Rapid Non-Destructive Analysis of Food Nutrient Content Using Swin-Nutrition
Abstract
1. Introduction
2. Materials and Methods
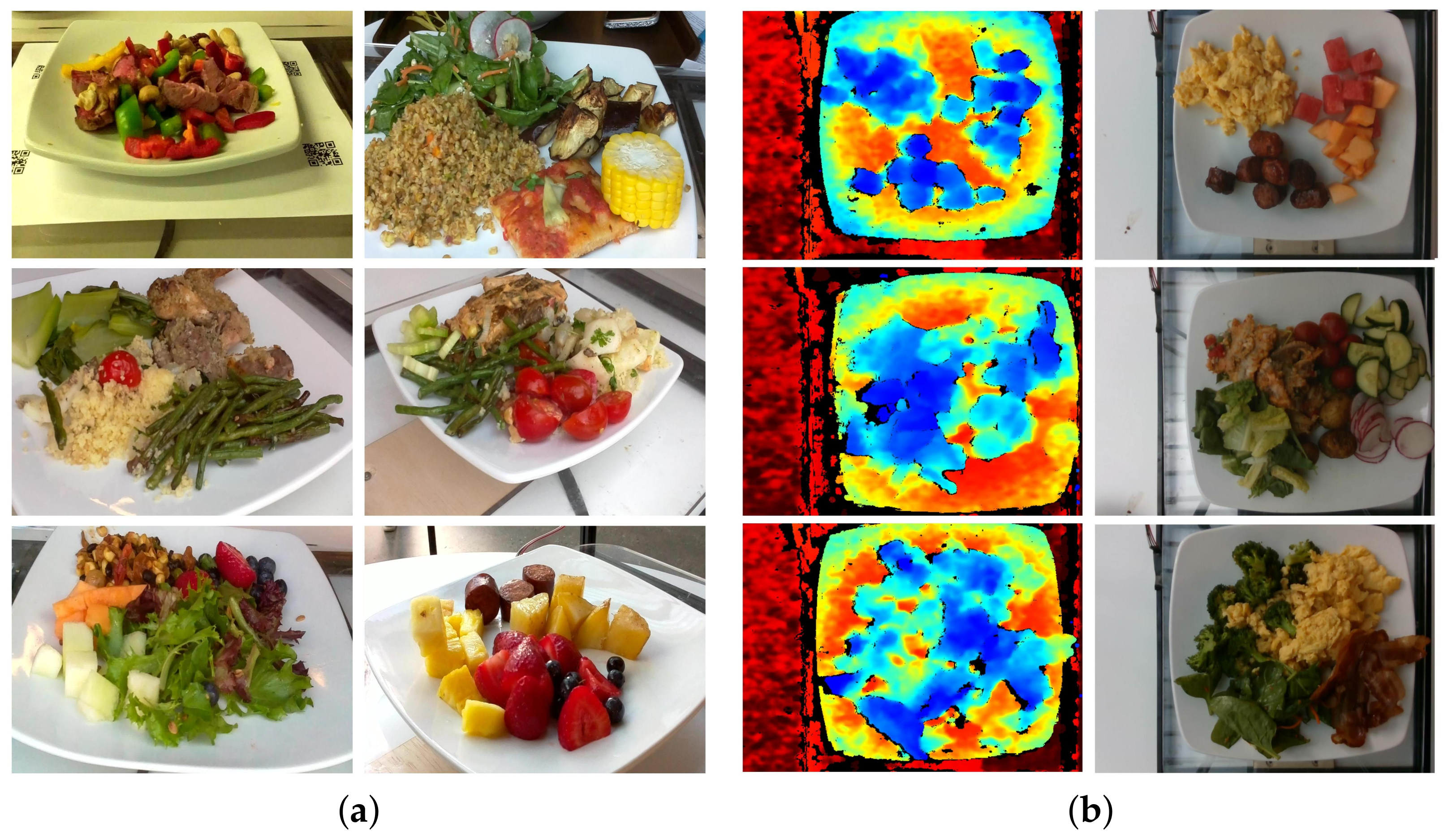
2.1. Data Preparation
2.2. Methods
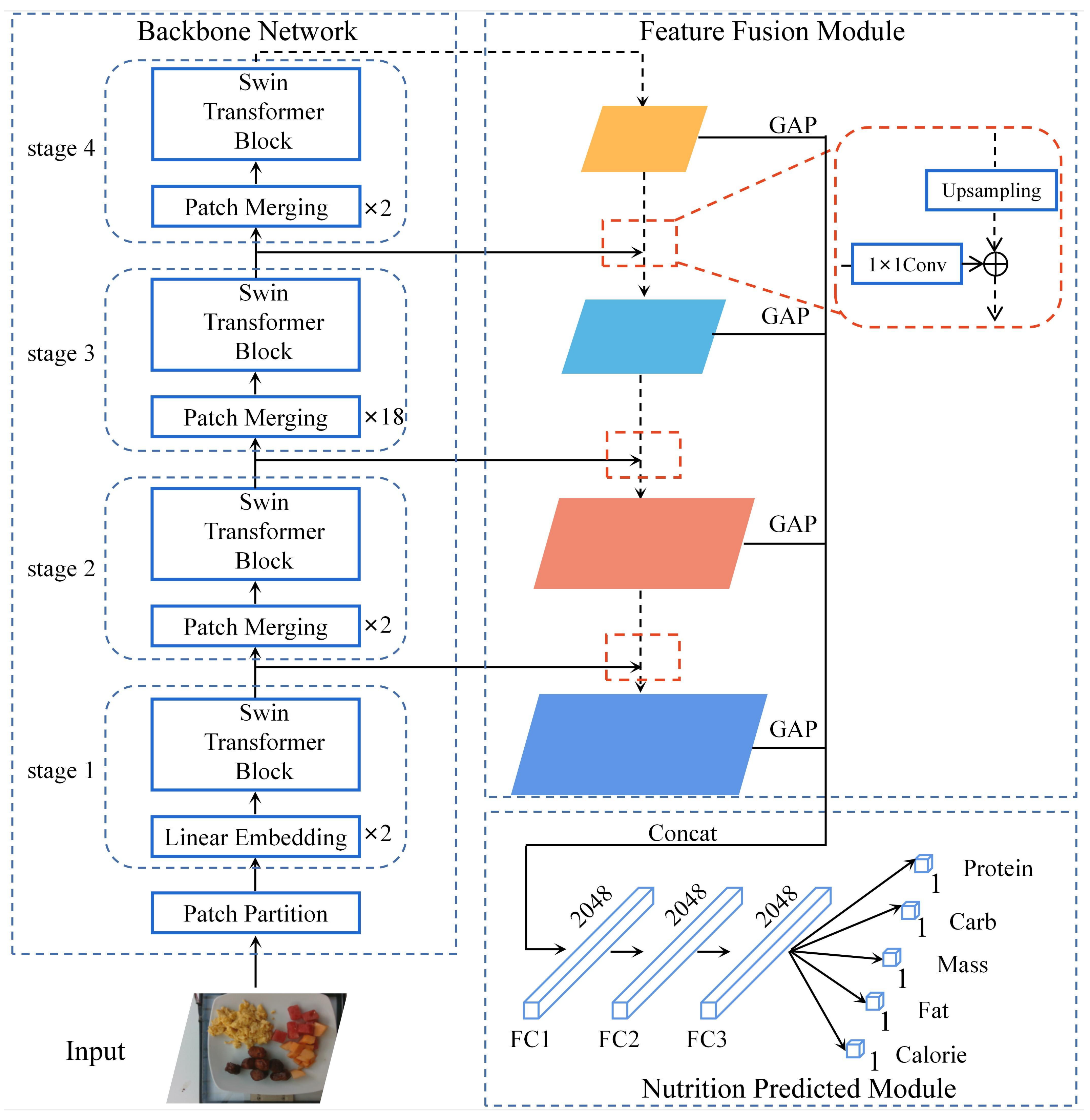
2.2.1. Overall Architecture of the Approach
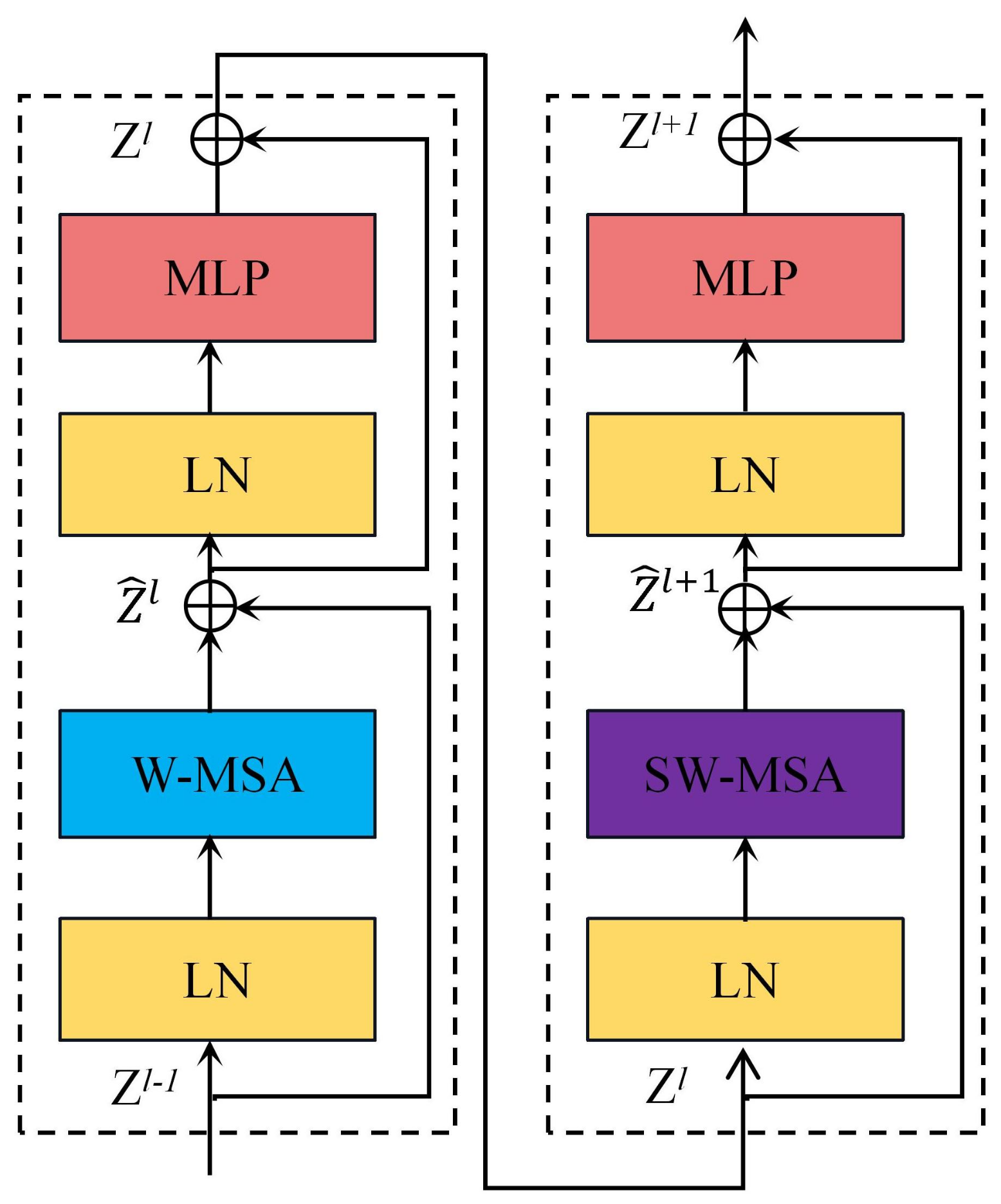
2.2.2. Swin-Nutrition’s Backbone Network
2.2.3. Feature Fusion Module
2.2.4. Multi-Task Loss Function
2.3. Evaluation Metrics
3. Results
3.1. Implementation Details
3.2. Comparison with Advanced NDDT Methods
- Inception V3 is an image classification network, which consists of 11 inception modules, each of which is composed of different convolutional layers combined by parallelism. Inception V3 introduces decomposed convolution, which is significant for enriching spatial features and increasing feature diversity.
- VGG-16 consists of thirteen convolutional layers and three fully connected layers, which simplify the structure of convolutional neural networks (using small convolutional kernels of 3 × 3 and maximum pooling layers of 2 × 2) and improve the performance of the network by constantly increasing the depth of the network.
- ResNet is a deeper network structure, which is composed of residual blocks and residual connections. The ResNet-101 network is mainly composed of four residual blocks, which solve the problem of gradient disappearance and gradient explosion in the deeper network.
- T2T-Vit is based on transformer structure for the image classification task, which mainly includes tokens-to-tokens block, position encoding, encoder, and MLP block. The network solves the problem that visual transformer is difficult to learn richer features by tokens-to-tokens block. The module models the local information of surrounding tokens by integrating adjacent tokens.
- Swin Transformer effectively combines the respective advantages of CNN and transformer. The method is designed for the structure of the transformer based on the hierarchical structure of CNN, and the multi-scale features for model prediction are generated by this blocked transformer module. The Swin Transformer can be widely applied to various fields of computer vision, such as image classification, image segmentation, and object detection.
3.3. Performance Analysis of the Swin-Nutrition
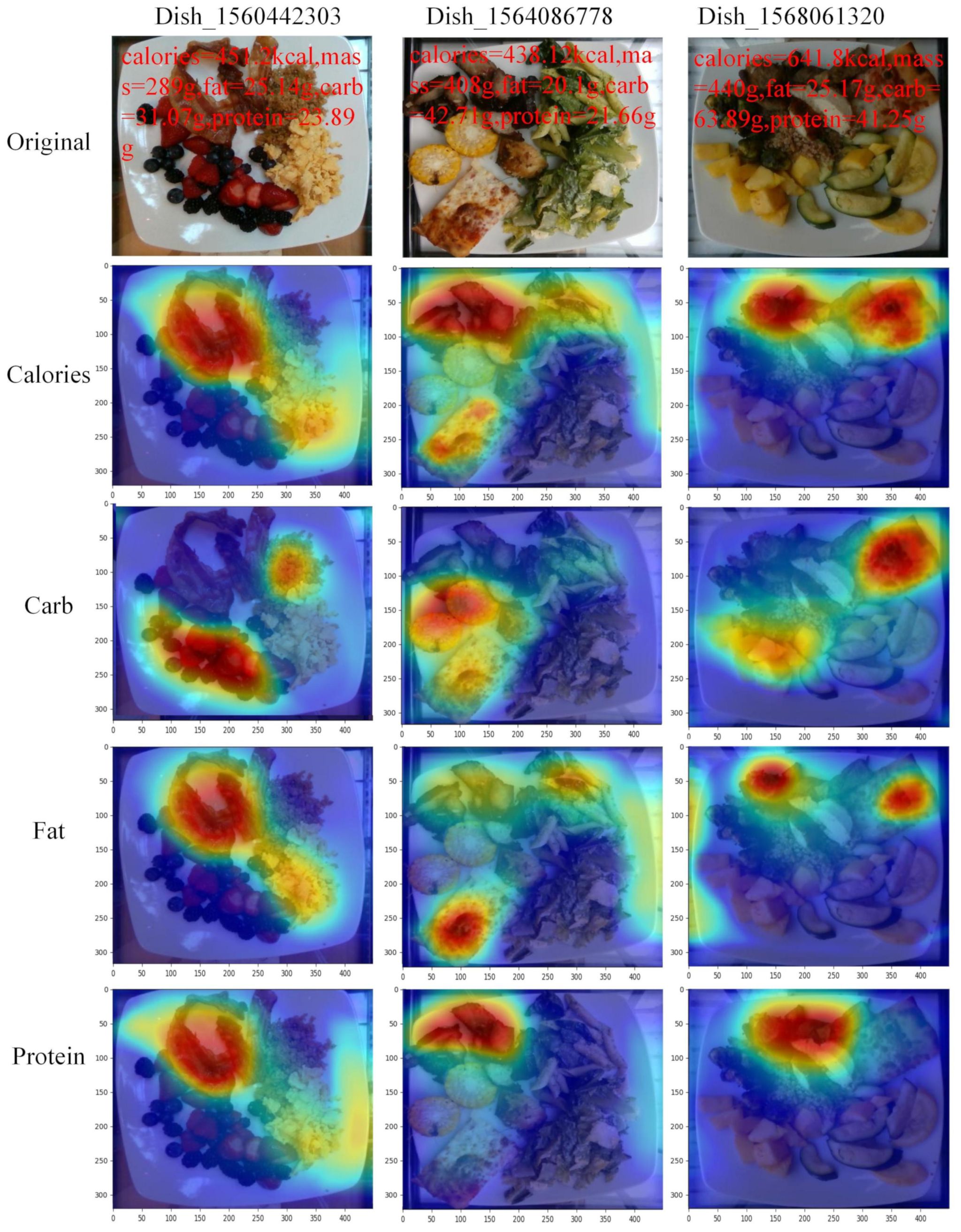
3.4. Visualization and Analysis of Nutrient Prediction Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Q.; Zhang, C.; Zhao, J.; Ouyang, Q. Recent advances in emerging imaging techniques for non-destructive detection of food quality and safety. Trends Anal. Chem. 2013, 52, 261–274. [Google Scholar] [CrossRef]
- Abasi, S.; Minaei, S.; Jamshidi, B.; Fathi, D. Dedicated non-destructive devices for food quality measurement: A review. Trends Food Sci. Technol. 2018, 78, 197–205. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, C.; Liu, F.; Qiu, Z.; He, Y. Application of deep learning in food: A review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1793–1811. [Google Scholar] [CrossRef] [PubMed]
- Min, W.; Jiang, S.; Liu, L.; Rui, Y.; Jain, R. A survey on food computing. ACM Comput. Surv. 2020, 52, 1–36. [Google Scholar] [CrossRef]
- Jamshidi, B.; Mohajerani, E.; Jamshidi, J.; Minaei, S.; Sharifi, A. Non-destructive detection of pesticide residues in cucumber using visible/near-infrared spectroscopy. Food Addit. Contam. Part A 2015, 32, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Hassoun, A.; Sahar, A.; Lakhal, L.; Aït-Kaddour, A. Fluorescence spectroscopy as a rapid and non-destructive method for monitoring quality and authenticity of fish and meat products: Impact of different preservation conditions. LWT Food Sci. Technol. 2019, 103, 279–292. [Google Scholar] [CrossRef]
- Oto, N.; Oshita, S.; Makino, Y.; Kawagoe, Y.; Sugiyama, J.; Yoshimura, M. Non-destructive evaluation of ATP content and plate count on pork meat surface by fluorescence spectroscopy. Meat Sci. 2013, 93, 579–585. [Google Scholar] [CrossRef]
- Melendez-Martinez, A.J.; Stinco, C.M.; Liu, C.; Wang, X.D. A simple HPLC method for the comprehensive analysis of cis/trans (Z/E) geometrical isomers of carotenoids for nutritional studies. Food Chem. 2013, 138, 1341–1350. [Google Scholar] [CrossRef]
- Maphosa, Y.; Jideani, V.A. Dietary fiber extraction for human nutrition—A review. Food Rev. Int. 2016, 32, 98–115. [Google Scholar] [CrossRef]
- Foster, E.; Bradley, J. Methodological considerations and future insights for 24-hour dietary recall assessment in children. Nutr. Res. 2018, 51, 1–11. [Google Scholar] [CrossRef]
- El-Mesery, H.S.; Mao, H.; Abomohra, A.E.F. Applications of non-destructive technologies for agricultural and food Products quality inspection. Sensors 2019, 19, 846. [Google Scholar] [CrossRef]
- Alexandrakis, D.; Downey, G.; Scannell, A.G. Rapid non-destructive detection of spoilage of intact chicken breast muscle using near-infrared and Fourier transform mid-infrared spectroscopy and multivariate statistics. Food Bioprocess Technol. 2012, 5, 338–347. [Google Scholar] [CrossRef]
- Xu, Q.; Wu, X.; Wu, B.; Zhou, H. Detection of apple varieties by near-infrared reflectance spectroscopy coupled with SPSO-PFCM. J. Food Process Eng. 2022, 45, e13993. [Google Scholar] [CrossRef]
- Liu, W.; Han, Y.; Wang, N.; Zhang, Z.; Wang, Q.; Miao, Y. Apple sugar content non-destructive detection device based on near-infrared multi-characteristic wavelength. J. Phys. Conf. Ser. 2022, 2221, 012012. [Google Scholar] [CrossRef]
- Liu, W.; Deng, H.; Shi, Y.; Xia, Y.; Liu, C.; Zheng, L. Application of multispectral imaging combined with machine learning methods for rapid and non-destructive detection of zearalenone (ZEN) in maize. Measurement 2022, 203, 111944. [Google Scholar] [CrossRef]
- Gong, Z.; Deng, D.; Sun, X.; Liu, J.; Ouyang, Y. Non-destructive detection of moisture content for Ginkgo biloba fruit with terahertz spectrum and image: A preliminary study. Infrared Phys. Technol. 2022, 120, 103997. [Google Scholar] [CrossRef]
- Xue, S.S.; Tan, J. Rapid and non-destructive composition analysis of cereal flour blends by front-face synchronous fluorescence spectroscopy. J. Cereal Sci. 2022, 106, 103494. [Google Scholar] [CrossRef]
- Min, W.; Wang, Z.; Liu, Y.; Luo, M.; Kang, L.; Wei, X.; Wei, X.; Jiang, S. Large scale visual food recognition. arXiv 2021, arXiv:2103.16107. [Google Scholar]
- Wang, W.; Min, W.; Li, T.; Dong, X.; Li, H.; Jiang, S. A review on vision-based analysis for automatic dietary assessment. Trends Food Sci. Technol. 2022, 122, 223–237. [Google Scholar] [CrossRef]
- Juan, T.M.; Benoit, A.; Jean-Pierre, D.; Marie-Claude, V. Precision nutrition: A review of personalized nutritional approaches for the prevention and management of metabolic syndrome. Nutrients 2017, 9, 913. [Google Scholar]
- Thames, Q.; Karpur, A.; Norris, W.; Xia, F.; Panait, L.; Weyand, T.; Sim, J. Nutrition5k: Towards Automatic Nutritional Understanding of Generic Food. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 8903–8911. [Google Scholar]
- Shim, J.S.; Oh, K.; Kim, H.C. Dietary assessment methods in epidemiologic studies. Epidemiol. Health 2014, 36, e2014009. [Google Scholar] [CrossRef]
- Illner, A.; Freisling, H.; Boeing, H.; Huybrechts, I.; Crispim, S.; Slimani, N. Review and evaluation of innovative technologies for measuring diet in nutritional epidemiology. Int. J. Epidemiol. 2012, 41, 1187–1203. [Google Scholar] [CrossRef] [PubMed]
- Gibney, M.; Allison, D.; Bier, D.; Dwyer, J. Uncertainty in human nutrition research. Nat. Food 2020, 1, 247–249. [Google Scholar] [CrossRef]
- Myers, A.; Johnston, N.; Rathod, V.; Korattikara, A.; Gorban, A.; Silberman, N.; Guadarrama, S.; Papandreou, G.; Huang, J.; Murphy, K. Im2Calories: Towards an automated mobile vision food diary. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1233–1241. [Google Scholar]
- Ege, T.; Yanai, K. Image-Based Food Calorie Estimation Using Knowledge on Food Categories, Ingredients and Cooking Directions. In Proceedings of the on Thematic Workshops of ACM Multimedia, Mountain View, CA, USA, 23–27 October 2017; pp. 367–375. [Google Scholar]
- Begum, N.; Goyal, A.; Sharma, S. Artificial Intelligence-Based Food Calories Estimation Methods in Diet Assessment Research. In Artificial Intelligence Applications in Agriculture and Food Quality Improvement; IGI Global: Hershey, PA, USA, 2022; pp. 276–290. [Google Scholar]
- Jaswanthi, R.; Amruthatulasi, E.; Bhavyasree, C.; Satapathy, A. A Hybrid Network Based on GAN and CNN for Food Segmentation and Calorie Estimation. In Proceedings of the International Conference on Sustainable Computing and Data Communication Systems (ICSCDS), Erode, India, 7–9 April 2022; pp. 436–441. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Kendall, A.; Gal, Y.; Cipolla, R. Multi-task learning using uncertainty to weigh losses for scene geometry and semantics. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7482–7491. [Google Scholar]
- De Myttenaere, A.; Golden, B.; Le Grand, B.; Rossi, F. Mean absolute percentage error for regression models. Neurocomputing 2016, 192, 38–48. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Yuan, L.; Chen, Y.; Wang, T.; Yu, W.; Shi, Y.; Jiang, Z.H.; Tay, F.E.; Feng, J.; Yan, S. Tokens-to-token vit: Training vision transformers from scratch on imagenet. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 558–567. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Zhang, T.; Wu, X.; Wu, B.; Dai, C.; Fu, H. Rapid authentication of the geographical origin of milk using portable near-infrared spectrometer and fuzzy uncorrelated discriminant transformation. J. Food Process Eng. 2022, 45, e14040. [Google Scholar] [CrossRef]
- Liu, H.; Ji, Z.; Liu, X.; Shi, C.; Yang, X. Non-destructive determination of chemical and microbial spoilage indicators of beef for freshness evaluation using front-face synchronous fluorescence spectroscopy. Food Chem. 2020, 321, 126628. [Google Scholar] [CrossRef]
| Method Types | Methods | Backbone | Pretain | Calories PMAE (%) | Mass PMAE (%) | Fat PMAE (%) | Carb. PMAE (%) | Protein PMAE (%) | Mean PMAE (%) |
|---|---|---|---|---|---|---|---|---|---|
| CNN-based methods | Inception V3 [35] | Inception V3 | N | 22.6 | 16.3 | 32.5 | 27.5 | 25.1 | 24.8 |
| VGG-16 [36] | VGG-16 | N | 21.9 | 16.0 | 32.1 | 28.0 | 24.6 | 24.5 | |
| ResNet-101+FPN [30] | ResNet-101 | N | 20.1 | 15.3 | 29.8 | 26.5 | 22.9 | 22.9 | |
| transformer- based methods | T2T-ViT [37] | T2T-ViT | N | 17.0 | 13.8 | 23.8 | 22.4 | 17.2 | 18.8 |
| Swin Transformer [29] | Swin-B | Y | 16.4 | 13.3 | 23.6 | 21.6 | 16.6 | 18.3 | |
| Swin-Nutrition | Swin-B | Y | 15.3 | 12.5 | 22.1 | 20.8 | 15.4 | 17.2 |
| Method | Nutrition5k | |||||||
|---|---|---|---|---|---|---|---|---|
| FFM | Multi-Task Loss | Pretrain-ing | Calories PMAE (%) | Mass PMAE (%) | Fat PMAE (%) | Carb. PMAE (%) | Protein PMAE (%) | Mean PMAE (%) |
| 17.1 | 14.6 | 24.3 | 22.3 | 17.2 | 19.1 | |||
| ✓ | 16.6 | 13.8 | 23.7 | 21.6 | 16.5 | 18.4 | ||
| ✓ | ✓ | 15.8 | 12.9 | 23.0 | 21.1 | 15.9 | 17.7 | |
| ✓ | ✓ | ✓ | 15.3 | 12.5 | 22.1 | 20.8 | 15.4 | 17.2 |
| Ingredient | Cal (kcal) | Fat (g) | Carb. (g) | Protein (g) |
|---|---|---|---|---|
| scrambled eggs | 111 | 8.25 | 1.2 | 7.5 |
| berries | 83.79 | 0.44 | 20.58 | 1.03 |
| quinoa | 44.4 | 0.70 | 7.77 | 1.63 |
| bacon | 205.58 | 15.96 | 0.53 | 14.06 |
| Sum | 444.77 | 25.35 | 30.08 | 24.22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, W.; Hou, S.; Jia, W.; Zheng, Y. Rapid Non-Destructive Analysis of Food Nutrient Content Using Swin-Nutrition. Foods 2022, 11, 3429. https://doi.org/10.3390/foods11213429
Shao W, Hou S, Jia W, Zheng Y. Rapid Non-Destructive Analysis of Food Nutrient Content Using Swin-Nutrition. Foods. 2022; 11(21):3429. https://doi.org/10.3390/foods11213429
Chicago/Turabian StyleShao, Wenjing, Sujuan Hou, Weikuan Jia, and Yuanjie Zheng. 2022. "Rapid Non-Destructive Analysis of Food Nutrient Content Using Swin-Nutrition" Foods 11, no. 21: 3429. https://doi.org/10.3390/foods11213429
APA StyleShao, W., Hou, S., Jia, W., & Zheng, Y. (2022). Rapid Non-Destructive Analysis of Food Nutrient Content Using Swin-Nutrition. Foods, 11(21), 3429. https://doi.org/10.3390/foods11213429