Metabolomics-Based Analysis of the Major Taste Contributors of Meat by Comparing Differences in Muscle Tissue between Chickens and Common Livestock Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Preparation
2.2. E-Tongue Analysis
2.3. LC-MS/MS Analysis
2.4. Data Treatment and Pre-Processing
2.5. Statistical Analysis
3. Results
3.1. Analysis of the Differences in Individual Tastes of Muscle Tissue of Four Different Livestock Species
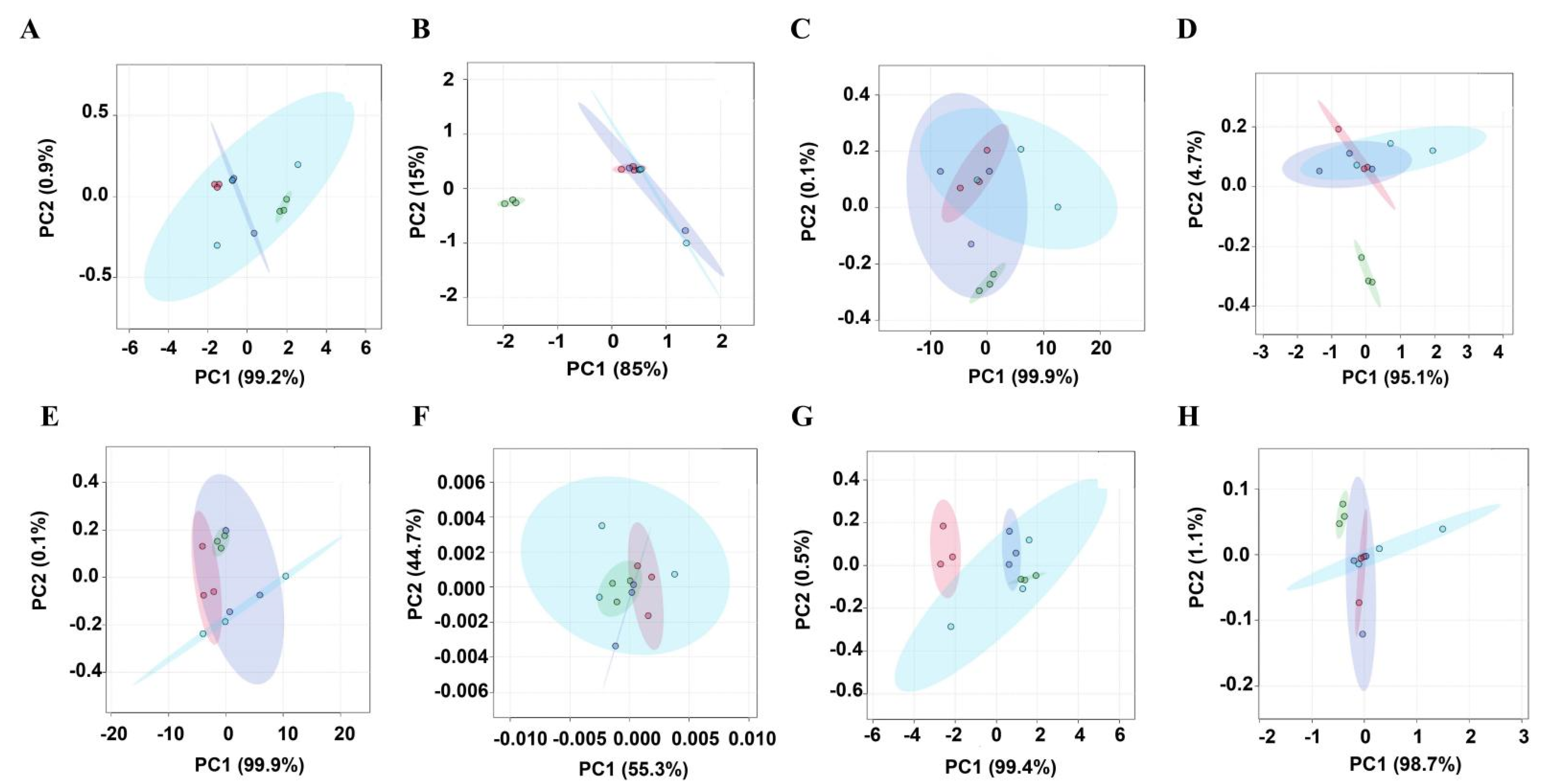
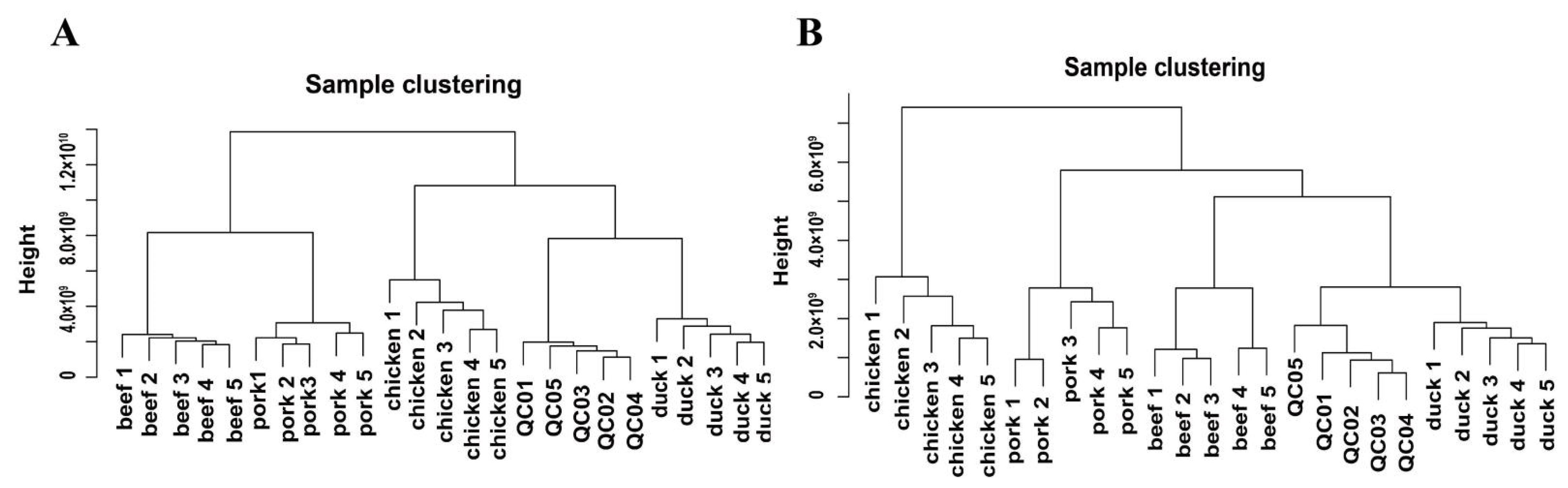
3.2. Clustering Analysis of Metabolites in Muscle Tissues of Four Different Livestock Species
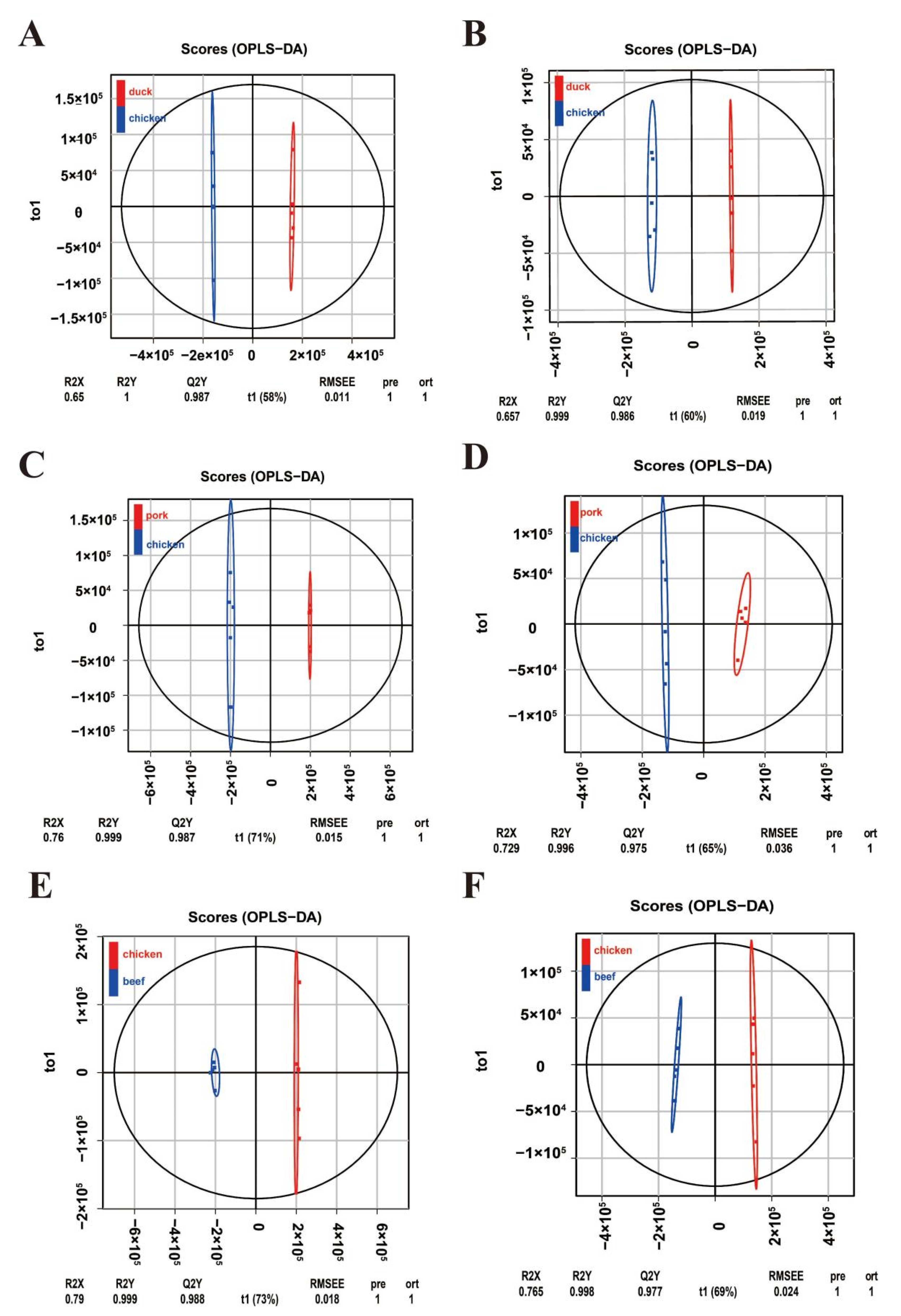
3.3. Analysis of Metabolic Differences between Chicken Versus Duck, Pork, and Beef
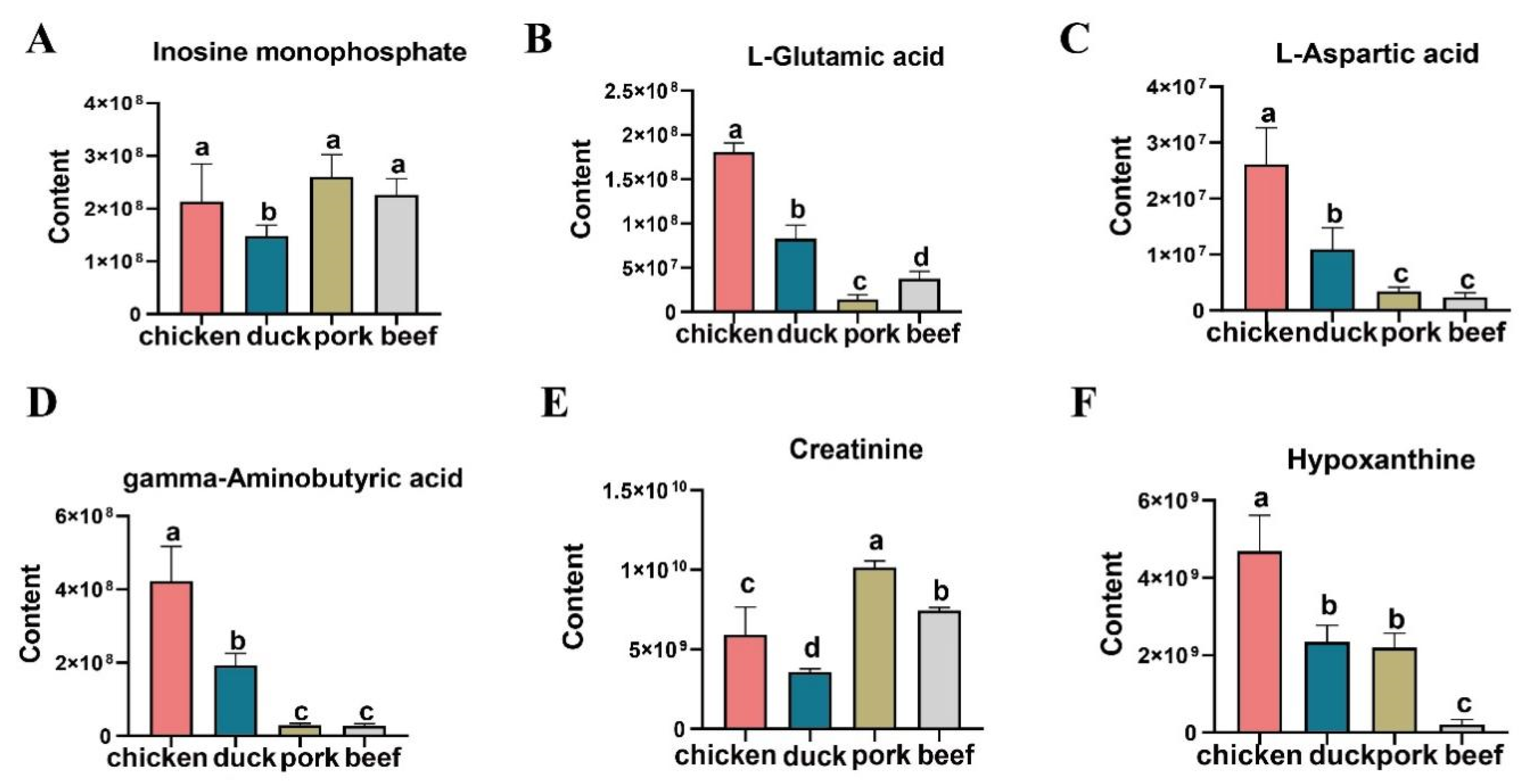
3.4. Screening for Important Taste Substances Contributing to the OPLS Model
3.5. KEGG Analysis of Differential Metabolites and Comparison of Important Taste Substances
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, P.; Chatli, M.K.; Mehta, N.; Singh, P.; Malav, O.P.; Verma, A.K. Meat analogues: Health promising sustainable meat substitutes. Crit. Rev. Food Sci. Nutr. 2017, 57, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.L.; Yu, X.Q.; Chen, J.; Tang, J.; Wang, L.; Li, Y.; Tang, C. Production of characteristic volatile markers and their relation to Staphylococcus aureus growth status in pork. Meat Sci. 2020, 160, 107956. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.Y.; Zhu, H.Z.; Wu, Z.-Y.; Song, W.-J.; Tang, C.-B.; Li, C.-B.; Ding, S.-J.; Zhou, G.-H. Evaluation of the effect of smooth muscle cells on the quality of cultured meat in a model for cultured meat. Food Res. Int. 2021, 150, 110786. [Google Scholar] [CrossRef] [PubMed]
- Taheri-Garavand, A.; Fatahi, S.; Omid, M.; Makino, Y. Meat quality evaluation based on computer vision technique: A review. Meat Sci. 2019, 156, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Wu, W.; Soladoye, O.P.; Aluko, R.E.; Bak, K.H.; Fu, Y.; Zhang, Y. Maillard reaction of food-derived peptides as a potential route to generate meat flavor compounds: A review. Food Res. Int. 2022, 151, 110823. [Google Scholar] [CrossRef]
- Huang, Q.; Dong, K.; Wang, Q.; Huang, X.; Wang, G.; An, F.; Luo, Z.; Luo, P. Changes in volatile flavor of yak meat during oxidation based on multi-omics. Food Chem. 2022, 371, 131103. [Google Scholar] [CrossRef]
- Yao, H.; Xu, Y.L.; Liu, W.; Lu, Y.; Gan, J.; Liu, Y.; Tao, N.; Wang, X.; Xu, C. Taste compounds generation and variation of broth in pork meat braised processing by chemical analysis and an electronic tongue system. J. Food Biochem. 2022, 46, e13766. [Google Scholar] [CrossRef]
- Du, C.; Qi, J.; Yang, C.; Zhang, Q.; Liu, D. Enrichment of taste and aroma perceptions in chicken meat stewed in braised soup used repeatedly. J. Food Sci. 2022, 87, 2563–2577. [Google Scholar] [CrossRef]
- Tu, T.; Wu, W.; Tang, X.; Ge, Q.; Zhan, J. Screening out important substances for distinguishing Chinese indigenous pork and hybrid pork and identifying different pork muscles by analyzing the fatty acid and nucleotide contents. Food Chem. 2021, 350, 129219. [Google Scholar] [CrossRef]
- Liu, Z.Q.; Zhang, L.; Sun, L.-H.; Li, X.-J.; Wan, N.-W.; Zheng, Y.-G. Enzymatic production of 5’-inosinic acid by a newly synthesised acid phosphatase/phosphotransferase. Food Chem. 2012, 134, 948–956. [Google Scholar] [CrossRef]
- Chen, X.; Luo, J.; Lou, A.; Wang, Y.; Yang, D.; Shen, Q.W. Duck breast muscle proteins, free fatty acids and volatile compounds as affected by curing methods. Food Chem. 2021, 338, 128138. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Yu, M.; Liu, Z.; Deng, D.; Cui, Y.; Tian, Z.; Wang, G. Effect of amino acids and their derivatives on meat quality of finishing pigs. J. Food Sci. Technol. 2020, 57, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Imanari, M.; Kadowaki, M.; Fujimura, S. Regulation of taste-active components of meat by dietary branched-chain amino acids; effects of branched-chain amino acid antagonism. Br. Poult. Sci. 2008, 49, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Han, F.; Aheto, J.H.; Rashed, M.M.A.; Pan, Z. Artificial bionic taste sensors coupled with chemometrics for rapid detection of beef adulteration. Food Sci. Nutr. 2021, 9, 5220–5228. [Google Scholar] [CrossRef]
- Khaled, A.Y.; Parrish, C.A.; Adedeji, A. Emerging nondestructive approaches for meat quality and safety evaluation-A review. Compr. Rev. Food Sci. Food Saf. 2021, 20, 3438–3463. [Google Scholar] [CrossRef]
- Liu, T.; Mo, Q.; Wei, J.; Zhao, M.; Tang, J.; Feng, F. Mass spectrometry-based metabolomics to reveal chicken meat improvements by medium-chain monoglycerides supplementation: Taste, fresh meat quality, and composition. Food Chem. 2021, 365, 130303. [Google Scholar] [CrossRef]
- Lana, A.; Longo, V.; Dalmasso, A.; D’Alessandro, A.; Bottero, M.T.; Zolla, L. Omics integrating physical techniques: Aged Piedmontese meat analysis. Food Chem. 2015, 172, 731–741. [Google Scholar] [CrossRef]
- Wen, D.; Liu, Y.; Yu, Q. Metabolomic approach to measuring quality of chilled chicken meat during storage. Poult. Sci. 2020, 99, 2543–2554. [Google Scholar] [CrossRef]
- Surányi, J.; Zaukuu, J.Z.; Friedrich, L.; Kovacs, Z.; Horváth, F.; Németh, C.; Kókai, Z. Electronic Tongue as a Correlative Technique for Modeling Cattle Meat Quality and Classification of Breeds. Foods 2021, 10, 2283. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Khan, M.I.; Jo, C.; Tariq, M.R. Meat flavor precursors and factors influencing flavor precursors—A systematic review. Meat Sci. 2015, 110, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Martini, S.; Conte, A.; Tagliazucchi, D. Comparative peptidomic profile and bioactivities of cooked beef, pork, chicken and turkey meat after in vitro gastro-intestinal digestion. J. Proteom. 2019, 208, 103500. [Google Scholar] [CrossRef] [PubMed]
- Ardö, Y. Flavour formation by amino acid catabolism. Biotechnol. Adv. 2006, 24, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Zalouk-Vergnoux, A.; Khalid, A.; Zalouk-Vergnoux, A.; Benali, S.; Mincheva, R.; Raquez, J.-M.; Bertrand, S.; Poirier, L. Are bio-based and biodegradable microplastics impacting for blue mussel (Mytilus edulis)? Mar. Pollut. Bull. 2021, 167, 112295. [Google Scholar] [CrossRef] [PubMed]
- Kunert, C.; Walker, A.; Hofmann, T. Taste modulating N-(1-methyl-4-oxoimidazolidin-2-ylidene) α-amino acids formed from creatinine and reducing carbohydrates. J. Agric. Food Chem. 2011, 59, 8366–8374. [Google Scholar] [CrossRef]
- Ichimura, S.; Nakamura, Y.; Yoshida, Y.; Hattori, A. Hypoxanthine enhances the cured meat taste. Anim. Sci. J. 2017, 88, 379–385. [Google Scholar] [CrossRef]
- Watanabe, G.; Kobayashi, H.; Shibata, M.; Kubota, M.; Kadowaki, M.; Fujimura, S. Reduction of dietary lysine increases free glutamate content in chicken meat and improves its taste. Anim. Sci. J. 2017, 88, 300–305. [Google Scholar] [CrossRef]
- Yu, Z.; Jiang, H.; Guo, R.; Yang, B.; You, G.; Zhao, M.; Liu, X. Taste, umami-enhance effect and amino acid sequence of peptides separated from silkworm pupa hydrolysate. Food Res. Int. 2018, 108, 144–150. [Google Scholar] [CrossRef]
- Tian, X.; Li, Z.J.; Chao, Y.Z.; Wu, Z.Q.; Zhou, M.X.; Xiao, S.T.; Zeng, J.; Zhe, J. Evaluation by electronic tongue and headspace-GC-IMS analyses of the flavor compounds in dry-cured pork with different salt content. Food Res. Int. 2020, 137, 109456. [Google Scholar] [CrossRef]
- Matsumoto, R.R. GABA receptors: Are cellular differences reflected in function? Brain Res. Brain Res. Rev. 1989, 14, 203–225. [Google Scholar] [CrossRef]
- Sarasa, S.B.; Mahendran, R.; Muthusamy, G.; Thankappan, B.; Selta, D.R.F.; Angayarkanni, J. A Brief Review on the Non-protein Amino Acid, Gamma-amino Butyric Acid (GABA): Its Production and Role in Microbes. Curr. Microbiol. 2020, 77, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Zaukuu, J.Z.; Gillay, Z.; Kovacs, Z. Standardized Extraction Techniques for Meat Analysis with the Electronic Tongue: A Case Study of Poultry and Red Meat Adulteration. Sensors 2021, 21, 481. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Cai, Y.; Su, Y.; Wang, D.; Pan, X.; Zhi, X. Study of meat quality and flavour in different cuts of Duroc-Bamei binary hybrid pigs. Vet. Med. Sci. 2021, 7, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.F.; Liu, G.H.; Cai, H.Y.; Chang, W.H.; Ma, J.S.; Zheng, A.J.; Zhang, S. Attempts to increase inosinic acid in broiler meat by using feed additives. Poult. Sci. 2014, 93, 2802–2808. [Google Scholar] [CrossRef]
- Guo, Q.; Kong, X.; Hu, C.; Zhou, B.; Wang, C.; Shen, Q.W. Fatty Acid Content, Flavor Compounds, and Sensory Quality of Pork Loin as Affected by Dietary Supplementation with l-arginine and Glutamic Acid. J. Food Sci. 2019, 84, 3445–3453. [Google Scholar] [CrossRef]
- Maughan, C.; Martini, S. Identification and quantification of flavor attributes present in chicken, lamb, pork, beef, and turkey. J. Food Sci. 2012, 77, S115–S121. [Google Scholar] [CrossRef]
- Tikk, M.; Tikk, K.; Tørngren, M.A.; Meinert, L.; Aaslyng, M.D.; Karlsson, A.H.; Andersen, H.J. Development of inosine monophosphate and its degradation products during aging of pork of different qualities in relation to basic taste and retronasal flavor perception of the meat. J. Agric. Food Chem. 2006, 54, 7769–7777. [Google Scholar] [CrossRef]

| Pathway | ID | Name |
|---|---|---|
| Histidine metabolism | POS00077 | 3-Methylhistidine |
| POS00395 | Formiminoglutamic acid | |
| NEG00068 | 3-Methylhistidine | |
| NEG00083 | L-Glutamic acid | |
| NEG00088 | Anserine | |
| NEG00241 | L-Aspartic acid | |
| NEG02183 | - | |
| Neuroactive ligand–receptor interaction | POS00078 | Taurine |
| POS00195 | Gamma-aminobutyric acid | |
| POS00505 | 2-Acetyl-1-alkyl-sn-glycero-3-phosphocholine | |
| NEG00060 | Gamma-aminobutyric acid | |
| NEG00083 | L-Glutamic acid | |
| NEG00241 | L-Aspartic acid | |
| GABAergic synapse | POS00195 | Gamma-aminobutyric acid |
| NEG00060 | Gamma-aminobutyric acid | |
| NEG00083 | L-Glutamic acid | |
| Nicotine addiction | POS00195 | Gamma-aminobutyric acid |
| NEG00060 | Gamma-aminobutyric acid | |
| NEG00083 | L-Glutamic acid | |
| Alanine, aspartate, and glutamate metabolism | POS00195 | Gamma-aminobutyric acid |
| POS04633 | - | |
| NEG00060 | Gamma-aminobutyric acid | |
| NEG00083 | L-Glutamic acid | |
| NEG00241 | L-Aspartic acid | |
| Protein digestion and absorption | NEG00083 | L-Glutamic acid |
| NEG00241 | L-Aspartic acid | |
| ABC transporters | POS00078 | Taurine |
| NEG00083 | L-Glutamic acid | |
| NEG00241 | L-Aspartic acid | |
| Cyanoamino acid metabolism | NEG00241 | L-Aspartic acid |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Liu, X.; Wang, Y.; Zhao, G.; Wen, J.; Cui, H. Metabolomics-Based Analysis of the Major Taste Contributors of Meat by Comparing Differences in Muscle Tissue between Chickens and Common Livestock Species. Foods 2022, 11, 3586. https://doi.org/10.3390/foods11223586
Wang Y, Liu X, Wang Y, Zhao G, Wen J, Cui H. Metabolomics-Based Analysis of the Major Taste Contributors of Meat by Comparing Differences in Muscle Tissue between Chickens and Common Livestock Species. Foods. 2022; 11(22):3586. https://doi.org/10.3390/foods11223586
Chicago/Turabian StyleWang, Yanke, Xiaojing Liu, Yongli Wang, Guiping Zhao, Jie Wen, and Huanxian Cui. 2022. "Metabolomics-Based Analysis of the Major Taste Contributors of Meat by Comparing Differences in Muscle Tissue between Chickens and Common Livestock Species" Foods 11, no. 22: 3586. https://doi.org/10.3390/foods11223586
APA StyleWang, Y., Liu, X., Wang, Y., Zhao, G., Wen, J., & Cui, H. (2022). Metabolomics-Based Analysis of the Major Taste Contributors of Meat by Comparing Differences in Muscle Tissue between Chickens and Common Livestock Species. Foods, 11(22), 3586. https://doi.org/10.3390/foods11223586




