Symbiotic Husbandry of Chickens and Pigs Does Not Increase Pathogen Transmission Risk
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design (Sampling)
2.1.1. Pre Sampling
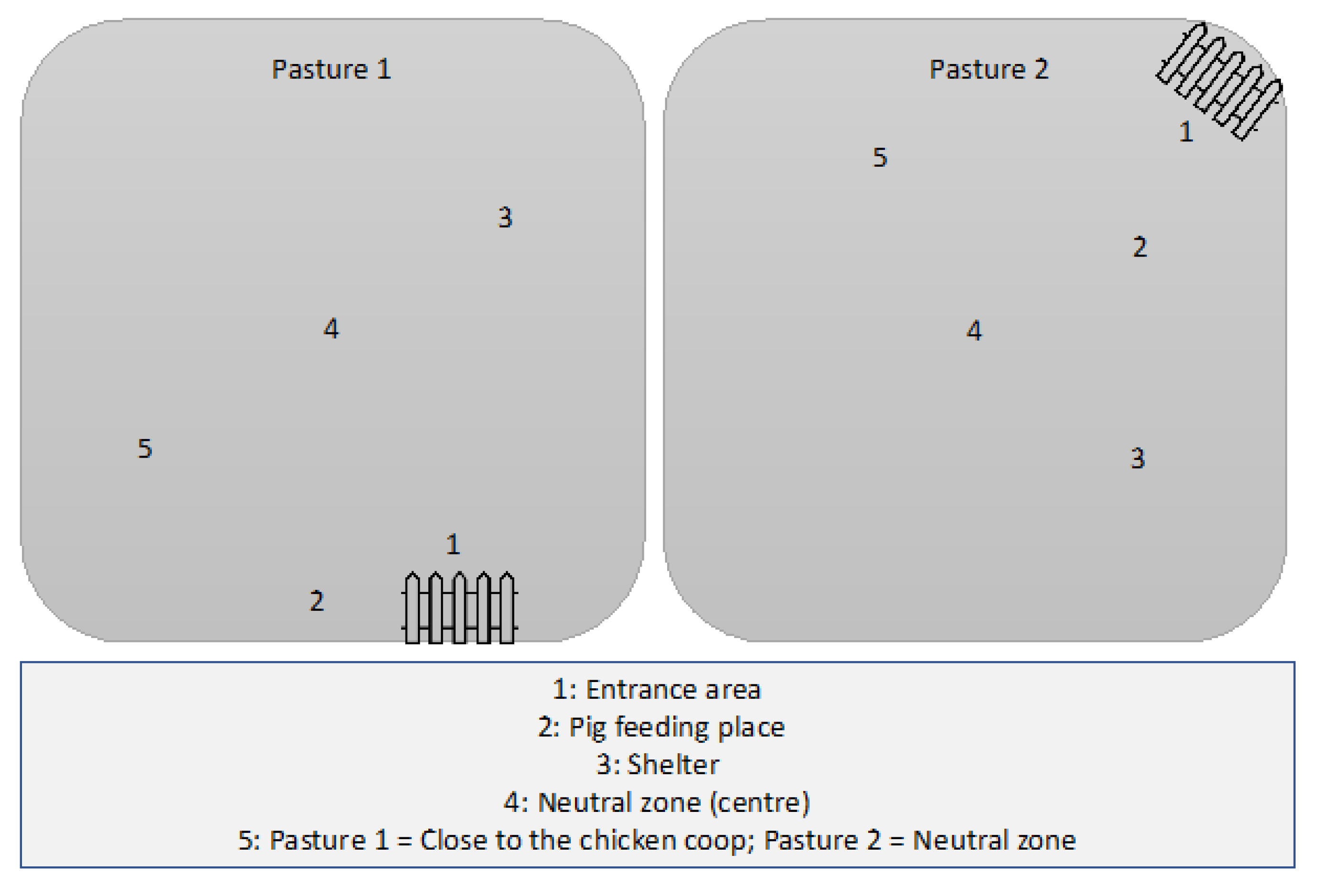
2.1.2. Forms of Husbandry
2.2. Sample Collection
2.2.1. Animals
2.2.2. Soil
2.3. Sample Preparation
2.3.1. Animal Samples
2.3.2. Soil Samples
2.4. Bacteriological Investigation
2.4.1. Isolation of Escherichia coli
2.4.2. Isolation of Salmonella spp.
2.4.3. Isolation of Campylobacter spp.
2.4.4. Species Identification by MALDI-TOF MS
2.4.5. FT-IR
2.5. Analysis for Similarities
2.6. Statistical Analysis
2.6.1. Pearson´s Correlation
2.6.2. Chi-Square
3. Results
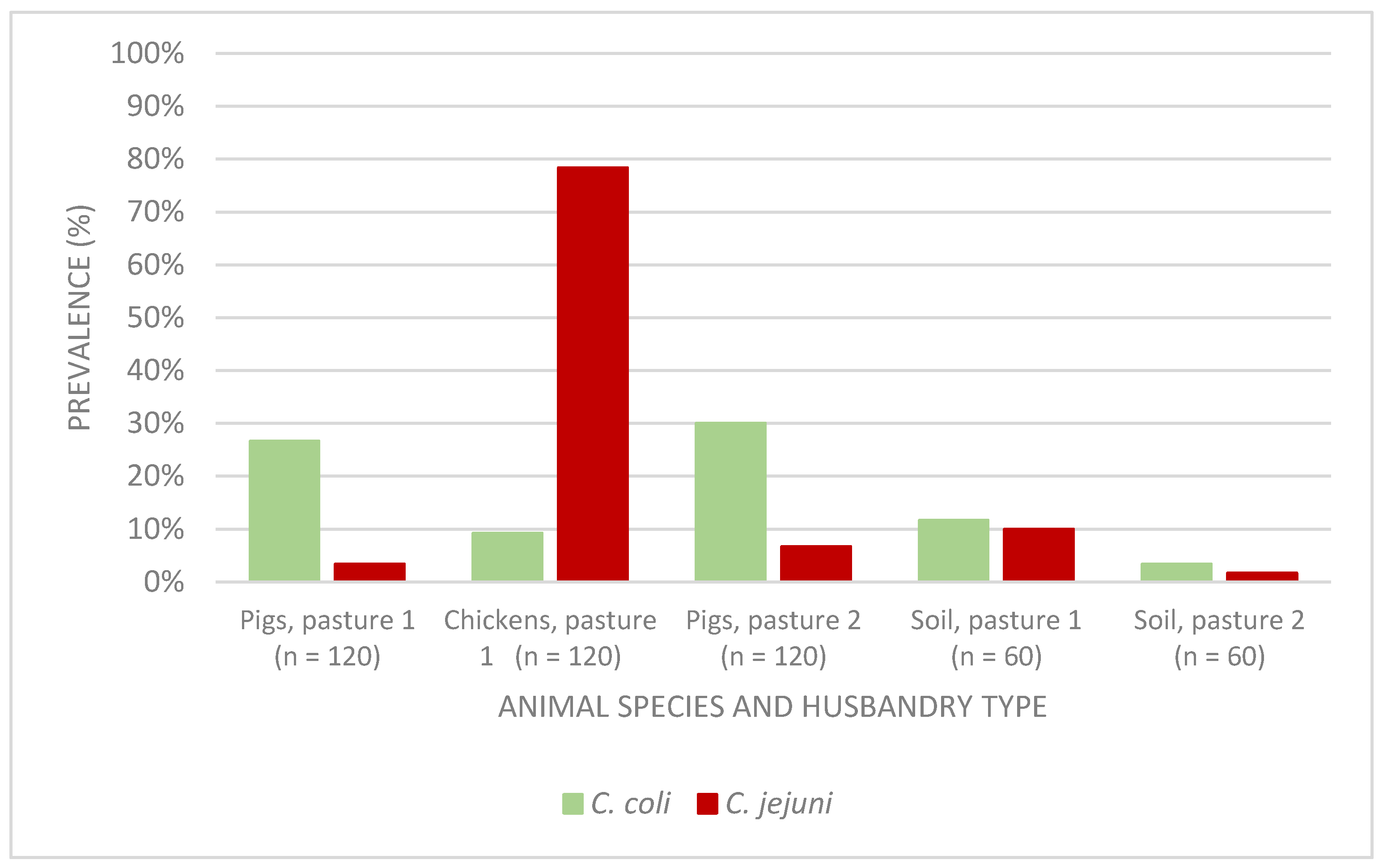
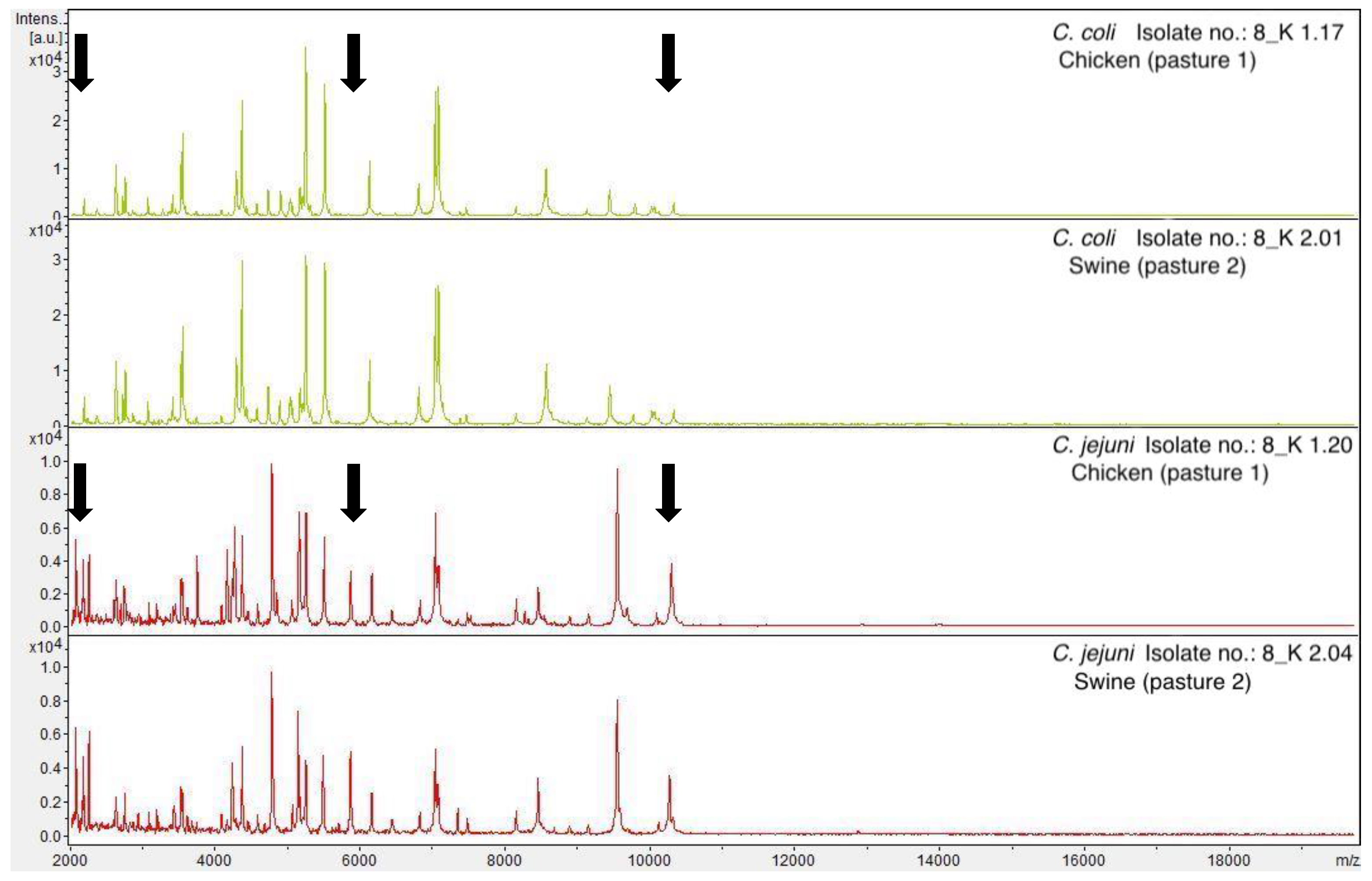
3.1. Detection and Similarity Analysis of Campylobacter spp.
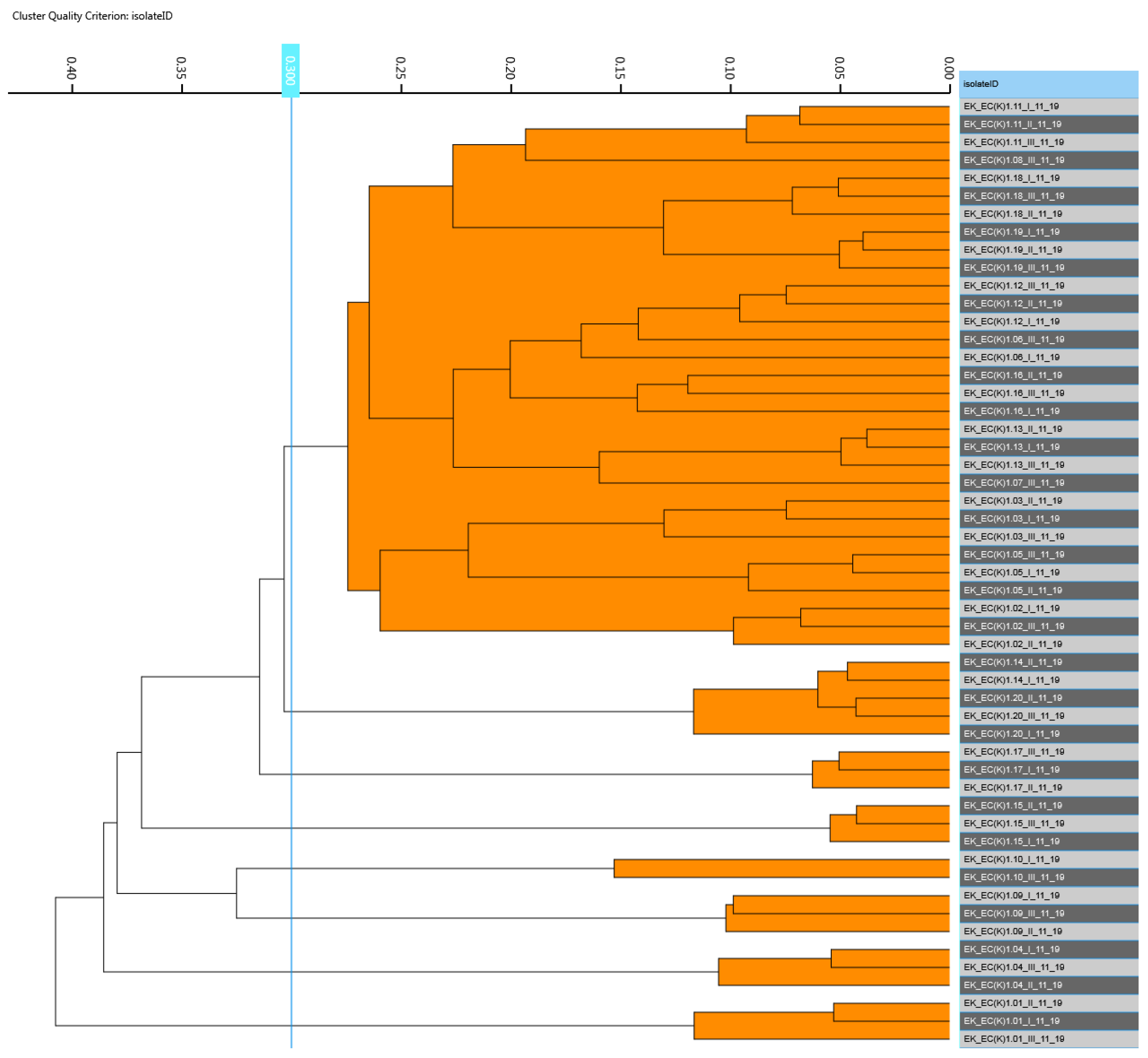
3.2. Detection and Similarity Analysis of Escherichia coli
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christoph-Schulz, I. SocialLab—Nutztierhaltung im Spiegel der Gesellschaft. J. Consum. Prot. Food Saf. 2018, 13, 145–236. [Google Scholar] [CrossRef]
- Jochemsen, H. An ethical foundation for careful animal husbandry. NJAS—Wagening. J. Life Sci. 2013, 66, 55–63. [Google Scholar] [CrossRef][Green Version]
- Brumfiel, G. Animal-rights activists invade Europe. Nature 2008, 451, 1034–1036. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Albernaz-Gonçalves, R.; Olmos, G.; Hötzel, M. My pigs are ok, why change?–Animal welfare accounts of pig farmers. Animal 2021, 15, 100154. [Google Scholar] [CrossRef] [PubMed]
- Wissenschaftlicher Beirat Agrarpolitik beim BMEL. Wege zu Einer Gesell-Schaftlich Akzeptierten Nutztierhaltung. Kurzfassung des Gutachtens; Wissenschaftlicher Beirat Agrarpolitik beim BMEL: Berlin, Germany, 2015. [Google Scholar]
- Eurobarometer, S. Attitudes of EU Citizens towards Animal Welfare; European Commission: Brussels, Belgium, 2007. [Google Scholar]
- Bundesministerium für Ernährung und Landwirtschaft. Deutschland, Wie es Isst. Available online: https://www.bmel.de/SharedDocs/Downloads/Broschueren/Ernaehrungsreport2017.pdf?__blob=publicationFile (accessed on 25 February 2022).
- Tallentire, C.W.; Edwards, S.A.; Van Limbergen, T.; Kyriazakis, I. The challenge of incorporating animal welfare in a social life cycle assessment model of European chicken production. Int. J. Life Cycle Assess. 2019, 24, 1093–1104. [Google Scholar] [CrossRef]
- Bundesanstalt für Landwirtschaft und Ernährung. Ökologische Tierhaltung. Available online: https://www.praxis-agrar.de/tier/artikel/oekologische-tierhaltung/ (accessed on 25 February 2022).
- Aerts, S.; Lips, D.; Spencer, S.; Decuypere, E.; De Tavernier, J. A new framework for the assessment of animal welfare: Integrating existing knowledge from a practical ethics perspective. J. Agric. Environ. Ethics 2006, 19, 67–76. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, e06971. [Google Scholar]
- European Food Safety Authority. The European Union One Health 2019 Zoonoses Report; EFSA: Parma, Italy, 2021.
- Robert-Koch-Institut. Campylobacter-Enteritis. Available online: https://www.rki.de/DE/Content/Infekt/EpidBull/Merkblaetter/Ratgeber_Campylobacter.html (accessed on 1 April 2022).
- Friedman, C.R.; Hoekstra, R.M.; Samuel, M.; Marcus, R.; Bender, J.; Shiferaw, B.; Reddy, S.; Ahuja, S.D.; Helfrick, D.L.; Hardnett, F.; et al. Risk Factors for Sporadic Campylobacter Infection in the United States: A Case-Control Study in FoodNet Sites. Clin. Infect. Dis. 2004, 38, S285–S296. [Google Scholar] [CrossRef]
- Endtz, H.P. 50—Campylobacter Infections. In Hunter’s Tropical Medicine and Emerging Infectious Diseases, 10th ed.; Ryan, E.T., Hill, D.R., Solomon, T., Aronson, N.E., Endy, T.P., Eds.; Elsevier: London, UK, 2020; pp. 507–511. [Google Scholar]
- Møller Nielsen, E.; Engberg, J.; Madsen, M. Distribution of serotypes of Campylobacter jejuni and C. coli from Danish patients, poultry, cattle and swine. FEMS Immunol. Med. Microbiol. 1997, 19, 47–56. [Google Scholar] [CrossRef]
- Alter, T.; Gaull, F.; Kasimir, S.; Gürtler, M.; Mielke, H.; Linnebur, M.; Fehlhaber, K. Prevalences and transmission routes of Campylobacter spp. strains within multiple pig farms. Vet. Microbiol. 2005, 108, 251–261. [Google Scholar] [CrossRef]
- Robert-Koch-Institut. Infektionsepidemiologisches Jahrbuch meldepflichtiger Krankheiten für 2020; RKI: Berlin, Germany, 2021. [Google Scholar]
- Duijkeren, E.V.; Wannet, W.J.B.; Houwers, D.J.; Pelt, W.V. Serotype and Phage Type Distribution of Salmonella Strains Isolated from Humans, Cattle, Pigs, and Chickens in The Netherlands from 1984 to 2001. J. Clin. Microbiol. 2002, 40, 3980–3985. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shivaprasad, H. Fowl typhoid and pullorum disease. Rev. Sci. Tech. 2000, 19, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Calva, E.; Maloy, S. One Health and Food-Borne Disease: Salmonella Transmission between Humans, Animals, and Plants. Microbiol. Spectr. 2014, 2, 26082128. [Google Scholar] [CrossRef] [PubMed]
- Ellington, C.; Hebron, C.; Crespo, R.; Machado, G. Unraveling the Contact Network Patterns between Commercial Turkey Operation in North Carolina and the Distribution of Salmonella Species. Pathogens 2021, 10, 1539. [Google Scholar] [CrossRef]
- Robert-Koch-Institut. Salmonellose (Salmonellen-Gastroenteritis). RKI-Ratgeber Ärzte 2016, 13. [Google Scholar] [CrossRef]
- Allocati, N.; Masulli, M.; Alexeyev, M.F.; Di Ilio, C. Escherichia coli in Europe: An Overview. Int. J. Environ. Res. Public Health 2013, 10, 6235–6254. [Google Scholar] [CrossRef] [PubMed]
- Kayser, F.H. Medical Microbiology; Thieme: Stuttgart, Germany, 2005; p. 292. [Google Scholar]
- Schwaiger, K.; Harms, K.; Hölzel, C.; Meyer, K.; Karl, M.; Bauer, J. Tetracycline in liquid manure selects for co-occurrence of the resistance genes tet(M) and tet(L) in Enterococcus faecalis. Vet. Microbiol. 2009, 139, 386–392. [Google Scholar] [CrossRef]
- Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen. Probenahme und Untersuchung am 21.9./3.11.2011 in der Umgebung der Deponie Eyller Berg; LANUV/NRW: Essen, Germany, 2011. [Google Scholar]
- Lauer, W.F.; Martinez, F.L.; Patel, A. Validation of RAPID’E. coli 2 for Enumeration and Differentiation of Escherichia coli and Other Coliform Bacteria in Selected FoodsPerformance-Tested MethodSM 050601. J. AOAC Int. 2007, 90, 1284–1315. [Google Scholar] [CrossRef]
- BIORAD. RAPID’E.coli 2 for Water Testing for the Enumeration of Escherichia coli and Coliforms in Drinking Water for Human Consumption; Adgene Laboratoire: Le Hom, France, 2019. [Google Scholar]
- Bruker Daltonik GmbH. Compass 1.4 for FLEX Series User Manual, Doc No. 269834. User Manual—Volume 1; Bruker Daltonik GmbH: Bremen, Germany, 2015. [Google Scholar]
- Filip, Z.; Hermann, S.; Demnerová, K. FT-IR spectroscopic characteristics of differently cultivated Escherichia coli. Czech J. Food Sci. 2009, 26, 458–463. [Google Scholar] [CrossRef]
- Naumann, D.; Helm, D.; Labischinski, H. Microbiological characterizations by FT-IR spectroscopy. Nature 1991, 351, 81–82. [Google Scholar] [CrossRef] [PubMed]
- Ekruth, J.; Gottschalk, C.; Ulrich, S.; Gareis, M.; Schwaiger, K. Differentiation of S. chartarum (Ehrenb.) S. Hughes Chemotypes A and S via FT-IR Spectroscopy. Mycopathologia 2020, 185, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Bruker Daltonik GmbH. Biotyper IR. User Manual. Doc. No. 5022573 ed; Bruker Daltonik GmbH: Bremen, Germany, 2017. [Google Scholar]
- Herrmannsdorfer Landwerkstätten Glonn GmbH & Co., KG. Weideschweine und Symbiotische Landwirtschaft. Available online: https://www.herrmannsdorfer.de/landwirtschaft/symbiotisch/ (accessed on 11 March 2022).
- Eisler, M.C.; Lee, M.R.; Tarlton, J.F.; Martin, G.B.; Beddington, J.; Dungait, J.A.; Greathead, H.; Liu, J.; Mathew, S.; Miller, H. Agriculture: Steps to sustainable livestock. Nature 2014, 507, 32–34. [Google Scholar] [CrossRef]
- Elzen, B.; Bos, B. The RIO approach: Design and anchoring of sustainable animal husbandry systems. Technol. Forecast. Soc. Change 2019, 145, 141–152. [Google Scholar] [CrossRef]
- Murgueitio, E.; Barahona, R.; Chará, J.; Flores, M.; Mauricio, R.; Molina, J. The intensive silvopastoral systems in Latin America sustainable alternative to face climatic change in animal husbandry. Cuba. J. Agric. Sci. 2016, 49, 541–554. [Google Scholar]
- Bundesministerium für Wirtschaftliche Zusammenarbeit und Entwicklung. Nachhaltige Landwirtschaft. Available online: https://www.bmz.de/de/entwicklungspolitik/ernaehrungssicherung/nachhaltige-landwirtschaft#anc=id_51074_51074 (accessed on 27 April 2022).
- Stadig, L.M.; Rodenburg, T.B.; Ampe, B.; Reubens, B.; Tuyttens, F.A.M. Effects of shelter type, early environmental enrichment and weather conditions on free-range behaviour of slow-growing broiler chickens. Animal 2017, 11, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Landwirtschaft, B.L.F. Evaluierung Alternativer Haltungsformen für Legehennen; LfL: Thüringen, Germany, 2004. [Google Scholar]
- Dawkins, M.S.; Cook, P.A.; Whittingham, M.J.; Mansell, K.A.; Harper, A.E. What makes free-range broiler chickens range? In Situ measurement of habitat preference. Anim. Behav. 2003, 66, 151–160. [Google Scholar] [CrossRef]
- Bundesamt für Verbraucherschutz und Lebensmittelsicherheit (BVL). Zoonosen-Monitoring 2020; BVL: Osceola, FL, USA, 2021. [Google Scholar]
- Parry, S.; Palmer, S.; Slader, J.; Humphrey, T.; Group, S.E.W.I.D.L. Risk factors for salmonella food poisoning in the domestic kitchen–a case control study. Epidemiol. Infect. 2002, 129, 277–285. [Google Scholar] [CrossRef]
- Leotta, G.A.; Suzuki, K.; Alvarez, F.; Nuñez, L.; Silva, M.; Castro, L.; Faccioli, M.; Zarate, N.; Weiler, N.; Alvarez, M. Prevalence of Salmonella spp. in backyard chickens in Paraguay. Int. J. Poult. Sci. 2010, 6, 533–536. [Google Scholar] [CrossRef]
- Bailey, J.; Cosby, D. Salmonella prevalence in free-range and certified organic chickens. J. Food Prot. 2005, 68, 2451–2453. [Google Scholar] [CrossRef]
- Staley, M.; Conners, M.G.; Hall, K.; Miller, L.J. Linking stress and immunity: Immunoglobulin A as a non-invasive physiological biomarker in animal welfare studies. Horm. Behav. 2018, 102, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Bayerisches Landesamt für Gesundheit und Lebensmittelsicherheit. Prävalenz von Thermophilen Campylobacter spp. in Kotproben von Rindern, Schweinen und Geflügel Sowie in Lebensmittelproben. Available online: https://www.lgl.bayern.de/forschung/forschung_interdisziplinaer/fp_campylobacter_kotproben_lebensmittelproben.htm (accessed on 16 March 2022).
- Weljtens, M.J.B.M.; Bijker, P.G.H.; Van der Plas, J.; Urlings, H.A.P.; Biesheuvel, M.H. Prevalence of campylobacter in pigs during fattening; an epidemiological study. Vet. Q. 1993, 15, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Boes, J.; Nersting, L.; Nielsen, E.M.; Kranker, S.; Enøe, C.; Wachmann, H.C.; Baggesen, D.L. Prevalence and Diversity of Campylobacter jejuni in Pig Herds on Farms with and without Cattle or Poultry. J. Food Prot. 2005, 68, 722–727. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dong, Y.; Deng, F.; Liu, D.; Yao, H.; Zhang, Q.; Shen, J.; Liu, Z.; Gao, Y.; Wu, C.; et al. Species shift and multidrug resistance of Campylobacter from chicken and swine, China, 2008–14. J. Antimicrob. Chemother. 2015, 71, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Dobrindt, U. (Patho-)Genomics of Escherichia coli. Int. J. Med. Microbiol. 2005, 295, 357–371. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.D.; Redfield, R.J. Non-canonical CRP sites control competence regulons in Escherichia coli and many other γ-proteobacteria. Nucleic Acids Res. 2006, 34, 6001–6014. [Google Scholar] [CrossRef] [PubMed]
- Dubois, D.; Leyssene, D.; Chacornac, J.P.; Kostrzewa, M.; Schmit, P.O.; Talon, R.; Bonnet, R.; Delmas, J. Identification of a Variety of Staphylococcus Species by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2010, 48, 941–945. [Google Scholar] [CrossRef]
- Ilina, E.N.; Borovskaya, A.D.; Serebryakova, M.V.; Chelysheva, V.V.; Momynaliev, K.T.; Maier, T.; Kostrzewa, M.; Govorun, V.M. Application of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for the study of Helicobacter pylori. Rapid Commun. Mass Spectrom. 2010, 24, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Nagy, E.; Maier, T.; Urban, E.; Terhes, G.; Kostrzewa, M. Species identification of clinical isolates of Bacteroides by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 2009, 15, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Sabença, C.; de Sousa, T.; Oliveira, S.; Viala, D.; Théron, L.; Chambon, C.; Hébraud, M.; Beyrouthy, R.; Bonnet, R.; Caniça, M.; et al. Next-Generation Sequencing and MALDI Mass Spectrometry in the Study of Multiresistant Processed Meat Vancomycin-Resistant Enterococci (VRE). Biology 2020, 9, 89. [Google Scholar] [CrossRef] [PubMed]
- Fadeev, E.; Cardozo-Mino, M.G.; Rapp, J.Z.; Bienhold, C.; Salter, I.; Salman-Carvalho, V.; Molari, M.; Tegetmeyer, H.E.; Buttigieg, P.L.; Boetius, A. Comparison of two 16S rRNA primers (V3–V4 and V4–V5) for studies of arctic microbial communities. Front. Microbiol. 2021, 12, 637526. [Google Scholar] [CrossRef] [PubMed]
- Hansson, I.; Persson, M.; Svensson, L.; Engvall, E.O.; Johansson, K.-E. Identification of nine sequence types of the 16S rRNA genes of Campylobacter jejuni subsp. jejuni isolated from broilers. Acta Vet. Scand. 2008, 50, 10. [Google Scholar] [CrossRef]
- DLG. DLG Merkblatt 406: Haltung von Masthühnern; DLG Verlag: Frankfurt, Germany, 2017. [Google Scholar]
- Schwaiger, K.; Storch, J.; Bauer, C.; Bauer, J. Development of selected bacterial groups of the rectal microbiota of healthy calves during the first week postpartum. J. Appl. Microbiol. 2020, 128, 366–375. [Google Scholar] [CrossRef] [PubMed]

| Total | Value | Degree of Freedom | Asymptomatic Significance z (Two-Sided) | Exact Significance z (Two-Sided) | Exact Significance z (One-Sided) |
|---|---|---|---|---|---|
| Pearson’s chi-square test | 0.000 | 1 | 0.984 | ||
| Continuity correction | 0.000 | 1 | 1.000 | ||
| Likelihood-ratio test | 0.000 | 1 | 0.984 | ||
| Fisher’s exact test | 1.000 | 0.551 | |||
| Linear correlation | 0.000 | 1 | 0.984 | ||
| Number of valid cases | 225 |
| Total | Value | Degree of Freedom | Asymptomatic Significance z (Two-Sided) | Exact Significance z (Two-Sided) | Exact Significance z (One-Sided) |
|---|---|---|---|---|---|
| Pearson’s chi-square test | 1.153 | 1 | 0.283 | ||
| Continuity correction | 0.868 | 1 | 0.351 | ||
| Likelihood-ratio test | 1.154 | 1 | 0.283 | ||
| Fisher’s exact test | 0.321 | 0.176 | |||
| Linear correlation | 1.148 | 1 | 0.284 | ||
| Number of valid cases | 231 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaeder, E.; Dorn-In, S.; Gareis, M.; Schwaiger, K. Symbiotic Husbandry of Chickens and Pigs Does Not Increase Pathogen Transmission Risk. Foods 2022, 11, 3126. https://doi.org/10.3390/foods11193126
Kaeder E, Dorn-In S, Gareis M, Schwaiger K. Symbiotic Husbandry of Chickens and Pigs Does Not Increase Pathogen Transmission Risk. Foods. 2022; 11(19):3126. https://doi.org/10.3390/foods11193126
Chicago/Turabian StyleKaeder, Emma, Samart Dorn-In, Manfred Gareis, and Karin Schwaiger. 2022. "Symbiotic Husbandry of Chickens and Pigs Does Not Increase Pathogen Transmission Risk" Foods 11, no. 19: 3126. https://doi.org/10.3390/foods11193126
APA StyleKaeder, E., Dorn-In, S., Gareis, M., & Schwaiger, K. (2022). Symbiotic Husbandry of Chickens and Pigs Does Not Increase Pathogen Transmission Risk. Foods, 11(19), 3126. https://doi.org/10.3390/foods11193126






