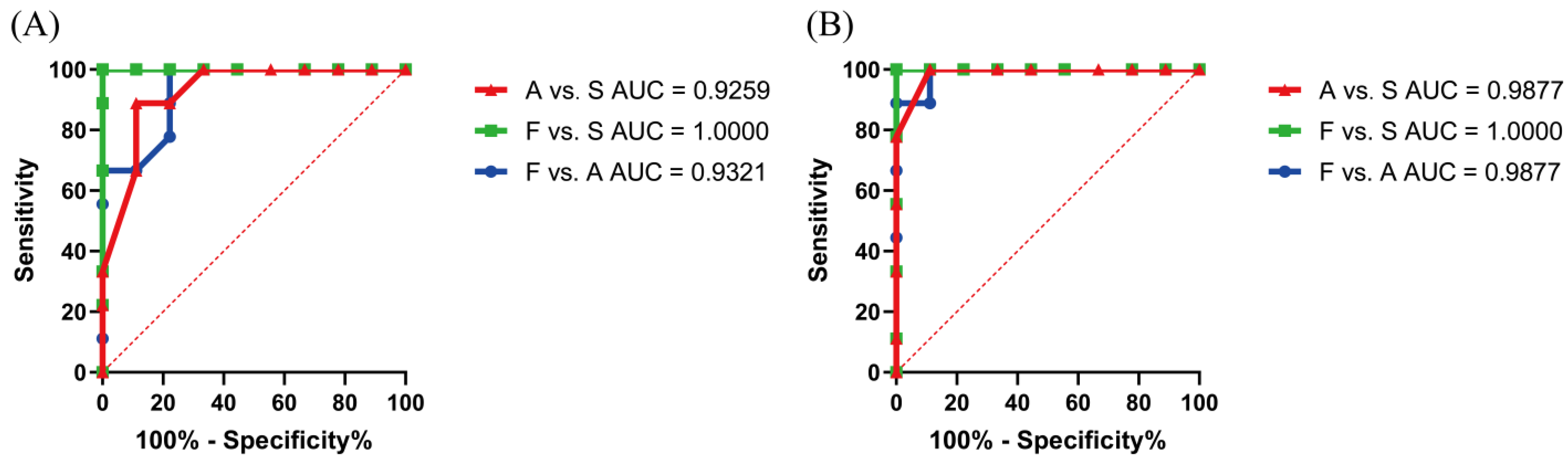
3.8. Verification of Potential Peptide Markers by Parallel Reaction Monitoring (PRM) Determination and Real-Life Sample Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, X.; Sun, X.; Wu, V.C.H.; Xie, J.; Pan, Y.; Zhao, Y.; Malakar, P.K. Predicting shelf-life of chilled pork sold in China. Food Control 2013, 32, 334–340. [Google Scholar] [CrossRef]
- Katiyo, W.; de Kock, H.L.; Coorey, R.; Buys, E.M. Sensory implications of chicken meat spoilage in relation to microbial and physicochemical characteristics during refrigerated storage. LWT 2020, 128, 109468. [Google Scholar] [CrossRef]
- Nychas, G.J.; Skandamis, P.N.; Tassou, C.C.; Koutsoumanis, K.P. Meat spoilage during distribution. Meat Sci. 2008, 78, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Zhao, J.; Luo, Z.; Lin, Q.; Luo, F.; Yang, T. Systematic evaluation of the physicochemical properties and the volatile flavors of yak meat during chilled and controlled freezing-point storage. J. Food Sci. Technol. 2020, 57, 1351–1361. [Google Scholar] [CrossRef]
- Lytou, A.E.; Nychas, G.-J.E.; Panagou, E.Z. Effect of pomegranate based marinades on the microbiological, chemical and sensory quality of chicken meat: A metabolomics approach. Int. J. Food Microbiol. 2018, 267, 42–53. [Google Scholar] [CrossRef]
- Fu, L.; Wang, A.; Zhang, H.; Zhou, Q.; Chen, F.; Su, W.; Yu, A.; Ji, Z.; Liu, Q. Analysis of chicken breast meat freshness with an electrochemical approach. J. Electroanal. Chem. 2019, 855, 113622. [Google Scholar] [CrossRef]
- Argyri, A.A.; Jarvis, R.M.; Wedge, D.; Xu, Y.; Panagou, E.Z.; Goodacre, R.; Nychas, G.-J.E. A comparison of Raman and FT-IR spectroscopy for the prediction of meat spoilage. Food Control 2013, 29, 461–470. [Google Scholar] [CrossRef]
- Chang, W.C.-W.; Wu, H.-Y.; Yeh, Y.; Liao, P.-C. Untargeted foodomics strategy using high-resolution mass spectrometry reveals potential indicators for fish freshness. Anal. Chim. Acta 2020, 1127, 98–105. [Google Scholar] [CrossRef]
- Aru, V.; Pisano, M.B.; Savorani, F.; Engelsen, S.B.; Cosentino, S.; Marincola, F.C. Metabolomics analysis of shucked mussels’ freshness. Food Chem. 2016, 205, 58–65. [Google Scholar] [CrossRef]
- Li, X.; Chen, Y.; Cai, L.; Xu, Y.; Yi, S.; Zhu, W.; Mi, H.; Li, J.; Lin, H. Freshness assessment of turbot (Scophthalmus maximus) by Quality Index Method (QIM), biochemical, and proteomic methods. LWT 2017, 78, 172–180. [Google Scholar] [CrossRef]
- Pellissery, A.J.; Vinayamohan, P.G.; Amalaradjou, M.A.R.; Venkitanarayanan, K. Spoilage bacteria and meat quality. In Meat Quality Analysis; Academic Press Ltd.: London, UK, 2020; pp. 307–334. [Google Scholar]
- Schrader, M. Origins, Technological Development, and Applications of Peptidomics. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2018; Volume 1719, pp. 3–39. [Google Scholar]
- Stachniuk, A.; Sumara, A.; Montowska, M.; Fornal, E. Peptide markers for distinguishing guinea fowl meat from that of other species using liquid chromatography-mass spectrometry. Food Chem. 2021, 345, 128810. [Google Scholar] [CrossRef] [PubMed]
- Kominami, Y.; Hayashi, T.; Tokihiro, T.; Ushio, H. Peptidomic analysis characterising proteolysis in thaw-aging of beef short plate. Food Chem. 2021, 3, 100051. [Google Scholar] [CrossRef] [PubMed]
- Bauchart, C.; Morzel, M.; Chambon, C.; Mirand, P.P.; Reynes, C.; Buffiere, C.; Remond, D. Peptides reproducibly released by in vivo digestion of beef meat and trout flesh in pigs. Br. J. Nutr. 2007, 98, 1187–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dallas, D.C.; Guerrero, A.; Khaldi, N.; Castillo, P.A.; Martin, W.F.; Smilowitz, J.T.; Bevins, C.L.; Barile, D.; German, J.B.; Lebrilla, C.B. Extensive in vivo Human Milk Peptidomics Reveals Specific Proteolysis Yielding Protective Antimicrobial Peptides. J. Proteome Res. 2013, 12, 2295–2304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mora, L.; Escudero, E.; Toldra, F. A peptidomic approach to study the contribution of added casein proteins to the peptide profile in Spanish dry-fermented sausages. Int. J. Food Microbiol. 2015, 212, 41–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montowska, M. Using Peptidomics to Determine the Authenticity of Processed Meat; Academic Press Ltd.: London, UK, 2017; pp. 225–240. [Google Scholar]
- Gallego, M.; Mora, L.; Toldrá, F. Peptidomics as a tool for quality control in dry-cured ham processing. J. Proteom. 2016, 147, 98–107. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-P.; Zhang, H.-W.; Zhang, X.-M.; Yu, F.-Q.-H.; Zhang, F.; Xue, C.-H.; Xue, Y.; Tang, Q.-J.; Li, Z.-J. Identification of potential peptide markers for the shelf-life of Pacific oysters (Crassostrea gigas) during anhydrous preservation via mass spectrometry-based peptidomics. LWT 2020, 134, 109922. [Google Scholar] [CrossRef]
- Hunt, M.C.; King, A. AMSA Meat Color Measurement Guidelines; American Meat Science Association: Champaign, IL, USA, 2012. [Google Scholar]
- Ye, K.; Ding, D.; Zhu, X.; Wu, Z.; Hu, Q.; Li, R. Modified atmosphere packaging with a small change in gas ratio could maintain pork quality during −3 degrees C storage. Food Control 2020, 109, 106943. [Google Scholar] [CrossRef]
- Lan, Y.; Shang, Y.; Song, Y.; Dong, Q. Changes in the quality of superchilled rabbit meat stored at different temperatures. Meat Sci. 2016, 117, 173–181. [Google Scholar] [CrossRef]
- Bassey, A.P.; Chen, Y.; Zhu, Z.; Odeyemi, O.A.; Frimpong, E.B.; Ye, K.; Li, C.; Zhou, G. Assessment of quality characteristics and bacterial community of modified atmosphere packaged chilled pork loins using 16S rRNA amplicon sequencing analysis. Food Res. Int. 2021, 145, 110412. [Google Scholar] [CrossRef]
- Azkargorta, M.; Escobes, I.; Iloro, I.; Elortza, F. Mass spectrometric identification of endogenous peptides. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2018; Volume 1719, pp. 59–70. [Google Scholar]
- Miao, Z.; Ding, K.; Jin, S.; Dai, L.; Dai, C.; Li, X. Using serum peptidomics to discovery the diagnostic marker for different stage of ulcerative colitis. J. Pharm. Biomed 2021, 193, 113725. [Google Scholar] [CrossRef] [PubMed]
- Clerens, S.; Thomas, A.; Gathercole, J.; Plowman, J.E.; Yu, T.Y.; Grosvenor, A.J.; Deb-Choudhury, S. Proteomic and peptidomic differences and similarities between four muscle types from New Zealand raised Angus steers. Meat Sci. 2016, 121, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, W.; Zan, J.; Wu, C.; Tan, W. Untargeted lipidomics reveals progression of early Alzheimer’s disease in APP/PS1 transgenic mice. Sci. Rep. 2020, 10, 14509. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Yin, X.; Wang, Y.; Chen, Q.; Kong, B. The prediction of specific spoilage organisms in Harbin red sausage stored at room temperature by multivariate statistical analysis. Food Control 2021, 123, 107701. [Google Scholar] [CrossRef]
- Liu, P.; Wang, S.; Zhang, H.; Wang, H.; Kong, B. Influence of glycated nitrosohaemoglobin prepared from porcine blood cell on physicochemical properties, microbial growth and flavour formation of Harbin dry sausages. Meat Sci. 2019, 148, 96–104. [Google Scholar] [CrossRef]
- Anas, M.; Ahmad, S.; Malik, A. Microbial Escalation in Meat and Meat Products and Its Consequences; Springer International Publishing AG: Cham, Switzerland, 2019; pp. 29–49. [Google Scholar]
- Zhou, C.; Wang, J.; Li, R.; Ye, K. High-throughput sequencing analysis of the bacterial community for assessing the differences in extraction methods of bacteria separation from chilled pork. LWT 2020, 134, 110213. [Google Scholar] [CrossRef]
- Ding, D.; Zhou, C.; Ge, X.; Ye, K.; Wang, P.; Bai, Y.; Zhou, G. The effect of different degrees of superchilling on shelf life and quality of pork during storage. J. Food Process Pres. 2020, 44, e14394. [Google Scholar] [CrossRef] [Green Version]
- Bhat, Z.F.; Pathak, V. Quality evaluation of mutton Harrisa during one week refrigerated storage. J. Food Sci. Tech. Mys. 2012, 49, 620–625. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Lee, S.; Lee, K.; Baek, S.; Seo, J. Development of a pH indicator composed of high moisture-absorbing materials for real-time monitoring of chicken breast freshness. Food Sci. Biotechnol 2017, 26, 37–42. [Google Scholar] [CrossRef]
- Koziol, K.; Maj, D.; Bieniek, J. Changes in the color and pH of rabbit meat in the aging process. Med. Weter 2015, 71, 104–108. [Google Scholar]
- Bassey, A.P.; Chen, Y.; Zhu, Z.; Odeyemi, O.A.; Gao, T.; Olusola, O.O.; Ye, K.; Li, C.; Zhou, G. Evaluation of spoilage indexes and bacterial community dynamics of modified atmosphere packaged super-chilled pork loins. Food Control. 2021, 130, 108383. [Google Scholar] [CrossRef]
- Liu, Q.; Kong, B.; Han, J.; Chen, Q.; He, X. Effects of superchilling and cryoprotectants on the quality of common carp (Cyprinus carpio) surimi: Microbial growth, oxidation, and physiochemical properties. LWT 2014, 57, 165–171. [Google Scholar] [CrossRef]
- Senapati, M.; Sahu, P.P. Meat quality assessment using Au patch electrode Ag-SnO2/SiO2/Si MIS capacitive gas sensor at room temperature. Food Chem. 2020, 324, 126893. [Google Scholar] [CrossRef] [PubMed]
- Strupat, K. Molecular weight determination of peptides and proteins by ESI and MALDI. In Methods in Enzymology; Springer International Publishing AG: Cham, Switzerland, 2005; Volume 405, pp. 1–36. [Google Scholar]
- Bhat, Z.F.; Morton, J.D.; Mason, S.L.; Bekhit, A.E.-D.A. Role of calpain system in meat tenderness: A review. Food Sci. Hum. Well 2018, 7, 196–204. [Google Scholar] [CrossRef]
- Pomponio, L.; Ertbjerg, P. The effect of temperature on the activity of μ- and m-calpain and calpastatin during post-mortem storage of porcine longissimus muscle. Meat Sci. 2012, 91, 50–55. [Google Scholar] [CrossRef]
- Xin, J.; Li, Z.; Li, X.; Li, M.; Li, G.; Rao, W.; Zhang, D. Meat color stability of ovine muscles is related to glycolytic dehydrogenase activities. J. Food Sci. 2018, 83, 2432–2438. [Google Scholar] [CrossRef]
- Rauniyar, N. Parallel Reaction Monitoring: A targeted experiment performed using high resolution and high mass accuracy mass spectrometry. Int. J. Mol. Sci. 2015, 16, 28566–28581. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Goo, J.M.; Jo, C.-H. Receiver operating characteristic (ROC) curve: Practical review for radiologists. Korean J. Radiol. 2004, 5, 11–18. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Z.; Dai, C.; Bassey, A.P.; Tang, C.; Han, Y.; Wang, C.; Zhou, G. Identification of Potential Peptide Marker(s) for Evaluating Pork Meat Freshness via Mass Spectrometry-Based Peptidomics during Storage under Different Temperatures. Foods 2022, 11, 1144. https://doi.org/10.3390/foods11081144
Wei Z, Dai C, Bassey AP, Tang C, Han Y, Wang C, Zhou G. Identification of Potential Peptide Marker(s) for Evaluating Pork Meat Freshness via Mass Spectrometry-Based Peptidomics during Storage under Different Temperatures. Foods. 2022; 11(8):1144. https://doi.org/10.3390/foods11081144
Chicago/Turabian StyleWei, Zhenqian, Chen Dai, Anthony P. Bassey, Changbo Tang, Yu Han, Chong Wang, and Guanghong Zhou. 2022. "Identification of Potential Peptide Marker(s) for Evaluating Pork Meat Freshness via Mass Spectrometry-Based Peptidomics during Storage under Different Temperatures" Foods 11, no. 8: 1144. https://doi.org/10.3390/foods11081144
APA StyleWei, Z., Dai, C., Bassey, A. P., Tang, C., Han, Y., Wang, C., & Zhou, G. (2022). Identification of Potential Peptide Marker(s) for Evaluating Pork Meat Freshness via Mass Spectrometry-Based Peptidomics during Storage under Different Temperatures. Foods, 11(8), 1144. https://doi.org/10.3390/foods11081144