Improved Light and In Vitro Digestive Stability of Lutein-Loaded Nanoparticles Based on Soy Protein Hydrolysates via Pepsin
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of SPI and Its Hydrolysates by Pepsin and Alcalase (PSPI and ASPI)
2.3. Characteristics of SPI, PSPI, and ASPI
2.3.1. Degree of Hydrolysis (DH) Measurement
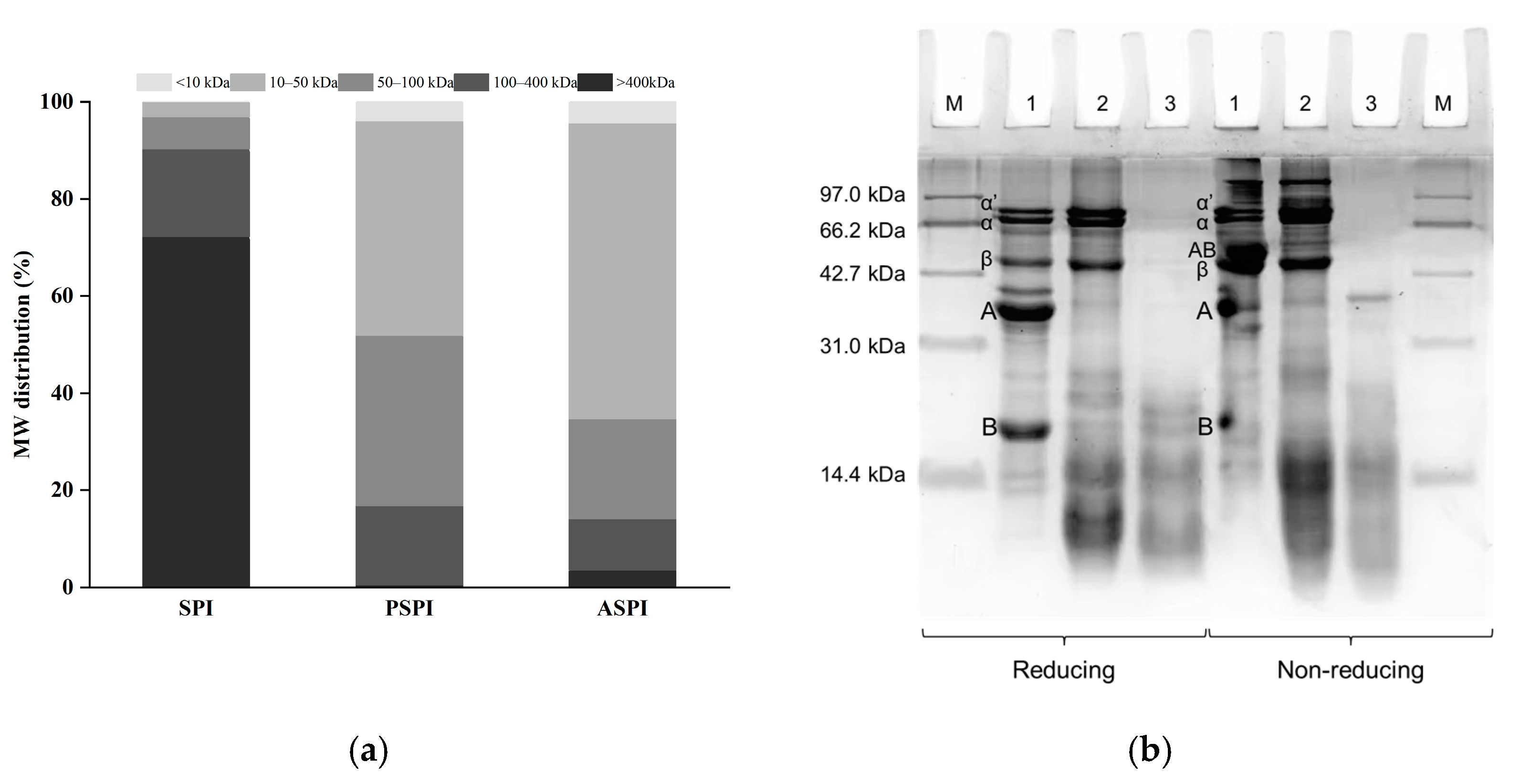
2.3.2. Molecular Weight (MW) Distribution
2.3.3. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.3.4. Surface Hydrophobicity Determination
2.4. Preparation of Lutein-Loaded Nanoparticles
2.5. Encapsulation Efficiency (EE) and Loading Capacity (LC) of Lutein
2.6. Particle Characteristics
2.6.1. Particle Size, Polydispersity Index (PDI) and ζ-Potential
2.6.2. Atomic Force Microscopy (AFM) Images
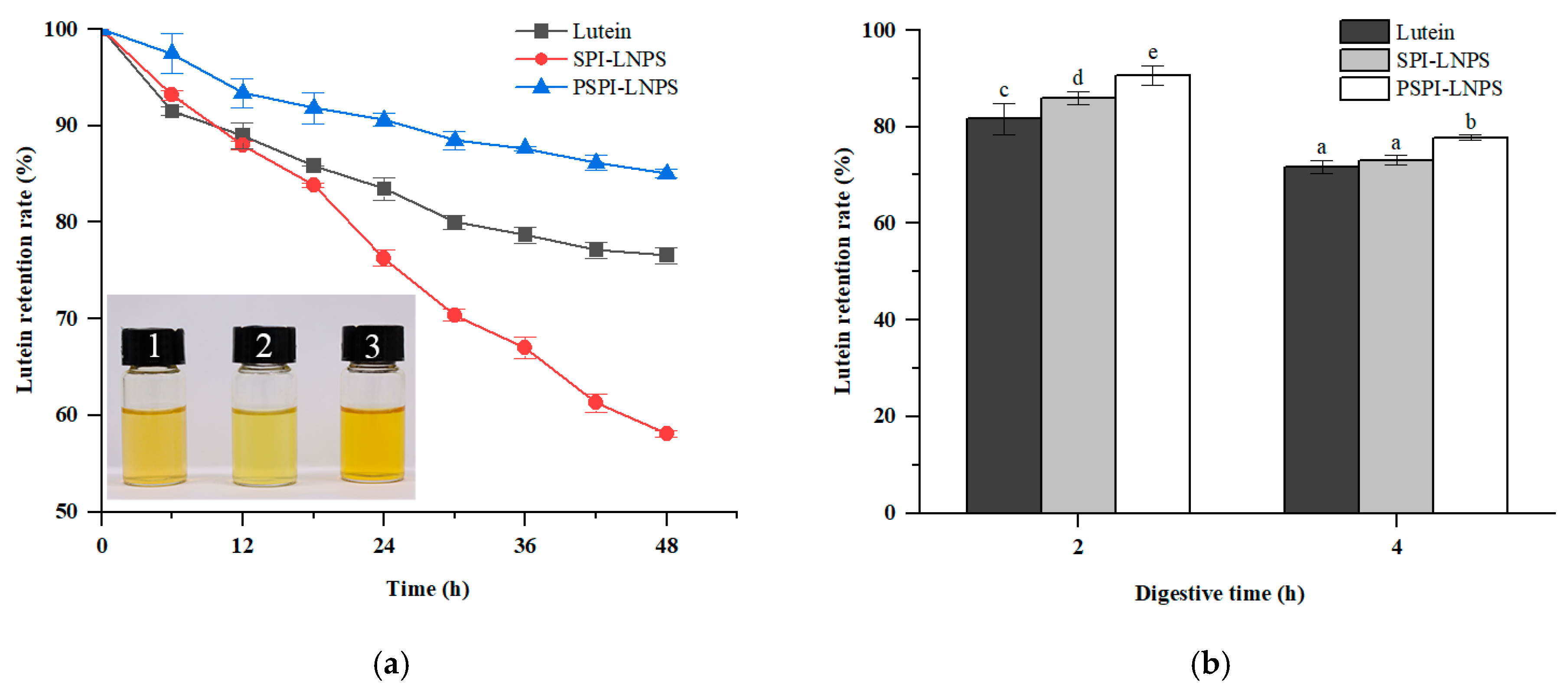
2.7. Light Stability
2.8. In Vitro Gastrointestinal Digestion
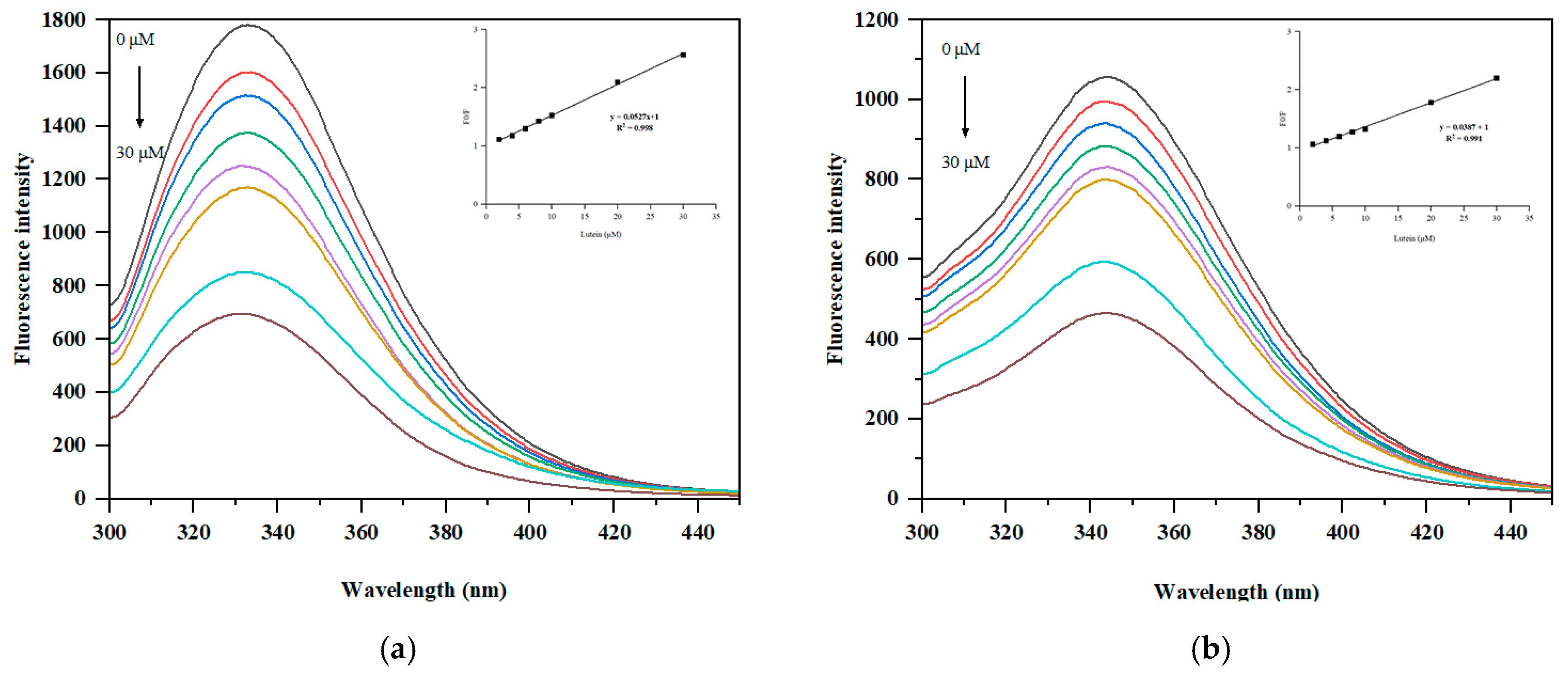
2.9. Measurement of Intrinsic Fluorescence
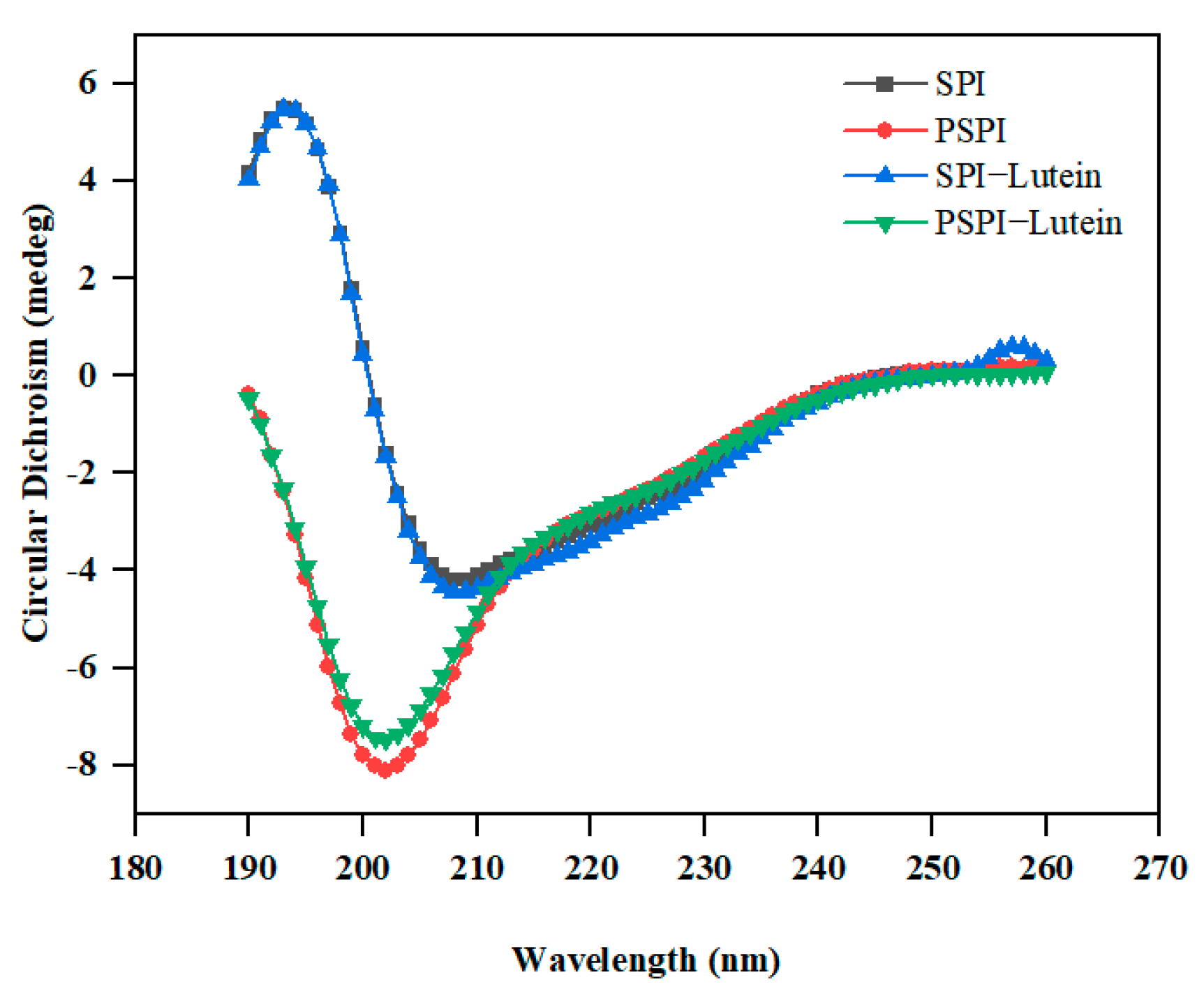
2.10. Circular Dichroism (Cd) Analysis
2.11. The Fourier Transform Infrared Spectra (FTIR) and X-ray Diffraction (XRD) Analysis
2.12. Statistical Analysis
3. Results and Discussion
3.1. Characteristics of SPI, PSPI, and ASPI
3.2. Encapsulation Properties and Particle Characteristics
3.3. Effect of Light and Digestive Environment on the Stability of Lutein-Loaded Nanoparticles
3.4. Fluorescence Spectroscopy Analysis
3.5. CD Analysis
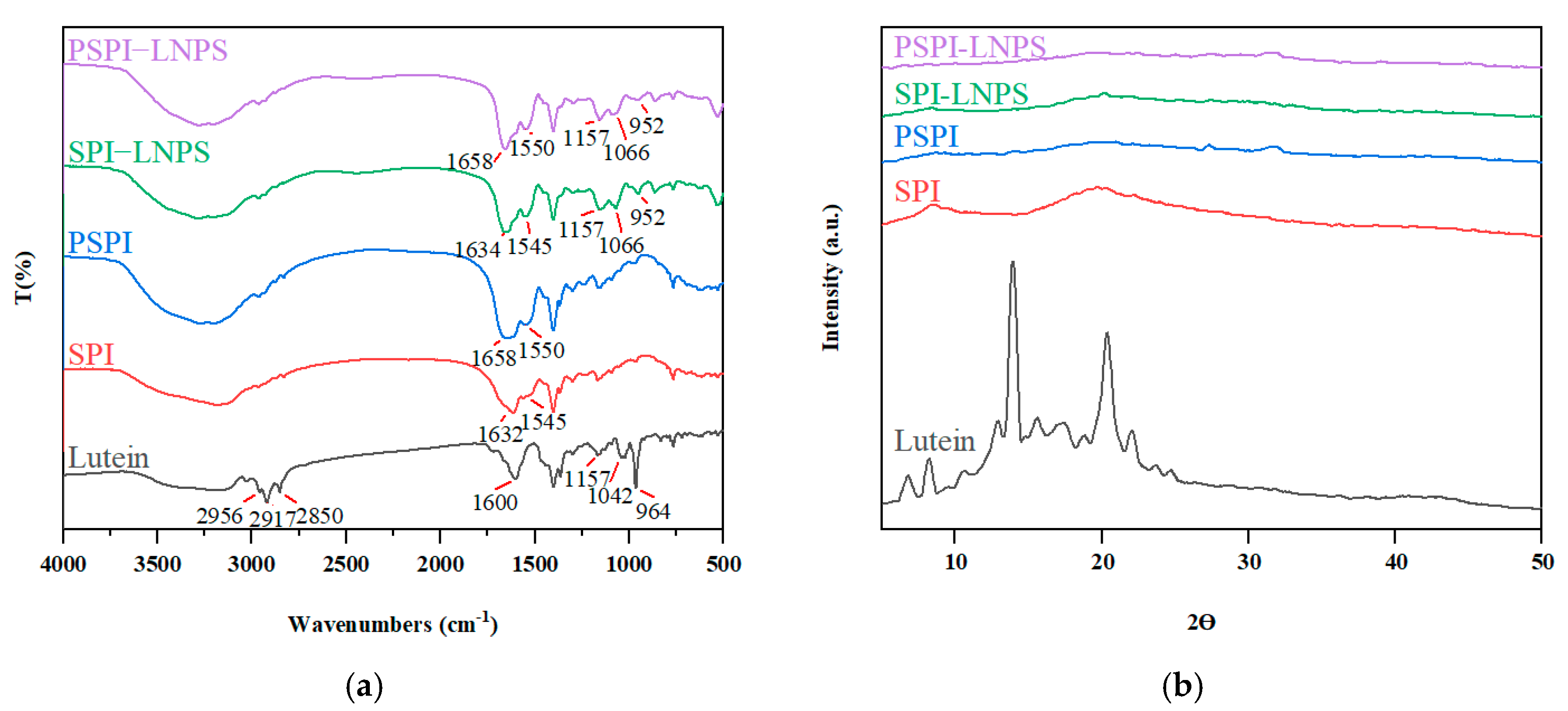
3.6. FT-IR and XRD Analysis
3.7. Morphology Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Johra, F.T.; Bepari, A.K.; Bristy, A.T.; Reza, H.M. A mechanistic review of beta-carotene, lutein, and zeaxanthin in eye health and disease. Antioxidants 2020, 9, 1046. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Rauf, A.; Tareq, A.M.; Jahan, S.; Bin Emran, T.; Shahriar, T.G.; Dhama, K.; Alhumaydhi, F.A.; Aljohani, A.S.M.; Rebezov, M.; et al. Potential health benefits of carotenoid lutein: An updated review. Food Chem. Toxicol. 2021, 154, 112328. [Google Scholar] [CrossRef] [PubMed]
- Boon, C.S.; McClements, D.J.; Weiss, J.; Decker, E.A. Factors influencing the chemical stability of carotenoids in foods. Crit. Rev. Food Sci. Nutr. 2010, 50, 515–532. [Google Scholar] [CrossRef]
- Teo, A.; Lee, S.J.; Goh, K.K.T.; Wolber, F.M. Kinetic stability and cellular uptake of lutein in WPI-stabilised nanoemulsions and emulsions prepared by emulsification and solvent evaporation method. Food Chem. 2017, 221, 1269–1276. [Google Scholar] [CrossRef]
- McClements, D.J. Nanotechnology approaches for improving the healthiness and sustainability of the modern food supply. ACS Omega 2020, 5, 29623–29630. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.Y.; Zhu, X.L.; Abbas, S.; Karangwa, E.; Zhang, X.M.; Xia, S.Q.; Feng, B.A.; Jia, C.S. Stable nanoparticles prepared by heating electrostatic complexes of whey protein isolate-dextran conjugate and chondroitin sulfate. J. Agric. Food Chem. 2015, 63, 4179–4189. [Google Scholar] [CrossRef] [PubMed]
- Muhoza, B.; Zhang, Y.T.; Xia, S.Q.; Cai, J.B.; Zhang, X.M.; Su, J.K. Improved stability and controlled release of lutein-loaded micelles based on glycosylated casein via Maillard reaction. J. Funct. Food 2018, 45, 1–9. [Google Scholar] [CrossRef]
- Jiao, Y.; Zheng, X.Q.; Chang, Y.; Li, D.J.; Sun, X.H.; Liu, X.L. Zein-derived peptides as nanocarriers to increase the water solubility and stability of lutein. Food Funct. 2018, 9, 117–123. [Google Scholar] [CrossRef]
- Ma, X.Y.; Chen, X.X.; Ma, M.Y.; Xu, Y.; Wu, X.M.; Mu, G.Q.; Zhu, X.M. Lutein transport systems loaded with rice protein-based self-assembled nanoparticles. Food Biosci. 2021, 42, 101061. [Google Scholar] [CrossRef]
- Tang, C.H. Strategies to utilize naturally occurring protein architectures as nanovehicles for hydrophobic nutraceuticals. Food Hydrocoll. 2021, 112, 106344. [Google Scholar] [CrossRef]
- Peng, S.F.; Zhou, L.; Cai, Q.Z.; Zou, L.Q.; Liu, C.M.; Liu, W.; McClements, D.J. Utilization of biopolymers to stabilize curcumin nanoparticles prepared by the pH-shift method: Caseinate, whey protein, soy protein and gum Arabic. Food Hydrocoll. 2020, 107, 105963. [Google Scholar] [CrossRef]
- Chen, N.N.; Lin, L.Z.; Sun, W.Z.; Zhao, M.M. Stable and pH-sensitive protein nanogels made by self-assembly of heat denatured soy protein. J. Agric. Food Chem. 2014, 62, 9553–9561. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Li, X.T.; Zhang, N.; Tang, C.H. Novel soy β-conglycinin nanoparticles by ethanol-assisted disassembly and reassembly: Outstanding nanocarriers for hydrophobic nutraceuticals. Food Hydrocoll. 2019, 91, 246–255. [Google Scholar] [CrossRef]
- Xiang, H.; Sun-waterhouse, D.; Cui, C.; Wang, W.; Dong, K.M. Modification of soy protein isolate by glutaminase for nanocomplexation with curcumin. Food Chem. 2018, 268, 504–512. [Google Scholar] [CrossRef]
- Yuan, D.; Zhou, F.B.; Shen, P.H.; Zhang, Y.H.; Lin, L.Z.; Zhao, M.M. Self-assembled soy protein nanoparticles by partial enzymatic hydrolysis for pH-driven encapsulation and delivery of hydrophobic cargo curcumin. Food Hydrocoll. 2021, 120, 106759. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Zhao, M.M.; Ning, Z.X.; Yu, S.J.; Tang, N.; Zhou, F.B. Development of a sono-assembled, bifunctional soy peptide nanoparticle for cellular delivery of hydrophobic active cargoes. J. Agric. Food Chem. 2018, 66, 4208–4218. [Google Scholar] [CrossRef]
- Du, Y.Z.; Bao, C.; Huang, J.; Jiang, P.; Jiao, L.L.; Ren, F.Z.; Li, Y. Improved stability, epithelial permeability and cellular antioxidant activity of beta-carotene via encapsulation by self-assembled alpha-lactalbumin micelles. Food Chem. 2019, 271, 707–714. [Google Scholar] [CrossRef]
- Feng, J.; Xiong, Y.L. Interaction and, functionality of mixed myofibrillar and enzyme-hydrolyzed soy proteins. J. Food Sci. 2003, 68, 803–809. [Google Scholar] [CrossRef]
- Shen, P.H.; Zhou, F.B.; Zhang, Y.H.; Yuan, D.; Zhao, Q.Z.; Zhao, M.M. Formation and characterization of soy protein nanoparticles by controlled partial enzymatic hydrolysis. Food Hydrocoll. 2020, 105, 105844. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Determination of the degree of hydrolysis of food protein hydrolysates by trinitrobenzenesulfonic acid. J. Agric. Food Chem. 1979, 27, 1256–1262. [Google Scholar] [CrossRef]
- Jiang, Y.Q.; Wang, Z.J.; He, Z.Y.; Zeng, M.M.; Qin, F.; Chen, J. Effect of heat-induced aggregation of soy protein isolate on protein-glutaminase deamidation and the emulsifying properties of deamidated products. LWT-Food Sci. Technol. 2022, 154, 112328. [Google Scholar] [CrossRef]
- Haskard, C.A.; Li-Chan, E.C.Y. Hydrophobicity of bovine serum albumin and ovalbumin determined using uncharged (PRODAN) and anionic (ANS-) fluorescent probes. J. Agric. Food Chem. 1998, 46, 2671–2677. [Google Scholar] [CrossRef]
- Chen, F.P.; Li, B.S.; Tang, C.H. Nanocomplexation of soy protein isolate with curcumin: Influence of ultrasonic treatment. Food Res. Int. 2015, 75, 157–165. [Google Scholar] [CrossRef]
- Wu, Y.R.; Yin, Z.C.; Qie, X.J.; Chen, Y.; Zeng, M.M.; Wang, Z.J.; Qin, F.; Chen, J.; He, Z.Y. Interaction of soy protein isolate hydrolysates with cyanidin-3-O-glucoside and its effect on the in vitro Antioxidant Capacity of the Complexes under Neutral Condition. Molecules 2021, 26, 1721. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.L.; Qi, J.R.; Yin, S.W.; Wang, J.M.; Guo, J.; Weng, J.Y.; Liu, Q.R.; Yang, X.Q. Fabrication and characterization of stable soy beta-conglycinin-dextran core-shell nanogels prepared via a self-assembly approach at the isoelectric point. J. Agric. Food Chem. 2015, 63, 6075–6083. [Google Scholar] [CrossRef]
- Li, H.; Yuan, Y.K.; Zhu, J.X.; Wang, T.; Wang, D.F.; Xu, Y. Zein/soluble soybean polysaccharide composite nanoparticles for encapsulation and oral delivery of lutein. Food Hydrocoll. 2020, 103, 105715. [Google Scholar] [CrossRef]
- Liang, G.J.; Chen, W.P.; Qie, X.J.; Zeng, M.M.; Qin, F.; He, Z.Y.; Chen, J. Modification of soy protein isolates using combined pre-heat treatment and controlled enzymatic hydrolysis for improving foaming properties. Food Hydrocoll. 2020, 105, 105764. [Google Scholar] [CrossRef]
- Chen, W.P.; Liang, G.J.; Li, X.; He, Z.Y.; Zeng, M.M.; Gao, D.M.; Qin, F.; Goff, H.D.; Chen, J. Effects of soy proteins and hydrolysates on fat globule coalescence and meltdown properties of ice cream. Food Hydrocoll. 2019, 94, 279–286. [Google Scholar] [CrossRef]
- Yao, J.J.; Tanteeratarm, K.; Wei, L.S. Effects of maturation and storage on solubility, emulsion stability and gelation properties of isolated soy proteins. J. Am. Oil Chem. Soc. 1990, 67, 974–979. [Google Scholar] [CrossRef]
- Betancur-Ancona, D.; Martínez-Rosado, R.; Corona-Cruz, A.; Castellanos-Ruelas, A.; Jaramillo-Flores, M.E.; Chel-Guerrero, L. Functional properties of hydrolysates from Phaseolus lunatus seeds. Int. J. Food Sci. Technol. 2009, 44, 128–137. [Google Scholar] [CrossRef]
- Tsumura, K.; Saito, T.; Kugimiya, W.; Inouye, K. Selective proteolysis of the glycinin and β-conglycinin fractions in a soy protein isolate by pepsin and papain with controlled pH and temperature. J. Food Sci. 2010, 69, C363–C367. [Google Scholar] [CrossRef]
- Qi, X.; Xu, D.X.; Zhu, J.J.; Wang, S.J.; Peng, J.W.; Gao, W.; Cao, Y.P. Studying the interaction mechanism between bovine serum albumin and lutein dipalmitate: Multi-spectroscopic and molecular docking techniques. Food Hydrocoll. 2021, 113, 106513. [Google Scholar] [CrossRef]
- Gumus, C.E.; Davidov-Pardo, G.; McClements, D.J. Lutein-enriched emulsion-based delivery systems: Impact of maillard conjugation on physicochemical stability and gastrointestinal fate. Food Hydrocoll. 2016, 60, 38–49. [Google Scholar] [CrossRef]
- Vekshin, N.L. Division of tyrosine and tryptophan fluorescent components by synchronous scanning method. Biofizika 1996, 41, 1176–1179. [Google Scholar]
- Vorob’ev, M.M.; Vogel, V.; Guler, G.; Mantele, W. Monitoring of demasking of peptide bonds during proteolysis by analysis of the apparent spectral shift of intrinsic protein fluorescence. Food Biophys. 2011, 6, 519–526. [Google Scholar] [CrossRef]
- Venyaminov, S.Y.; Braddock, W.D.; Prendergast, F.G. Spectroscopy of proteins in aqueous solution. Biophys. J. 1996, 70, SU187. [Google Scholar]
- Deng, J.J.; Li, M.L.; Zhang, T.; Chen, B.; Leng, X.J.; Zhao, G.H. Binding of proanthocyanidins to soybean (Glycine max) seed ferritin inhibiting protein degradation by protease in vitro. Food Res. Int. 2011, 44, 33–38. [Google Scholar] [CrossRef]
- Feng, J.; Wu, Y.H.; Zhang, L.X.; Li, Y.; Liu, S.B.; Wang, H.; Li, C.Y. Enhanced chemical stability, intestinal absorption, and intracellular antioxidant activity of cyanidin-3-O-glucoside by composite nanogel encapsulation. J. Agric. Food Chem. 2019, 67, 10432–10447. [Google Scholar] [CrossRef]
- Pujara, N.; Jambhrunkar, S.; Wong, K.Y.; McGuckin, M.; Popat, A. Enhanced colloidal stability, solubility and rapid dissolution of resveratrol by nanocomplexation with soy protein isolate. J. Colloid Interface Sci. 2017, 488, 303–308. [Google Scholar] [CrossRef]
- Yi, J.; Lam, T.I.; Yokoyama, W.; Cheng, L.W.; Zhong, F. Controlled release of beta-carotene in beta-lactoglobulin-dextran-conjugated nanoparticles’ in vitro digestion and transport with Caco-2 monolayers. J. Agric. Food Chem. 2014, 62, 8900–8907. [Google Scholar] [CrossRef]


| Sample | DH (%) | H0 |
|---|---|---|
| SPI | - | 86.24 ± 0.51 b |
| PSPI | 6.00 ± 0.05 a | 140.27 ± 1.76 c |
| ASPI | 14.96 ± 0.04 b | 20.78 ± 0.12 a |
| Sample | Average Particle Size (nm) | PDI | ζ-Potential (mV) |
|---|---|---|---|
| SPI | 59.5 ± 0.34 a | 0.55 ± 0.005 e | −25.30 ± 0.99 c |
| PSPI | 71.12 ± 0.37 b | 0.26 ± 0.003 b | −20.50 ± 0.71 a |
| ASPI | 76.23 ± 5.17 b | 0.52 ± 0.03 d | −23.10 ± 0.57 b |
| SPI–LNPS | 124.43 ± 1.85 d | 0.48 ± 0.01 c | −23.25 ± 0.92 b |
| PSPI–LNPS | 114.47 ± 1.45 c | 0.11 ± 0.009 a | −23.45 ± 0.78 b |
| ASPI–LNPS | 236.10 ± 8.20 e | 1.00 ± 0.00 f | −25.35 ± 0.07 c |
| kq/1012 M−1 s−1 | Ksv/104 M−1 | Ka/105 M−1 | n | |
|---|---|---|---|---|
| Lut-SPI | 5.27 ± 0.12 b | 5.27 ± 0.12 b | 0.63 ± 0.03 a | 1.02 ± 0.01 a |
| Lut-PSPI | 3.87 ± 0.17 a | 3.87 ± 0.17 a | 1.04 ± 0.12 b | 1.10 ± 0.03 b |
| Secondary Structure | α-Helix (%) | β-Sheet (%) | β-Turns (%) | Random Coil (%) |
|---|---|---|---|---|
| SPI | 16.24 ± 0.08 b | 32.84 ± 0.47 a | 22.24 ± 0.06 c | 28.69 ± 0.45 b |
| PSPI | 9.27 ± 0.05 c | 31.67 ± 0.27 b | 23.00 ± 0.02 a | 36.07 ± 0.28 a |
| SPI-Lut | 16.94 ± 0.51 a | 31.67 ± 0.68 b | 22.31 ± 0.09 c | 29.08 ± 0.11 b |
| PSPI-Lut | 8.74 ± 0.25 c | 32.92 ± 0.34 a | 22.78 ± 0.15 b | 35.56 ± 0.40 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, R.; Qie, X.; Wang, Z.; Chen, Q.; Zeng, M.; Chen, J.; Qin, F.; He, Z. Improved Light and In Vitro Digestive Stability of Lutein-Loaded Nanoparticles Based on Soy Protein Hydrolysates via Pepsin. Foods 2022, 11, 3635. https://doi.org/10.3390/foods11223635
Wu R, Qie X, Wang Z, Chen Q, Zeng M, Chen J, Qin F, He Z. Improved Light and In Vitro Digestive Stability of Lutein-Loaded Nanoparticles Based on Soy Protein Hydrolysates via Pepsin. Foods. 2022; 11(22):3635. https://doi.org/10.3390/foods11223635
Chicago/Turabian StyleWu, Renyi, Xuejiao Qie, Zhaojun Wang, Qiuming Chen, Maomao Zeng, Jie Chen, Fang Qin, and Zhiyong He. 2022. "Improved Light and In Vitro Digestive Stability of Lutein-Loaded Nanoparticles Based on Soy Protein Hydrolysates via Pepsin" Foods 11, no. 22: 3635. https://doi.org/10.3390/foods11223635
APA StyleWu, R., Qie, X., Wang, Z., Chen, Q., Zeng, M., Chen, J., Qin, F., & He, Z. (2022). Improved Light and In Vitro Digestive Stability of Lutein-Loaded Nanoparticles Based on Soy Protein Hydrolysates via Pepsin. Foods, 11(22), 3635. https://doi.org/10.3390/foods11223635









