Geographical Origin Differentiation of Rice by LC–MS-Based Non-Targeted Metabolomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Sample Processing
2.3. Liquid Chromatography–Mass Spectrometry
2.4. Data Analysis
3. Results and Discussion
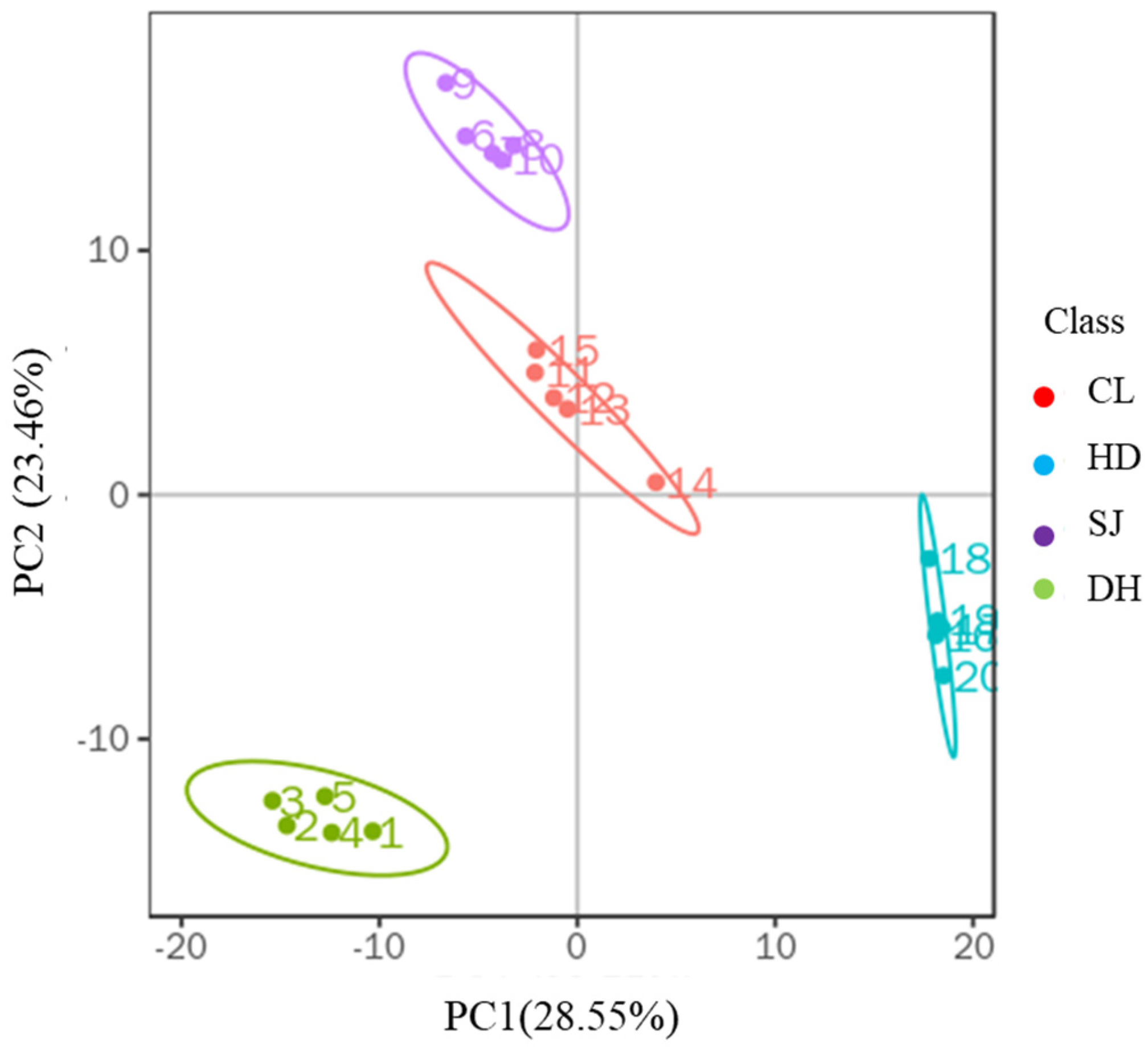
3.1. Principal Component Analysis
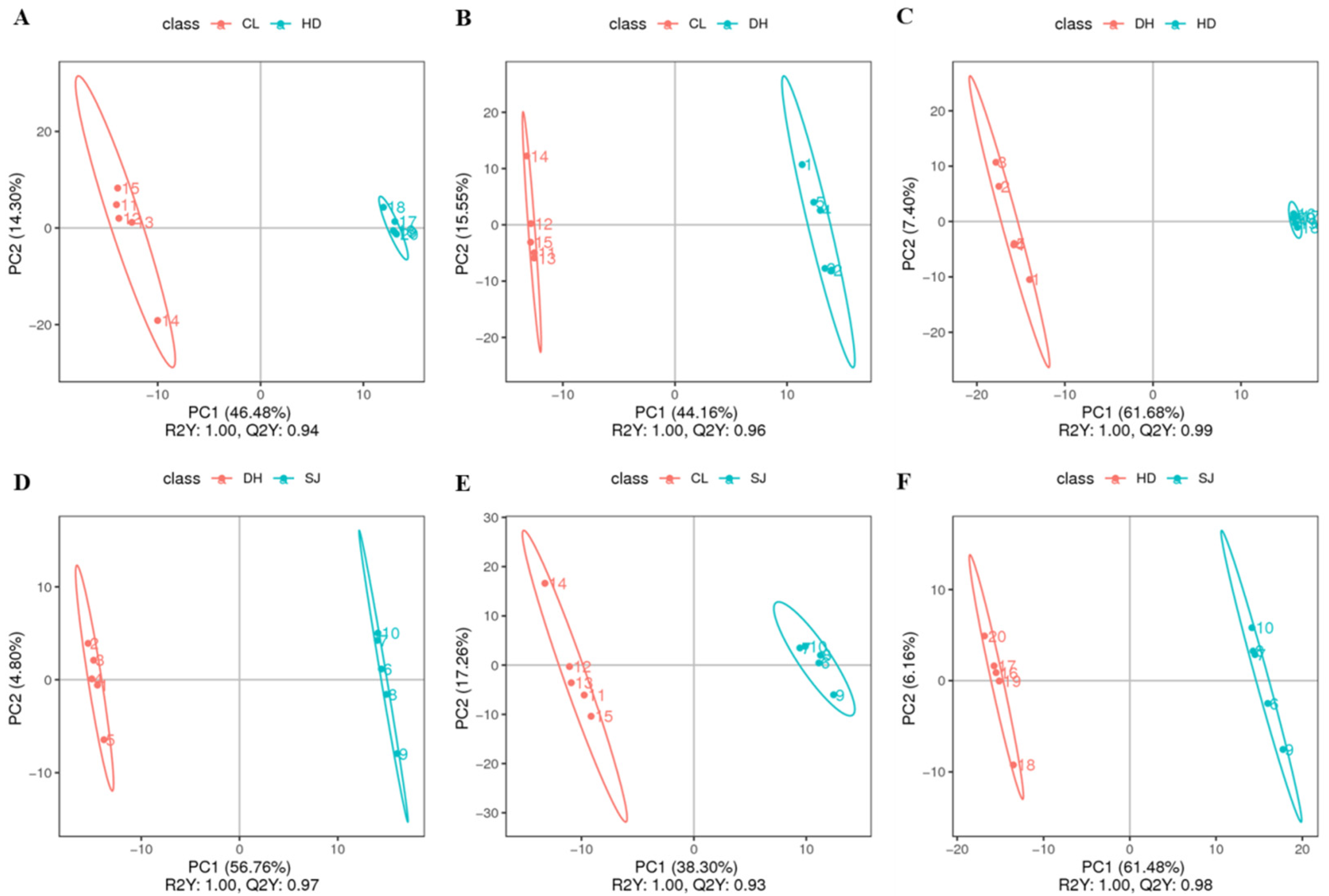
3.2. Partial Least Squares Discriminant Analysis
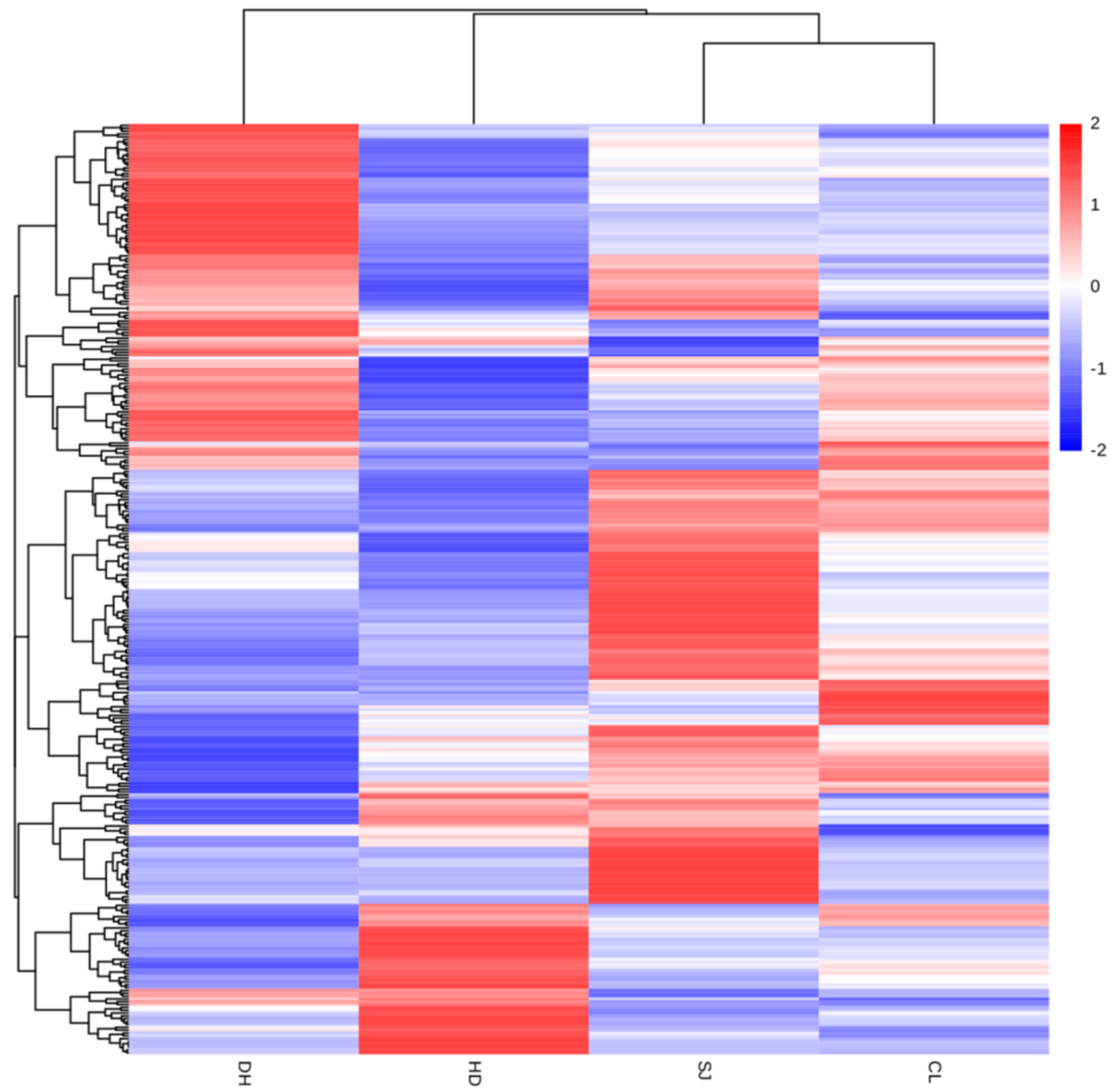
3.3. Cluster Analysis
3.4. Determination of Differential Metabolites
3.5. Annotation of Metabolites
3.6. Metabolite Pathway Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Arslan, M.; Zareef, M.; Tahir, H.E.; Zhang, J.; Ahmad, W.; Rakha, A.; Shi, J.; Xiaobo, Z.; Khan, M.R. Discrimination of basmati rice adulteration using colorimetric sensor array system. Food Control 2022, 132, 108513. [Google Scholar]
- Śliwińska-Bartel, M.; Burns, D.T.; Elliott, C. Rice fraud a global problem: A review of analytical tools to detect species, country of origin and adulterations. Trends Food Sci. Technol. 2021, 116, 36–46. [Google Scholar]
- Li, Q.; Zeng, J.; Lin, L.; Zhang, J.; Zhu, J.; Yao, L.; Wang, S.; Yao, Z.; Wu, Z. Low risk of category misdiagnosis of rice syrup adulteration in three botanical origin honey by ATR-FTIR and general model. Food Chem. 2020, 332, 127356. [Google Scholar]
- Le Nguyen Doan, D.; Nguyen, Q.C.; Marini, F.; Biancolillo, A. Authentication of rice (Oryza sativa L.) using near infrared spectroscopy combined with different chemometric classification strategies. Appl. Sci. 2021, 11, 362. [Google Scholar]
- Fang, C.J.; You, H.C.; Huang, Z.L.; Hsu, C.L.; Tsai, C.F.; Lin, Y.T.; Kao, Y.M.; Tseng, S.H.; Wang, D.Y.; Su, N.W. Simultaneous analysis of the stable carbon isotope ratios of acetoin and acetic acid by GC-C-IRMS for adulteration detection in brewed rice vinegar products. J. Agric. Food Chem. 2020, 68, 14252–14260. [Google Scholar]
- Uawisetwathana, U.; Karoonuthaisiri, N. Metabolomics for rice quality and traceability: Feasibility and future aspects. Curr. Opin. Food Sci. 2019, 28, 58–66. [Google Scholar]
- Xiao, R.; Ma, Y.; Zhang, D.; Qian, L. Discrimination of conventional and organic rice using untargeted LC-MS-based metabolomics. J. Cereal Sci. 2018, 82, 73–81. [Google Scholar]
- Asimi, S.; Ren, X.; Zhang, M.; Li, S.; Guan, L.; Wang, Z.; Liang, S.; Wang, Z. Fingerprinting of volatile organic compounds for the geographical discrimination of rice samples from northeast China. Foods 2022, 11, 1695. [Google Scholar]
- Wang, T.; Duedahl-Olesen, L.; Frandsen, H.L. Targeted and non-targeted unexpected food contaminants analysis by LC/HRMS: Feasibility study on rice. Food Chem. 2021, 338, 127957. [Google Scholar]
- Schütz, D.; Achten, E.; Creydt, M.; Riedl, J.; Fischer, M. Non-targeted LC-MS metabolomics approach towards an authentication of the geographical origin of grain maize (Zea mays L.) samples. Foods 2021, 10, 2160. [Google Scholar]
- Ryan, E.; Baxter, B.; Li, K.; Wolfe, L.; Yao, L.; Broecling, C.; Borreson, E.; Zhang, L.; Zarei, I.; Beale, M. Developing biomarkers of rice bran and navy bean intake via integrated metabolomics from infants, children and adults for association with gut health properties. Curr. Dev. Nutr. 2020, 4 (Suppl. 2), 463. [Google Scholar]
- Duan, H.; Tong, H.; Zhu, A.; Zhang, H.; Liu, L. Effects of heat, drought and their combined effects on morphological structure and physicochemical properties of rice (Oryza sativa L.) starch. J. Cereal Sci. 2020, 95, 103059. [Google Scholar]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 1–14. [Google Scholar]
- Valderrama, L.; Valderrama, P.; Carasek, E. A semi-quantitative model through PLS-DA in the evaluation of carbendazim in grape juices. Food Chem. 2022, 368, 130742. [Google Scholar]
- Mansuri, S.M.; Chakraborty, S.K.; Mahanti, N.K.; Pandiselvam, R. Effect of germ orientation during Vis-NIR hyperspectral imaging for the detection of fungal contamination in maize kernel using PLS-DA, ANN and 1D-CNN modelling. Food Control 2022, 139, 109077. [Google Scholar]
- Kotamreddy, J.N.R.; Hansda, C.; Mitra, A. Semi-targeted metabolomic analysis provides the basis for enhanced antioxidant capacities in pigmented rice grains. J. Food Meas. Charact. 2020, 14, 1183–1191. [Google Scholar]
- Sapozhnikova, Y.; Nuñez, A. Non-targeted analysis with liquid chromatography-high resolution mass spectrometry for the identification of food packaging migrants. J. Chromatogr. A 2022, 1676, 463215. [Google Scholar]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar]
- Kang, Y.; Lee, B.M.; Lee, E.M.; Kim, C.H.; Seo, J.A.; Choi, H.K.; Kim, Y.S.; Lee, D.Y. Unique metabolic profiles of Korean rice according to polishing degree, variety, and geo-environmental factors. Foods 2021, 10, 711. [Google Scholar]
- Lim, D.K.; Mo, C.; Lee, D.K.; Long, N.P.; Lim, J.; Kwon, S.W. Non-destructive profiling of volatile organic compounds using HS-SPME/GC–MS and its application for the geographical discrimination of white rice. J. Food Drug Anal. 2018, 26, 260–267. [Google Scholar]
- Kukusamude, C.; Sricharoen, P.; Limchoowong, N.; Kongsri, S. Heavy metals and probabilistic risk assessment via rice consumption in Thailand. Food Chem. 2021, 334, 127402. [Google Scholar]
- Bin Yusof, M.T.; Kershaw, M.J.; Soanes, D.M.; Talbot, N.J. FAR1 and FAR2 regulate the expression of genes associated with lipid metabolism in the rice blast fungus Magnaporthe oryzae. PLoS ONE 2014, 9, e99760. [Google Scholar]
- Chen, Z.; Du, Y.; Mao, Z.; Zhang, Z.; Li, P.; Cao, C. Grain starch, fatty acids, and amino acids determine the pasting properties in dry cultivation plus rice cultivars. Food Chem. 2022, 373, 131472. [Google Scholar]
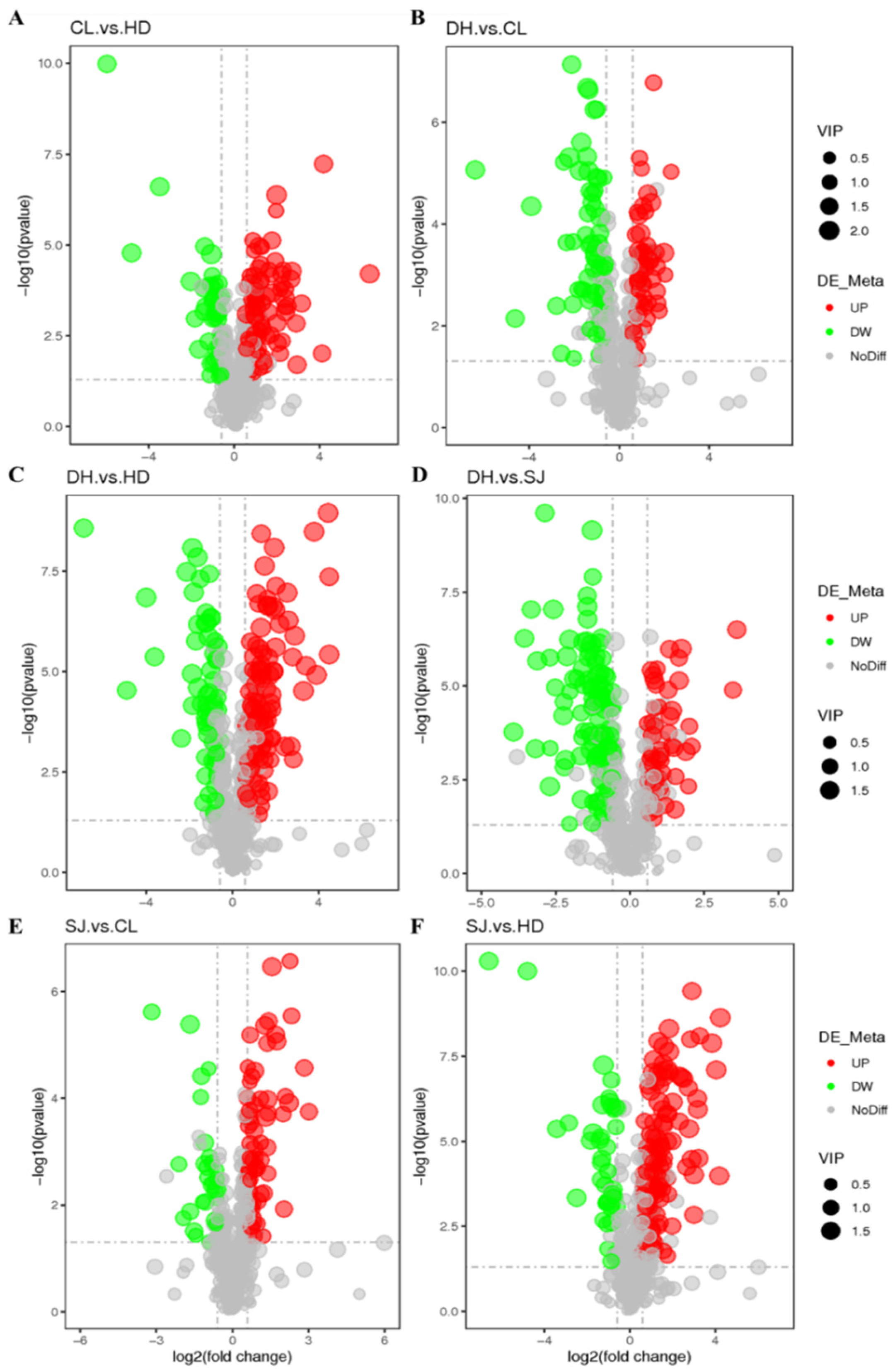

| Group | Number of Differential Metabolites | Up | Down |
|---|---|---|---|
| CL vs. HD | 133 | 93 | 40 |
| CL vs. DH | 126 | 54 | 72 |
| DH vs. SJ | 164 | 54 | 110 |
| DH vs. HD | 161 | 102 | 59 |
| SJ vs. CL | 94 | 66 | 28 |
| SJ vs. HD | 167 | 125 | 42 |
| Group | TOP3 Pathways | Number of Differential Metabolites |
|---|---|---|
| CL vs. HD | (a) Caffeine metabolic pathway composed of xanthine and 7—methyl xanthine (b) Biosynthesis pathway of terpenoid main chain composed of valproic acid; (c) The biosynthesis pathways of stilbenes, diarylheptanes and gingerols composed of chlorogenic acid. | 22 |
| CL vs. DH | (a) Biosynthesis pathway of panquinone and other terpene quinones composed of L-tyrosine, transcinnamic acid and 4-hydroxybenzoic acid; (b) Taurine and taurine metabolic pathways composed of sulfoacetic acid and taurine; (c) Plant hormone signal transduction pathway composed of jasmonic acid and salicylic acid | 32 |
| DH vs. SJ | (a) Fatty acid degradation pathway composed of glutaric acid and palmitic acid; (b) Lysine degradation pathway composed of glutaric acid and acetic acid; (c) Taurine and taurine metabolic pathways composed of sulfoacetic acid and taurine. | 39 |
| DH vs. HD | (a) Fatty acid degradation pathway composed of glutaric acid and palmitic acid; (b) Tryptophan metabolic pathway composed of N-formyl canine uridine and serotonin; (c) Biosynthesis pathway of pantothenic acid and coenzyme A composed of 3′-dephosphate-CoA and pantothenic acid | 35 |
| SJ vs. CL | (a) Caffeine metabolic pathway composed of 7-methylxanthine and xanthine; (b) Riboflavin metabolic pathway composed of vitamin B2; (c) Vitamin B6 metabolic pathway composed of 4-pyridoxine. | 18 |
| SJ vs. HD | (a) Ascorbic acid and aldoic acid metabolic pathway composed of d-glycoacid and L-ascorbic acid; (b) Valine, leucine and isoleucine degradation pathways composed of methylmalonic acid and acetoacetic acid; (c) Propionic acid metabolic pathway composed of methylmalonic acid and acetoacetic acid. | 39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Tan, M.; Deng, H.; Yang, X.; Yu, Y.; Zhou, D.; Dong, H. Geographical Origin Differentiation of Rice by LC–MS-Based Non-Targeted Metabolomics. Foods 2022, 11, 3318. https://doi.org/10.3390/foods11213318
Li Z, Tan M, Deng H, Yang X, Yu Y, Zhou D, Dong H. Geographical Origin Differentiation of Rice by LC–MS-Based Non-Targeted Metabolomics. Foods. 2022; 11(21):3318. https://doi.org/10.3390/foods11213318
Chicago/Turabian StyleLi, Zhanming, Mengmeng Tan, Huxue Deng, Xu Yang, Yue Yu, Dongren Zhou, and Hao Dong. 2022. "Geographical Origin Differentiation of Rice by LC–MS-Based Non-Targeted Metabolomics" Foods 11, no. 21: 3318. https://doi.org/10.3390/foods11213318
APA StyleLi, Z., Tan, M., Deng, H., Yang, X., Yu, Y., Zhou, D., & Dong, H. (2022). Geographical Origin Differentiation of Rice by LC–MS-Based Non-Targeted Metabolomics. Foods, 11(21), 3318. https://doi.org/10.3390/foods11213318







