Study on Rice Grain Mildewed Region Recognition Based on Microscopic Computer Vision and YOLO-v5 Model
Abstract
1. Introduction
2. Methods
2.1. Simulated Storage of Rice after Inoculation
- (1)
- Select a certain number of clean and mildew-free rice grains (indica rice, purchased from Huainan, Anhui Province, China). Put the sample grains into an oven and bake them at 80 °C for 4 h to kill the original field molds attached to the rice grains. Put the dried sample grains into a set of 90-mm round petri dishes (15 g grains for each petri dish).
- (2)
- Inoculate the three mold strains into the potato glucose agar (PDA) medium separately and activate them at 28 °C for 3 days. Elute the activated colonies with sterile distilled water to prepare spore suspension samples, measure the spore concentrations in the spore suspension samples using the plate counting method, and dilute the spore suspension samples of three molds to 1.5 × 104 CFU/mL.
- (3)
- Take 30 petri dishes containing 15 g of rice grains. Inoculate 1.5 mL of spore suspension to the sample grains in each petri dish (each kind of spore suspension will inoculate 10 Petri dishes). Add 1 mL of sterile water into each petri dish and shake the Petri dish to allow the grains to fully absorb the water. After inoculation, the moisture content of the rice grains will be greater than 20%, thus creating a suitable condition for simulating accelerated mold growth in rice grains in a highly humid environment. Then, place the inoculated sample grains in a constant temperature and humidity incubator and simulate rice storage under the conditions of 28 °C and 90% relative humidity. Take out five grain samples randomly from each petri dish every day to test the degree of mildew. The simulated storage will last 13 days until the grains reach a high mildew degree. During the course of the simulated storage, 1950 sample rice grains with different contamination levels of A. niger, P. citrinum, and A. cinerea are obtained (650 samples for each mold strain).
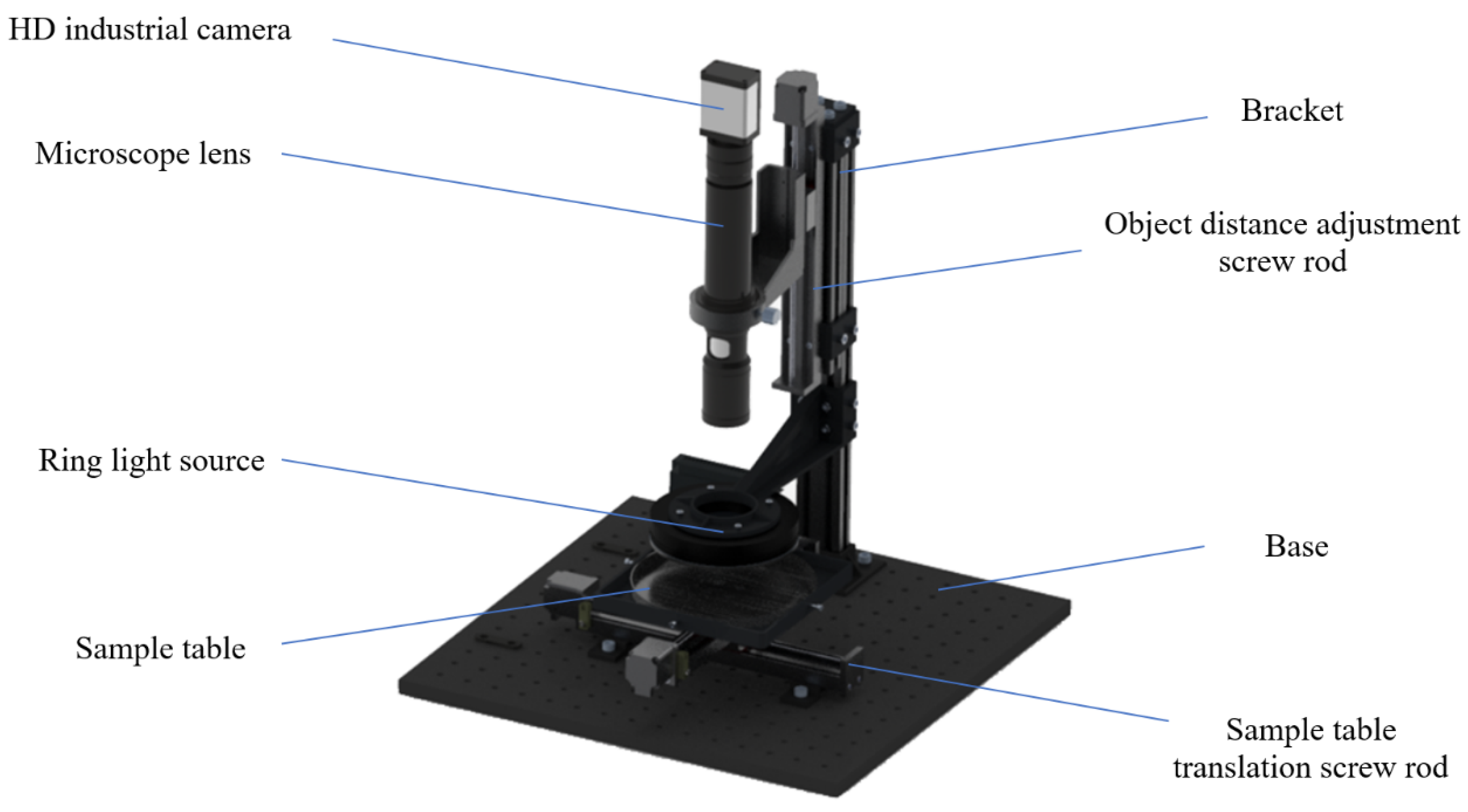
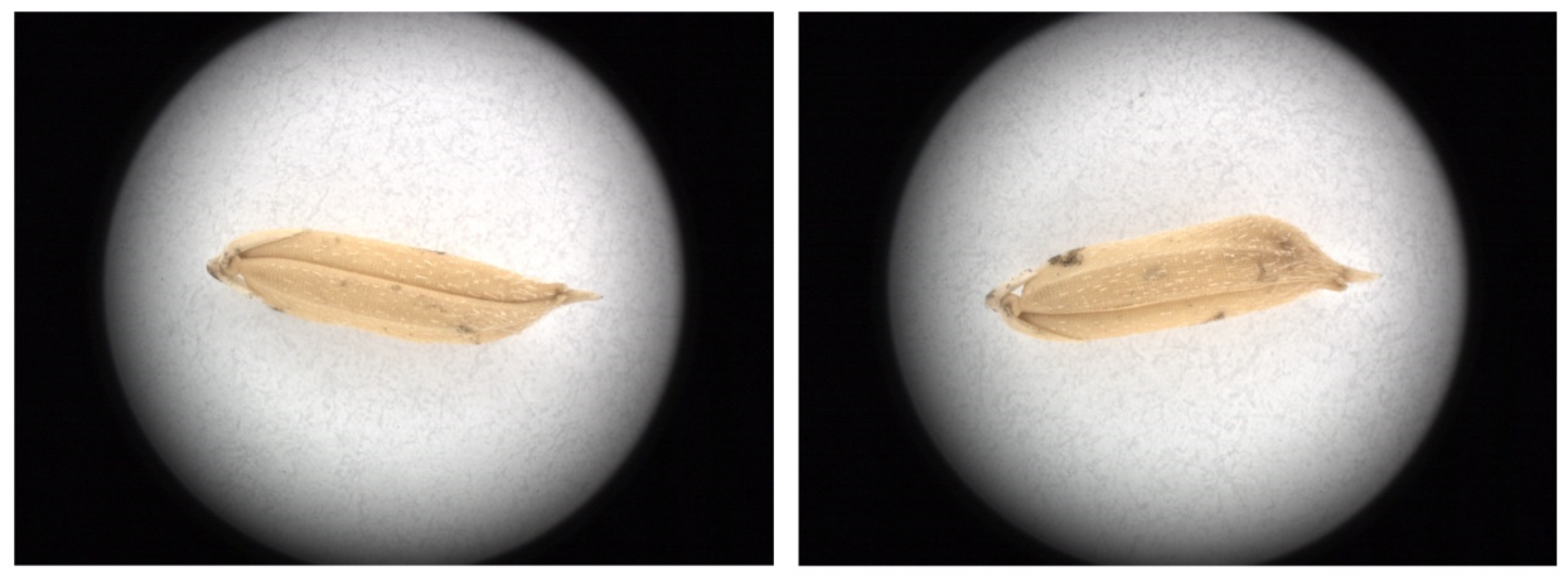
2.2. Acquisition of Rice Grain Microscopic Image
2.3. Pre-Process of Microscopic Images of Rice Grains
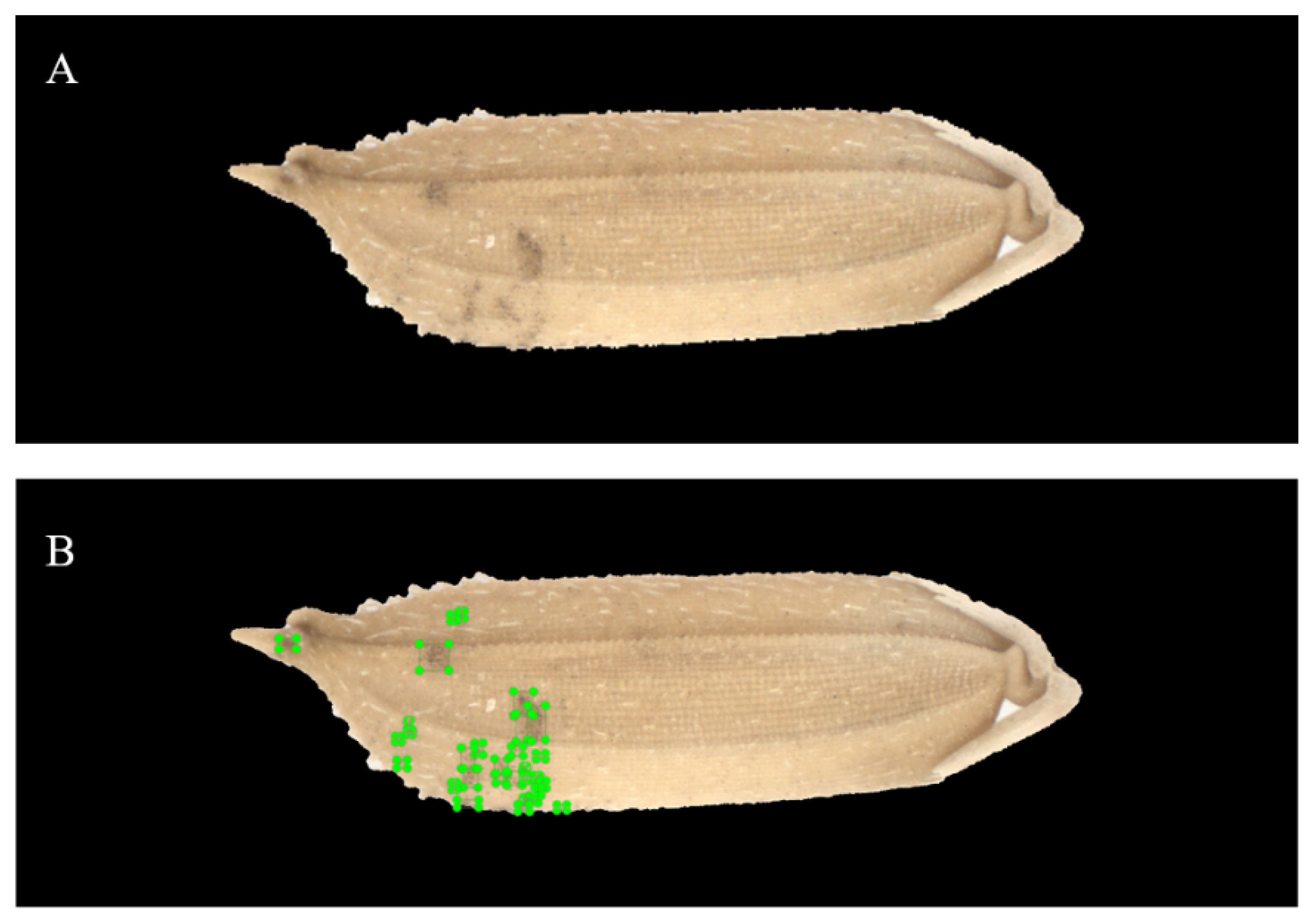
2.4. Image Marking
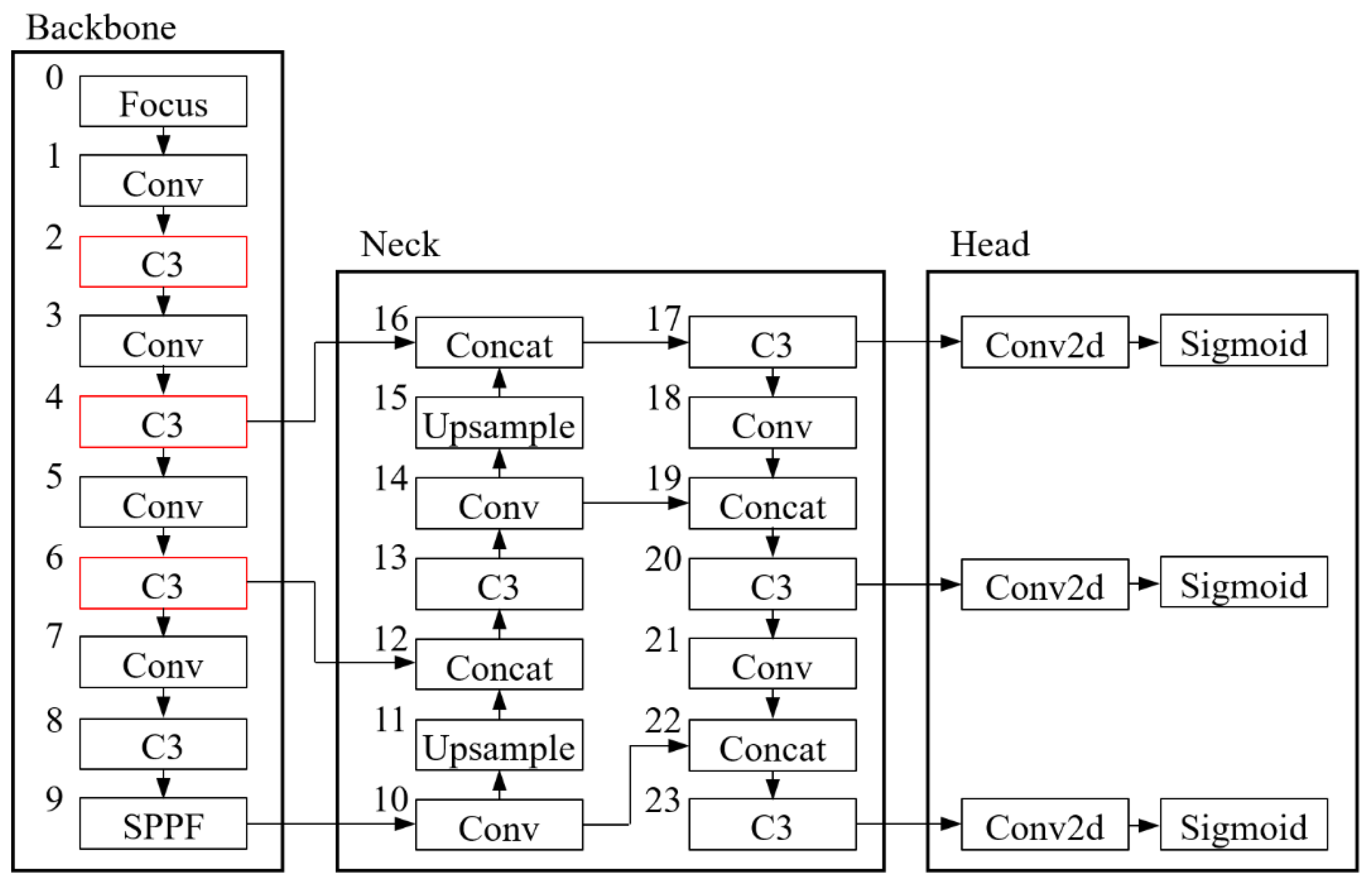
2.5. Model Establishment
2.6. Analysis of the Relationship between MAI and TVC of Rice Grain
3. Results
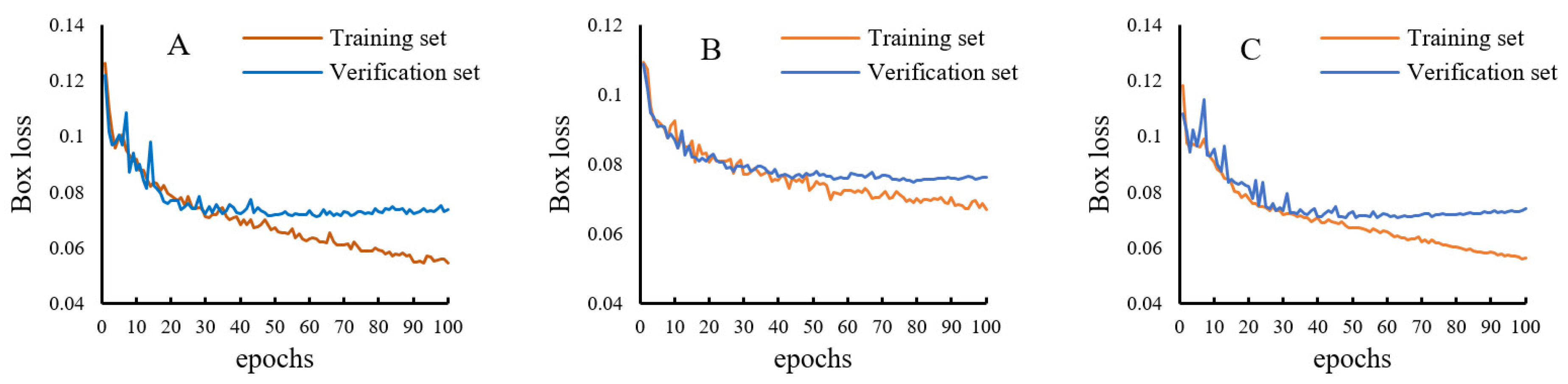
3.1. Variation of Box Loss during Model Training
3.2. Accuracy of Mildewed Region Detection Model
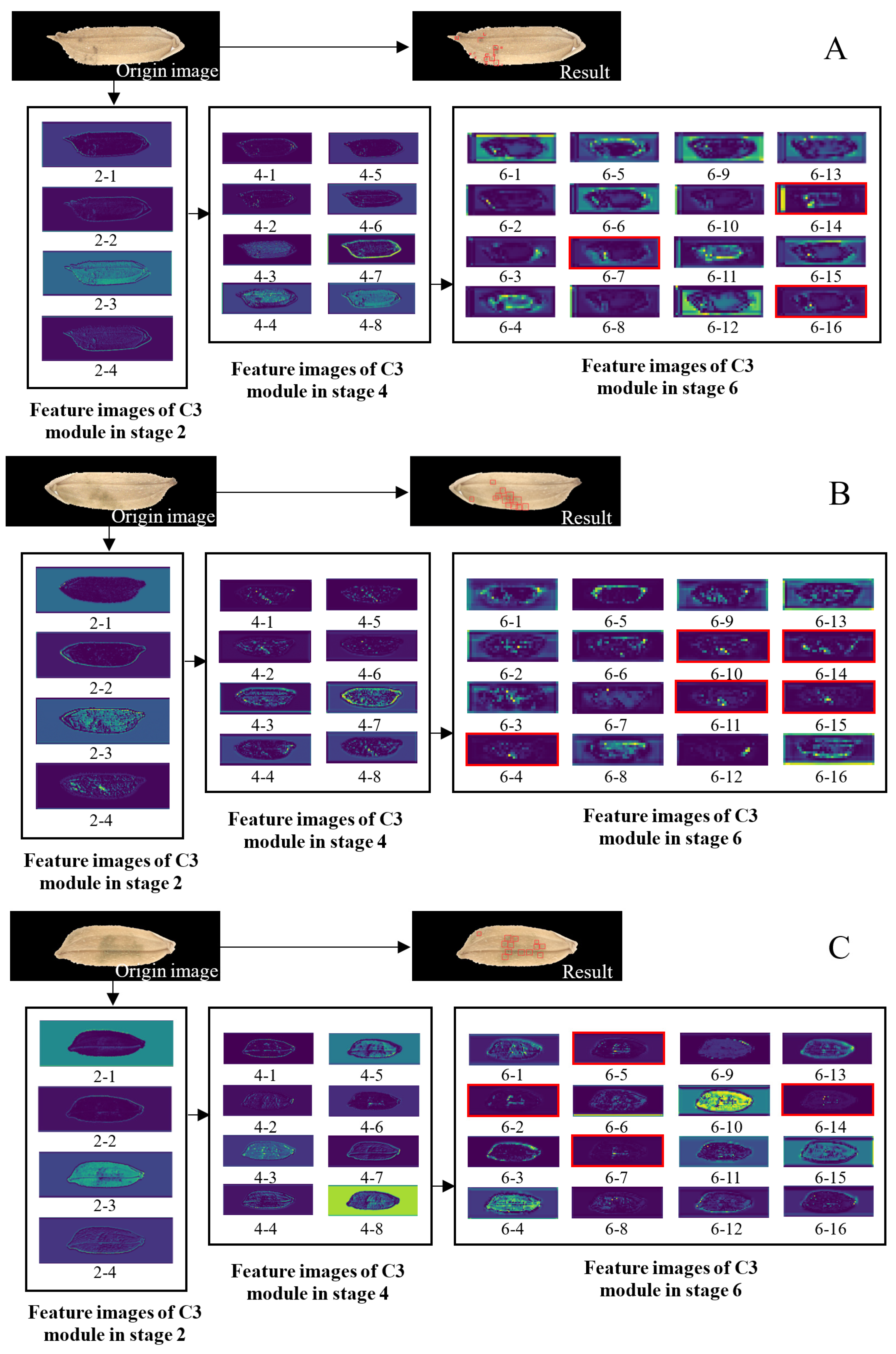
3.3. Analysis of Feature Images in the Middle Layer of the Model
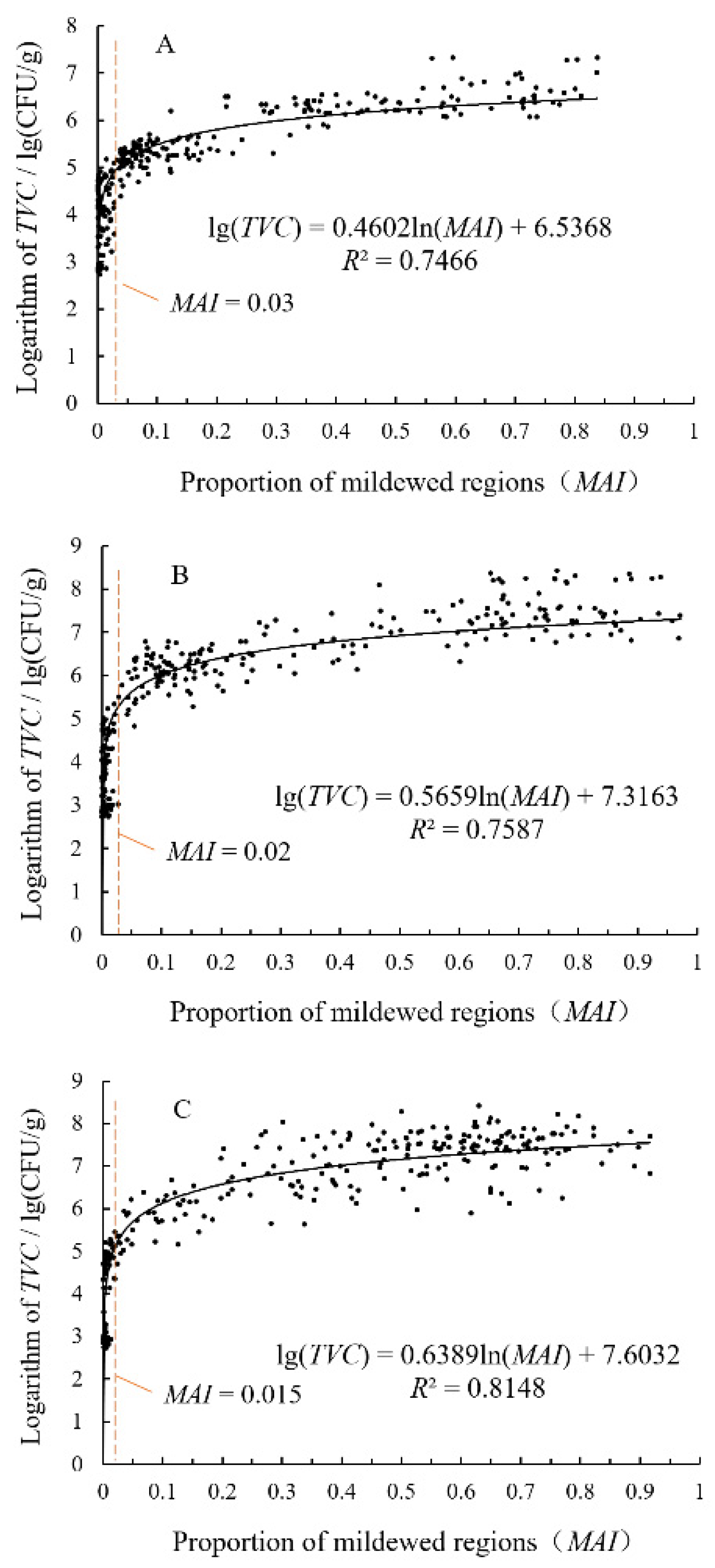
3.4. Analysis of Relationship between TVC and MAI of Rice Grain
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fredlund, E.; Thim, A.M.; Gidlund, A.; Brostedt, S.; Nyberg, M.; Olsen, M. Moulds and mycotoxins in rice from the Swedish retail market. Food Addit. Contam. 2009, 26, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Singh, C.B.; Jayas, D.S.; Paliwal, J.; White, N. Fungal damage detection in wheat using short-wave near-infrared hyperspectral and digital colour imaging. Int. J. Food Prop. 2012, 15, 11–24. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, C.; Sun, J.; Cui, Y.; Li, Q.; Jia, F.; Zheng, X. Rapid Non-destructive Detection for Molds Colony of Paddy Rice Based on Near Infrared Spectroscop. J. Northeast Agric. Univ. 2014, 21, 54–60. [Google Scholar]
- Siripatrawan, U.; Makino, Y. Monitoring fungal growth on brown rice grains using rapid and non-destructive hyperspectral imaging. Int. J. Food Microbiol. 2015, 199, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Barbedo, J.G.A.; Tibola, C.S.; Lima, M.I.P.L. Deoxynivalenol screening in wheat kernels using hyperspectral imaging. Biosyst. Eng. 2017, 155, 24–32. [Google Scholar] [CrossRef]
- Shen, F.; Wu, Q.; Tang, P.; Shao, X.; Jiang, D. Attenuated Total Reflectance-Fourier Transform Infrared Spectroscopy (ATR-FTIR) for Rapid Detection of Aflatoxin B1 in Brown Rice. J. Food Sci. 2016, 37, 187–191. [Google Scholar]
- Shen, F.; Wei, Y.; Zhang, B.; Shao, X.; Song, W.; Yang, H. Rapid Detection of Harmful Mold Infection in Rice by Near Infrared Spectroscopy. Spectrosc. Spect. Anal. 2018, 38, 3748–3752. [Google Scholar]
- Chu, X.; Wang, W.; Ni, X.; Li, C.; Li, Y. Classifying maize kernels naturally infected by fungi using near-infrared hyperspectral imaging. Infrared Phys. Technol. 2020, 105, 103242. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, K.; Du, L.; Yuan, J.; Tu, K.; Pan, L. Identification and classification of fungal colonies in moldy paddy based on computer vision. Trans. ASABE 2018, 61, 1497–1504. [Google Scholar] [CrossRef]
- Sun, K.; Wang, Z.; Tu, K.; Wang, S.; Pan, L. Recognition of mould colony on unhulled paddy based on computer vision using conventional machine-learning and deep learning techniques. Sci. Rep. 2016, 6, 37994. [Google Scholar] [CrossRef]
- Payman, S.H.; Bakhshipour, A.; Zareiforoush, H. Development of an expert vision-based system for inspecting rice quality indices. Qual. Assur. Saf. Crops 2018, 10, 103–114. [Google Scholar] [CrossRef]
- Chen, S.; Xiong, J.; Guo, W.; Bu, R.; Zheng, Z.; Chen, Y.; Yang, Z.; Lin, R. Colored rice quality inspection system using machine vision. J. Cereal Sci. 2019, 88, 87–95. [Google Scholar] [CrossRef]
- Shamim, M.I.; Pal, B.; Arora, A.S.; Pial, M.A. A deep convolutional neural network approach to rice grain purity analysis. Progress in Computing Analytics and Networking. In Proceedings of ICCAN 2019: Progress in Computing, Analytics and Networking; Springer: Singapore, 2020; Volume 1119, pp. 179–189. [Google Scholar]
- Jeyaraj, P.R.; Asokan, S.P.; Samuel Nadar, E.R. Computer-assisted real-time rice variety learning using deep learning network. Rice Sci. 2022, 29, 489–498. [Google Scholar] [CrossRef]
- Zhou, J.; Ju, X.; Sun, X.; Jin, H.; Yao, M.; Shen, H. Succession of Mould Flora for Paddy in Different Storage Conitions. J. Chin. Cereals Oils Assoc. 2008, 23, 133–136. [Google Scholar]
- Yang, S.; Cao, Y.; Zhao, H.; Fei, M. Research progress of rapid detection technology in grain mildew. J. Chin. Cereals Oils Assoc. 2018, 31, 21–23. [Google Scholar]
- Barsanti, L.; Birindelli, L.; Gualtieri, P. Water monitoring by means of digital microscopy identification and classification of microalgae. Environ. Sci. Process. Impacts 2021, 23, 1443–1457. [Google Scholar] [CrossRef] [PubMed]
- Suhail, K.; Brindha, D. A review on various methods for recognition of urine particles using digital microscopic images of urine sediments. Biomed. Signal Process. 2021, 68, 102806. [Google Scholar]
- Patericio, D.I.; Rieder, R. Computer vision and artificial intelligence in precision agriculture for grain crops: A systematic review. Comput. Elecron. Agric. 2018, 153, 69–81. [Google Scholar] [CrossRef]
- Velesaca, H.O.; Suarez, P.L.; Mira, R.; Sappa, A.D. Computer vision based food grain classification: A comprehensive survey. Comput. Elecron. Agric. 2021, 187, 106287. [Google Scholar] [CrossRef]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep learning for computer vision: A brief review. Comput. Intell. Neurosci. 2018, 2018, 7068349. [Google Scholar] [CrossRef]
- Mu, R.; Zeng, X. A review of deep learning research. KSII Trans. Internet Inf. Syst. 2019, 13, 1738–1764. [Google Scholar]
- Wang, Y.; Su, W. Convolutional neural networks in computer vision for grain crop phenotyping: A review. Agronomy 2022, 12, 2659. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Wang, Z.; Zhang, X.; Li, J.; Luan, K. A Yolo-based target detection model for offshore unmanned aerial vehicle data. Sustainability 2021, 13, 12980. [Google Scholar] [CrossRef]
- Ren, P.; Wang, L.; Fang, W.; Song, S.; Djahel, S. A novel squeeze YOLO-based real-time people counting approach. Int. J. Bio-Inspir. Comput. 2020, 16, 94–101. [Google Scholar] [CrossRef]
- Chen, W.; Huang, H.; Peng, S.; Zhou, C.; Zhang, C. YOLO-face: A real-time face detector. Vis. Comput. 2020, 37, 805–813. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, S. Broken Corn Detection Based on an Adjusted YOLO With Focal Loss. IEEE Access 2019, 7, 68281–68289. [Google Scholar] [CrossRef]
- Tang, F.; Cheng, S.; Wu, S. Growth of spoilage fungi in stored paddy. J. Chin. Cereals Oils Assoc. 2009, 24, 98–101. [Google Scholar]
- Atungulu, G.G.; Zhong, H.M.; Thote, S.; Okeyo, A.; Couch, A. Microbial prevalence on freshly-harvested long-grain pureline, hybrid, and medium-grain rice cultivars. Appl. Eng. Agric. 2015, 31, 949–956. [Google Scholar]
- Bhupendra; Mose, K.; Miglani, A.; Kankar, P.K. Deep CNN-based damage classification of milled rice grains using a high-magnification image dataset. Comput. Electron. Agric. 2022, 195, 106811. [Google Scholar] [CrossRef]
- Qi, C.; Gao, J.; Pearson, S.; Harman, H.; Chen, K.; Shu, L. Tea chrysanthemum detection under unstructured environments using the TC-YOLO model. Expert Syst. Appl. 2022, 193, 116473. [Google Scholar] [CrossRef]
- Assadzadeh, S.; Walker, C.K.; Panozzo, J.F. Deep learning segmentation in bulk grain images for prediction of grain market quality. Food Bioprocess Technol. 2022, 15, 1615–1628. [Google Scholar] [CrossRef]
- Yang, S.; Zheng, L.; He, P.; Wu, T.; Sun, S.; Wang, M. High-throughput soybean seeds phenotyping with convolutional neural networks and transfer learning. Plant Methods 2021, 17, 50. [Google Scholar] [CrossRef] [PubMed]


| Mildewed Region (×103 Pixels) | Normal Region (×103 Pixels) | Accuracy | Overall Accuracy | |||
|---|---|---|---|---|---|---|
| A. niger | Training set | Mildewed region | 40,886 | 4426 | 90.23% | 95.44% |
| Background area | 13,805 | 340,526 | 96.10% | |||
| Verification set | Mildewed region | 29,599 | 3562 | 89.26% | 94.93% | |
| Background area | 10,195 | 228,245 | 95.72% | |||
| P. citrinum | Training set | Mildewed region | 64,657 | 6015 | 91.45% | 93.73% |
| Background area | 21,477 | 346,273 | 94.16% | |||
| Verification set | Mildewed region | 49,429 | 4801 | 91.15% | 93.76% | |
| Background area | 15,015 | 233,549 | 93.96% | |||
| A. cinerea | Training set | Mildewed region | 62,591 | 6071 | 91.16% | 91.19% |
| Background area | 19,252 | 199,554 | 91.20% | |||
| Verification set | Mildewed region | 40,747 | 4431 | 90.19% | 91.52% | |
| Background area | 11,556 | 131,715 | 91.93% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, K.; Zhang, Y.-J.; Tong, S.-Y.; Tang, M.-D.; Wang, C.-B. Study on Rice Grain Mildewed Region Recognition Based on Microscopic Computer Vision and YOLO-v5 Model. Foods 2022, 11, 4031. https://doi.org/10.3390/foods11244031
Sun K, Zhang Y-J, Tong S-Y, Tang M-D, Wang C-B. Study on Rice Grain Mildewed Region Recognition Based on Microscopic Computer Vision and YOLO-v5 Model. Foods. 2022; 11(24):4031. https://doi.org/10.3390/foods11244031
Chicago/Turabian StyleSun, Ke, Yu-Jie Zhang, Si-Yuan Tong, Meng-Di Tang, and Chang-Bao Wang. 2022. "Study on Rice Grain Mildewed Region Recognition Based on Microscopic Computer Vision and YOLO-v5 Model" Foods 11, no. 24: 4031. https://doi.org/10.3390/foods11244031
APA StyleSun, K., Zhang, Y.-J., Tong, S.-Y., Tang, M.-D., & Wang, C.-B. (2022). Study on Rice Grain Mildewed Region Recognition Based on Microscopic Computer Vision and YOLO-v5 Model. Foods, 11(24), 4031. https://doi.org/10.3390/foods11244031





