The Fungal Microbiome of Wheat Flour Includes Potential Mycotoxin Producers
Abstract
:1. Introduction
2. Methods
2.1. Sampling and Study Overview
2.2. In-Vitro Isolation of Fungi and Molecular Identification
2.3. Metabarcoding Characterization of Fungal Communities
3. Results
3.1. Isolation and Identification of Fungi
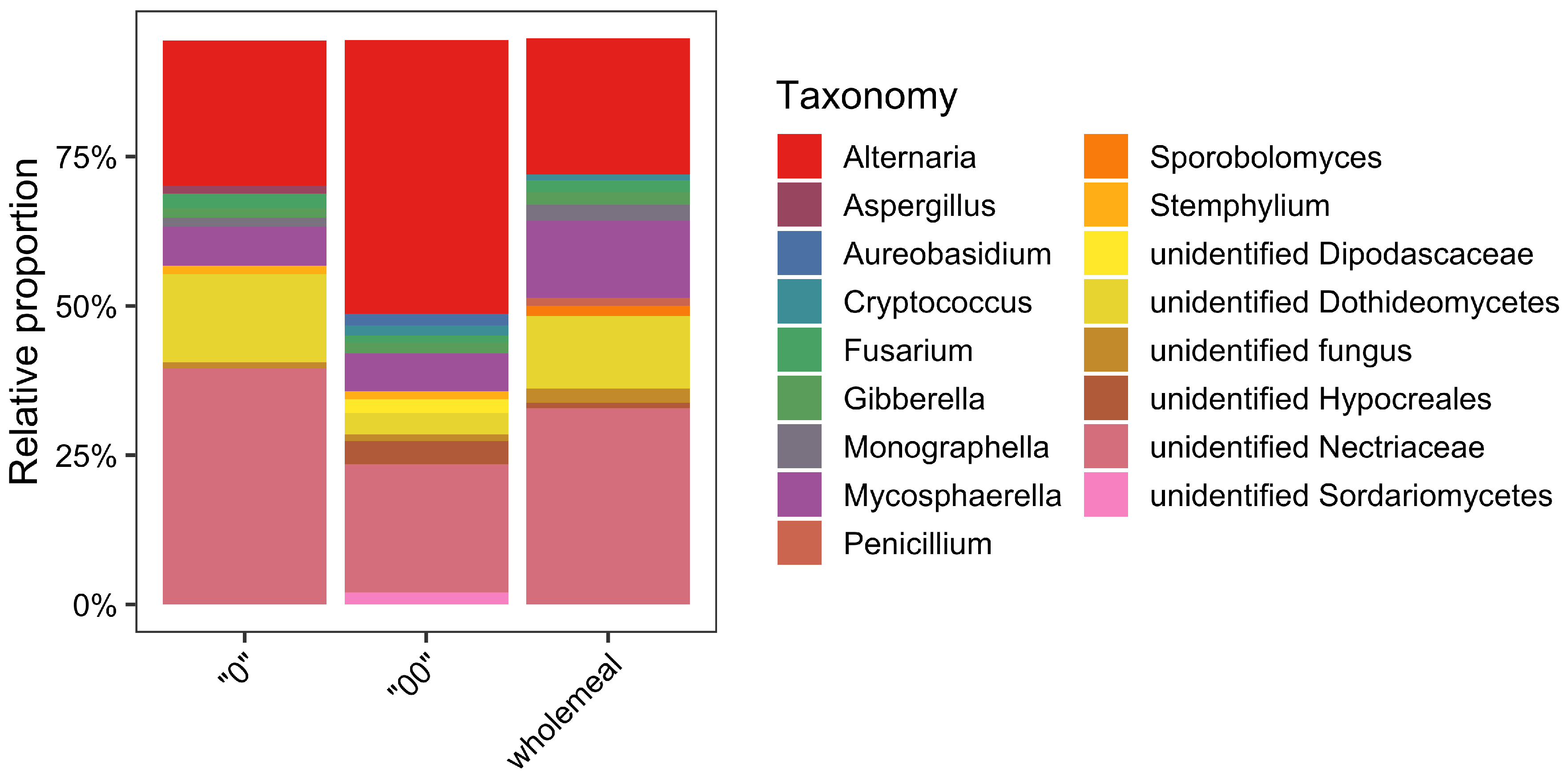
3.2. Metabarcoding Characterization of Fungal Communities
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Bullerman, L.B.; Bianchini, A. Food safety issues and the microbiology of cereals and cereal products. In Microbiologically Safe Foods; Wiley: Hoboken, NJ, USA, 2009; pp. 315–335. [Google Scholar]
- Doyle, M.P.; Diez-Gonzalez, F.; Hill, C. Food Microbiology: Fundamentals and Frontiers; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Miller, J.D. Fungi and mycotoxins in grain: Implications for stored product research. J. Stored Prod. Res. 1995, 31, 1–16. [Google Scholar] [CrossRef]
- Laca, A.; Mousia, Z.; Díaz, M.; Webb, C.; Pandiella, S.S. Distribution of microbial contamination within cereal grains. J. Food Eng. 2006, 72, 332–338. [Google Scholar] [CrossRef]
- Bhat, R.; Ramakrishna, Y.; Beedu, S.; Munshi, K. Outbreak of trichothecene mycotoxicosis associated with consumption of mould-damaged wheat products in Kashmir Valley, India. Lancet 1989, 333, 35–37. [Google Scholar] [CrossRef]
- Li, F.Q.; Luo, X.Y.; Yoshizawa, T. Mycotoxins (trichothecenes, zearalenone and fumonisins) in cereals associated with human red-mold intoxications stored since 1989 and 1991 in China. Nat. Toxins 1999, 7, 93–97. [Google Scholar] [CrossRef]
- Pestka, J.J.; Smolinski, A.T. Deoxynivalenol: Toxicology and potential effects on humans. J. Toxicol. Environ. Health Part B 2005, 8, 39–69. [Google Scholar] [CrossRef]
- Škrbić, B.; Živančev, J.; Đurišić-Mladenović, N.; Godula, M. Principal mycotoxins in wheat flour from the Serbian market: Levels and assessment of the exposure by wheat-based products. Food Control 2012, 25, 389–396. [Google Scholar] [CrossRef]
- Amirahmadi, M.; Shoeibi, S.; Rastegar, H.; Elmi, M.; Mousavi Khaneghah, A. Simultaneous analysis of mycotoxins in corn flour using LC/MS-MS combined with a modified QuEChERS procedure. Toxin Rev. 2018, 37, 187–195. [Google Scholar] [CrossRef]
- Dos Santos, I.D.; Pizzutti, I.R.; Dias, J.V.; Fontana, M.E.Z.; Souza, D.M.; Cardoso, C.D. Mycotoxins in wheat flour: Occurrence and co-occurrence assessment in samples from Southern Brazil. Food Addit. Contam. Part B 2021, 14, 151–161. [Google Scholar] [CrossRef]
- Cheli, F.; Pinotti, L.; Rossi, L.; Dell’Orto, V. Effect of milling procedures on mycotoxin distribution in wheat fractions: A review. LWT-Food Sci. Technol. 2013, 54, 307–314. [Google Scholar] [CrossRef]
- Palpacelli, V.; Beco, L.; Ciani, M. Vomitoxin and zearalenone content of soft wheat flour milled by different methods. J. Food Prot. 2007, 70, 509–513. [Google Scholar] [CrossRef]
- Boudra, H.; Le Bars, P.; Le Bars, J. Thermostability of ochratoxin A in wheat under two moisture conditions. Appl. Environ. Microbiol. 1995, 61, 1156–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, L.S.; Hlywka, J.J.; Senthil, K.R.; Bullerman, L.B.; Musser, S.M. Effects of time, temperature, and pH on the stability of fumonisin B1 in an aqueous model system. J. Agric. Food Chem. 1996, 44, 906–912. [Google Scholar] [CrossRef]
- Jackson, L.S.; Hlywka, J.J.; Senthil, K.R.; Bullerman, L.B. Effects of thermal processing on the stability of fumonisin B2 in an aqueous system. J. Agric. Food Chem. 1996, 44, 1984–1987. [Google Scholar] [CrossRef]
- Pineda-Valdes, G.; Bullerman, L.B. Thermal stability of moniliformin at varying temperature, pH, and time in an aqueous environment. J. Food Prot. 2000, 63, 1598–1601. [Google Scholar] [CrossRef] [PubMed]
- Ryu, D.; Hanna, M.A.; Eskridge, K.M.; Bullerman, L.B. Heat stability of zearalenone in an aqueous buffered model system. J. Agric. Food Chem. 2003, 51, 1746–1748. [Google Scholar] [CrossRef]
- Mesterházy, Á.; Oláh, J.; Popp, J. Losses in the grain supply chain: Causes and solutions. Sustainability 2020, 12, 2342. [Google Scholar] [CrossRef] [Green Version]
- Ntuli, V.; Mekbib, S.B.; Asita, A.; Molebatsi, N.; Makotoko, M.; Chatanga, P. Microbial and physicochemical characterization of maize and wheat flour from a milling company, Lesotho. Internet J. Food Saf. 2013, 15, 11–19. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Prigigallo, M.; Mosca, S.; Cacciola, S.; Cooke, D.; Schena, L. Molecular analysis of Phytophthora diversity in nursery-grown ornamental and fruit plants. Plant Pathol. 2015, 64, 1308–1319. [Google Scholar] [CrossRef] [Green Version]
- Abe, A.; Asano, K.; Sone, T. A molecular phylogeny-based taxonomy of the genus Rhizopus. Biosci. Biotechnol. Biochem. 2010, 74, 1325–1331. [Google Scholar] [CrossRef] [Green Version]
- Comby, M.; Lacoste, S.; Baillieul, F.; Profizi, C.; Dupont, J. Spatial and temporal variation of cultivable communities of co-occurring endophytes and pathogens in wheat. Front. Microbiol. 2016, 7, 403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irinyi, L.; Lackner, M.; De Hoog, G.S.; Meyer, W. DNA barcoding of fungi causing infections in humans and animals. Fungal Biol. 2016, 120, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Jayasiri, S.; Hyde, K.; Jones, E.; Jeewon, R.; Ariyawansa, H.; Bhat, J.; Camporesi, E.; Kang, J. Taxonomy and multigene phylogenetic evaluation of novel species in Boeremia and Epicoccum with new records of Ascochyta and Didymella (Didymellaceae). Mycosphere 2017, 8, 1080–1101. [Google Scholar] [CrossRef]
- Walther, G.; Pawłowska, J.; Alastruey-Izquierdo, A.; Wrzosek, M.; Rodriguez-Tudela, J.; Dolatabadi, S.; Chakrabarti, A.; De Hoog, G. DNA barcoding in Mucorales: An inventory of biodiversity. Pers. Mol. Phylogeny Evol. Fungi 2013, 30, 11–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crous, P.W.; Groenewald, J.Z. A phylogenetic re-evaluation of Arthrinium. IMA Fungus 2013, 4, 133–154. [Google Scholar] [CrossRef] [Green Version]
- Sandoval-Denis, M.; Guarnaccia, V.; Polizzi, G.; Crous, P. Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Pers.-Mol. Phylogeny Evol. Fungi 2018, 40, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; De Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS 2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Thrane, U.; Samson, R.A. Mycotoxin producers. In Food Mycology; CRC Press: Boca Raton, FL, USA, 2007; pp. 149–174. [Google Scholar]
- Moretti, A.; Logrieco, A.F.; Susca, A. Mycotoxins: An underhand food problem. In Mycotoxigenic Fungi; Springer: New York, NY, USA, 2017; pp. 3–12. [Google Scholar]
- Weidenbörner, M.; Wieczorek, C.; Appel, S.; Kunz, B. Whole wheat and white wheat flour—The mycobiota and potential mycotoxins. Food Microbiol. 2000, 17, 103–107. [Google Scholar] [CrossRef]
- Alhussaini, M.S. Mycobiota of wheat flour and detection of α-amylase and L-asparaginase enzymes. Life Sci. J. 2013, 10, 1112–1122. [Google Scholar]
- Hassan, Z.U.; Al-Thani, R.F.; Migheli, Q.; Jaoua, S. Detection of toxigenic mycobiota and mycotoxins in cereal feed market. Food Control 2018, 84, 389–394. [Google Scholar] [CrossRef]
- Alborch, L.; Bragulat, M.; Castellá, G.; Abarca, M.; Cabañes, F. Mycobiota and mycotoxin contamination of maize flours and popcorn kernels for human consumption commercialized in Spain. Food Microbiol. 2012, 32, 97–103. [Google Scholar] [CrossRef]
- Hafez, S.I.I.A.; Sater, M.A.A.; Hussein, N.A.G.; Amery, E.A.W. Fungal diversity associated with pearl millet (Pennisetum glaucum L.) grains from Taiz governorate, Yemen and their amylase production. J. Microbiol. Biotechnol. Food Sci. 2021, 2021, 118–123. [Google Scholar]
- Tralamazza, S.M.; Bemvenuti, R.H.; Zorzete, P.; de Souza Garcia, F.; Corrêa, B. Fungal diversity and natural occurrence of deoxynivalenol and zearalenone in freshly harvested wheat grains from Brazil. Food Chem. 2016, 196, 445–450. [Google Scholar] [CrossRef]
- Covarelli, L.; Beccari, G.; Prodi, A.; Generotti, S.; Etruschi, F.; Juan, C.; Ferrer, E.; Mañes, J. Fusarium species, chemotype characterisation and trichothecene contamination of durum and soft wheat in an area of central Italy. J. Sci. Food Agric. 2015, 95, 540–551. [Google Scholar] [CrossRef]
- Dos Santos, J.L.P.; Bernardi, A.O.; Morassi, L.L.P.; Silva, B.S.; Copetti, M.V.; Sant’Ana, A.S. Incidence, populations and diversity of fungi from raw materials, final products and air of processing environment of multigrain whole meal bread. Food Res. Int. 2016, 87, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Perrone, G.; Susca, A. Penicillium species and their associated mycotoxins. In Mycotoxigenic Fungi; Springer: New York, NY, USA, 2017; pp. 107–119. [Google Scholar]
- Cabañes, F.J.; Bragulat, M.R.; Castellá, G. Ochratoxin A producing species in the genus Penicillium. Toxins 2010, 2, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Perrone, G.; Gallo, A. Aspergillus species and their associated mycotoxins. In Mycotoxigenic Fungi; Springer: New York, NY, USA, 2017; pp. 33–49. [Google Scholar]
- Varga, J.; Frisvad, J.C.; Samson, R. Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Stud. Mycol. 2011, 69, 57–80. [Google Scholar] [CrossRef] [PubMed]
- Escrivá, L.; Oueslati, S.; Font, G.; Manyes, L. Alternaria mycotoxins in food and feed: An overview. J. Food Qual. 2017, 2017, 1569748. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.T.; Kim, J.; Jeon, S.J.; Lee, C.W.; Magan, N.; Lee, H.B. Mycotoxin production of Alternaria strains isolated from Korean barley grains determined by LC-MS/MS. Int. J. Food Microbiol. 2018, 268, 44–52. [Google Scholar] [CrossRef] [Green Version]
- Fernández Pinto, V.E.; Patriarca, A. Alternaria species and their associated mycotoxins. In Mycotoxigenic Fungi; Springer: New York, NY, USA, 2017; pp. 13–32. [Google Scholar]
- Ofek-Lalzar, M.; Gur, Y.; Ben-Moshe, S.; Sharon, O.; Kosman, E.; Mochli, E.; Sharon, A. Diversity of fungal endophytes in recent and ancient wheat ancestors Triticum dicoccoides and Aegilops sharonensis. FEMS Microbiol. Ecol. 2016, 92, fiw152. [Google Scholar] [CrossRef] [Green Version]
- Al-Sadi, A.M.; Al-Mazroui, S.; Phillips, A. Evaluation of culture-based techniques and 454 pyrosequencing for the analysis of fungal diversity in potting media and organic fertilizers. J. Appl. Microbiol. 2015, 119, 500–509. [Google Scholar] [CrossRef]
- van Elsas, J.D.; Duarte, G.F.; Keijzer-Wolters, A.; Smit, E. Analysis of the dynamics of fungal communities in soil via fungal-specific PCR of soil DNA followed by denaturing gradient gel electrophoresis. J. Microbiol. Methods 2000, 43, 133–151. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Li Destri Nicosia, M.G.; Cacciola, S.O.; Droby, S.; Schena, L. Metabarcoding analysis of fungal diversity in the phyllosphere and carposphere of olive (Olea europaea). PLoS ONE 2015, 10, e0131069. [Google Scholar] [CrossRef] [Green Version]
| Sequence Types (STs) | Associated Species | Flour Type |
|---|---|---|
| ALT1, ALT3 | Alternaria sp. | “0”, “00”, wholemeal |
| ALT2 | Alternaria infectoria | “00” |
| ARTH1 | Arthrinium arundinis | “0”, “00” |
| ASP1 | Aspergillus fasciculatus, A. kambarensis, A. oryzae | “00” |
| ASP2 | Aspergillus clavatus, A. apicalis | “00” |
| CHAET1 | Chaetomium globosum | “00” |
| CLA1, CLA2, CLA3, CLA4 | Cladosporium sp. | “0”, “00” |
| EPI1 | Epicoccum nigrum | “00” |
| FUS1 | Fusarium oxysporum | “00” |
| LICH1 | Lichtheimia corymbifera | “00” |
| MUC1, MUC3 | Mucor circinelloides | “00” |
| MUC2 | Mucor sp. | “00” |
| PEN1 | Penicillium sp. | “0”, wholemeal |
| PEN8 | Penicillium viridicatum, P. polonicum | “0” |
| PEN12 | Penicillium aurantiogriseum | “0”, “00”, wholemeal |
| PEN7 | Penicillium sp. | “0”, “00” |
| PEN9 | Penicillium albocoremium, P. thymicola | “00” |
| PEN6 | Penicillium verrucosum | “0”, “00” |
| PEN15 | Penicillium biforme, P. commune, P. solitum | “00” |
| PEN4 | Penicillium confertum, P. flavigenum | “00” |
| PEN2, PEN13 | Penicillium allii-sativi, P. chrysogenum | “00”, wholemeal |
| PEN10, PEN17 | Penicillium griseofulvum | “0”, “00”, wholemeal |
| PEN5, PEN16 | Penicillium sp. | “00”, wholemeal |
| PEN14 | Penicillium brevicompactum | “00” |
| PEN11 | Penicillium citrinum | “00” |
| RIZH1 | Rhizopus oryzae | “00” |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minutillo, S.A.; Ruano-Rosa, D.; Abdelfattah, A.; Schena, L.; Malacrinò, A. The Fungal Microbiome of Wheat Flour Includes Potential Mycotoxin Producers. Foods 2022, 11, 676. https://doi.org/10.3390/foods11050676
Minutillo SA, Ruano-Rosa D, Abdelfattah A, Schena L, Malacrinò A. The Fungal Microbiome of Wheat Flour Includes Potential Mycotoxin Producers. Foods. 2022; 11(5):676. https://doi.org/10.3390/foods11050676
Chicago/Turabian StyleMinutillo, Serena A., David Ruano-Rosa, Ahmed Abdelfattah, Leonardo Schena, and Antonino Malacrinò. 2022. "The Fungal Microbiome of Wheat Flour Includes Potential Mycotoxin Producers" Foods 11, no. 5: 676. https://doi.org/10.3390/foods11050676
APA StyleMinutillo, S. A., Ruano-Rosa, D., Abdelfattah, A., Schena, L., & Malacrinò, A. (2022). The Fungal Microbiome of Wheat Flour Includes Potential Mycotoxin Producers. Foods, 11(5), 676. https://doi.org/10.3390/foods11050676









