Explore the Interaction between Ellagic Acid and Zein Using Multi-Spectroscopy Analysis and Molecular Docking
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Ultraviolet-Visible (UV-Vis) Spectroscopy Analysis
2.3. Fluorescence Spectra Analysis
2.4. Quenching Behavior Modeling
2.5. Thermodynamic Parameters
2.6. Circular Dichroism (CD) Spectroscopy Determination
2.7. Molecular Docking Simulation
2.8. Statistical Analysis
3. Results and Discussion
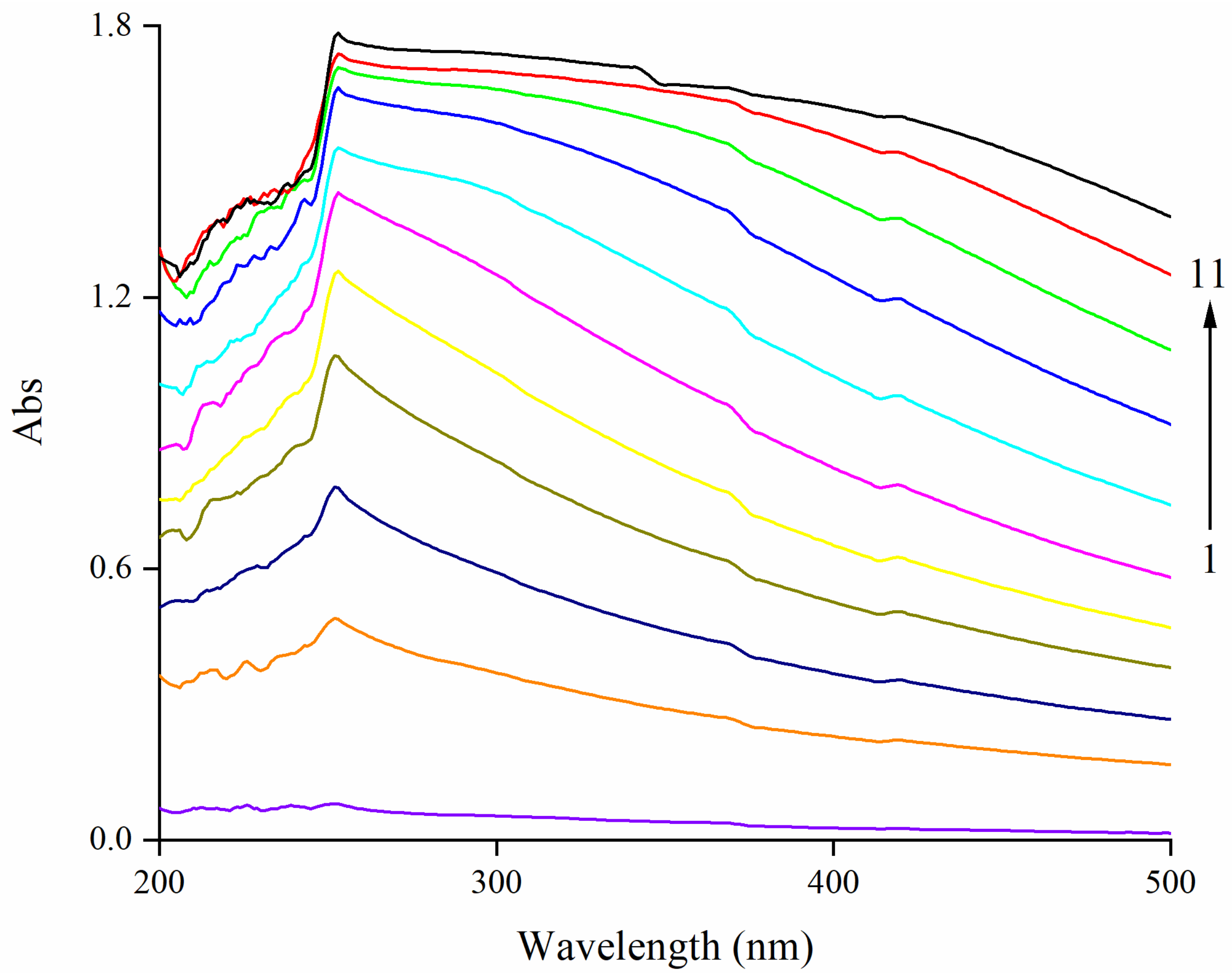
3.1. Effect of EA on UV–Vis Spectra of Zein
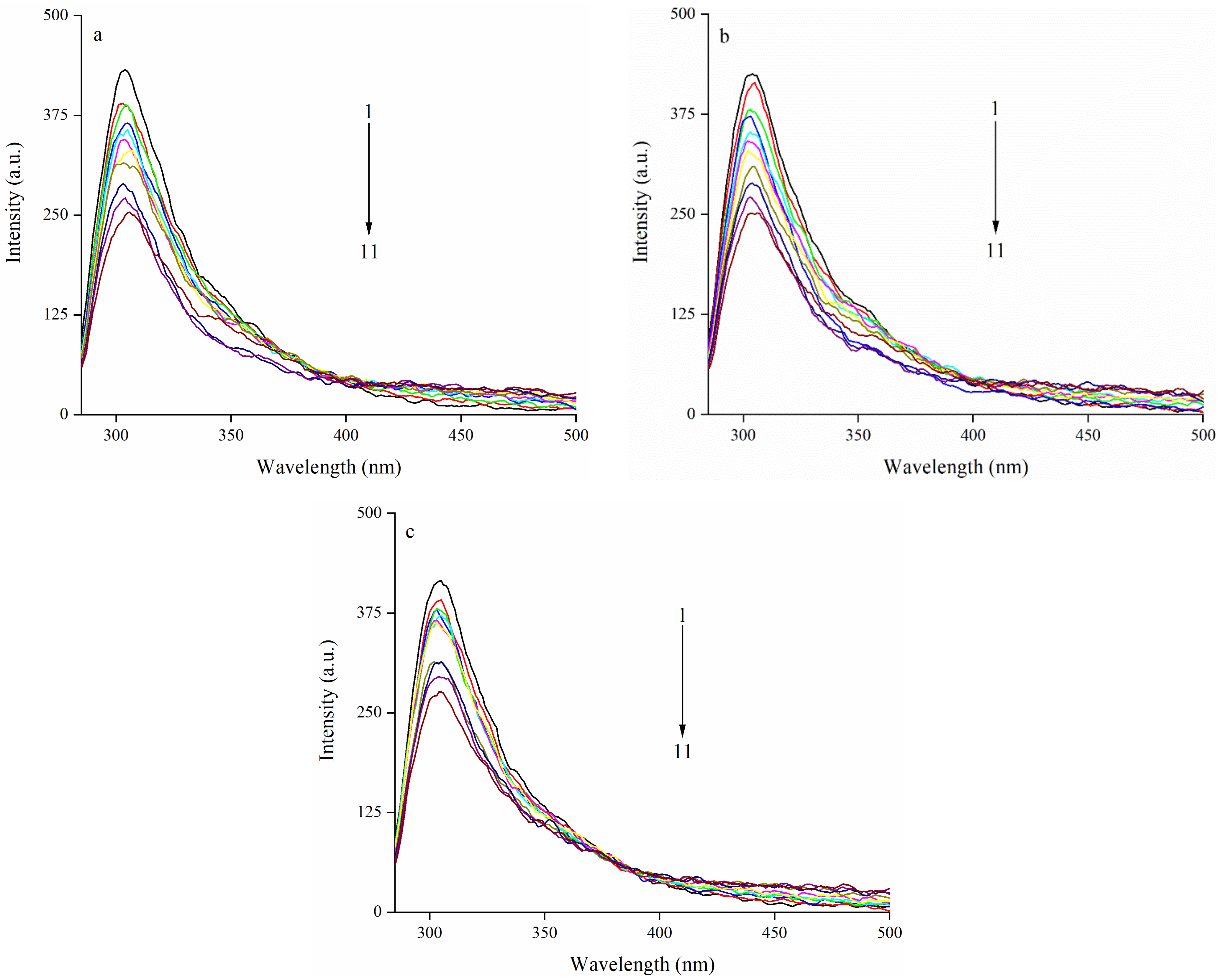
3.2. Effect of EA on Intrinsic Fluorescence Spectrum of Zein
3.2.1. The Quenching Effects of EA on the Fluorescence Emission Intensity of Zein
3.2.2. Fluorescence Quenching Behavior Modeling
3.2.3. Thermodynamic Parameters and Type of Quenching Force
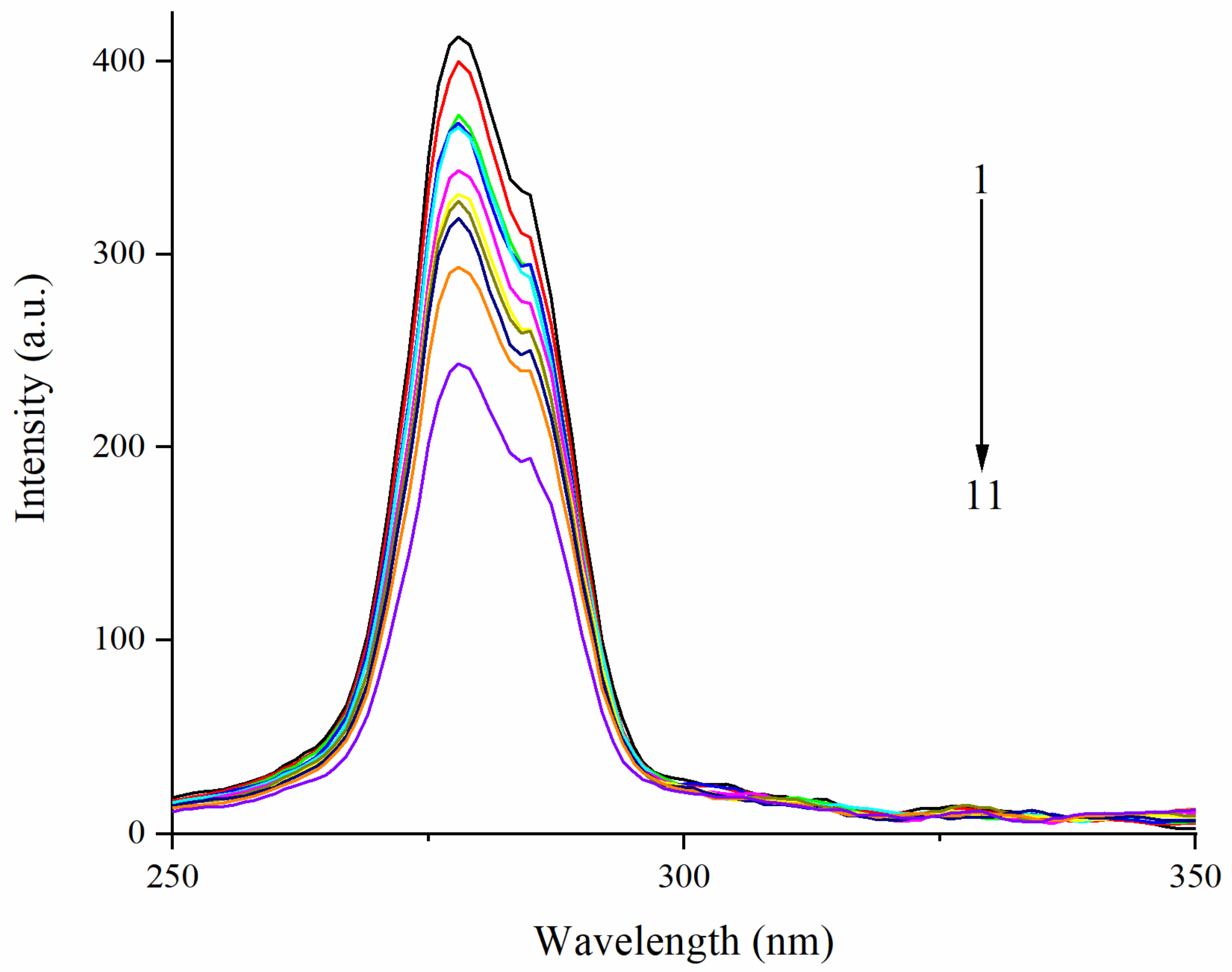
3.3. Effect of EA on Synchronous Fluorescence Spectra of Zein
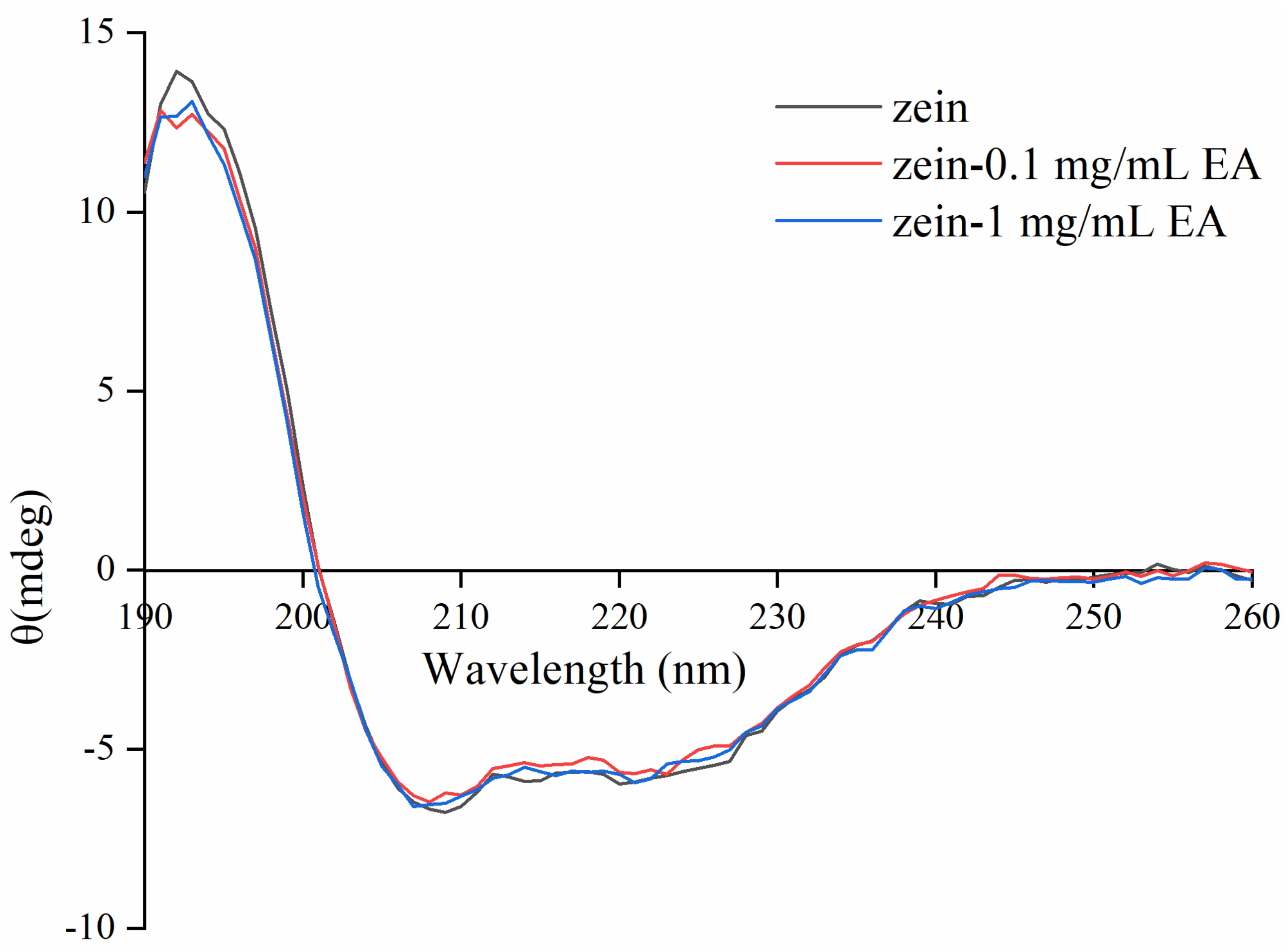
3.4. Effect of EA on Conformational Changes of Zein Determined by CD Spectroscopy
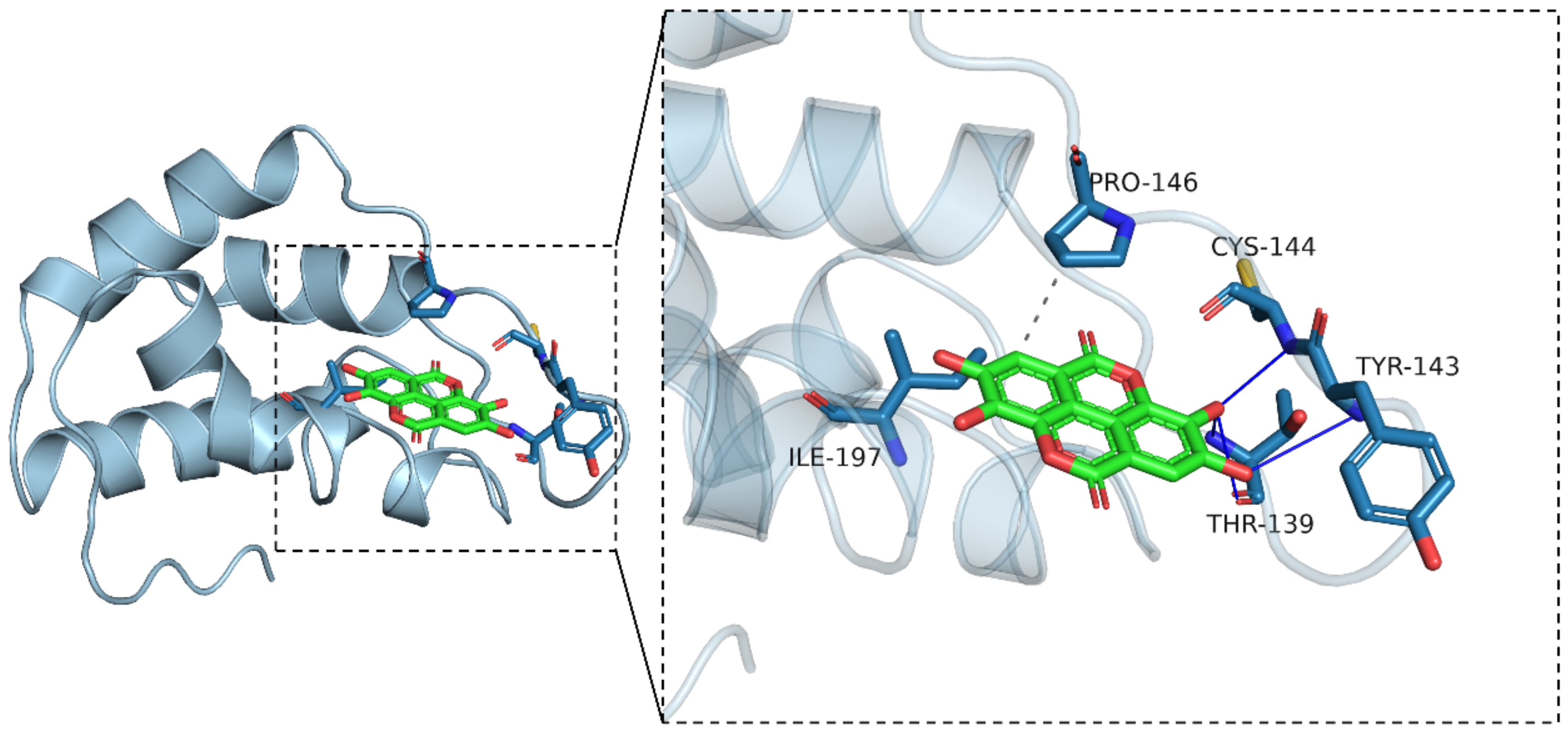
3.5. Molecular Docking Simulation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jiang, L.Z.; Liu, Y.J.; Li, L.; Qi, B.K.; Ju, M.N.; Xu, Y.; Zhang, Y.; Sui, X.N. Covalent conjugates of anthocyanins to soy protein: Unravelling their structure features and in vitro gastrointestinal digestion fate. Food Res. Int. 2019, 120, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Peng, N.; Chang, C.; McClements, D.J.; Su, Y.; Yang, Y. Fabrication of surface-active antioxidant food biopolymers: Conjugation of catechin polymers to egg white proteins. Food Biophys. 2017, 12, 198–210. [Google Scholar] [CrossRef]
- Le Bourvellec, C.; Renard, C.M.G.C. Interactions between polyphenols and macromolecules: Quantification methods and mechanisms. Crit. Rev. Food Sci. Nutr. 2012, 52, 213–248. [Google Scholar] [CrossRef]
- Bittner, S. When quinones meet amino acids: Chemical, physical and biological consequences. Amino Acids 2006, 30, 205–224. [Google Scholar] [CrossRef]
- Kanakis, C.D.; Hasni, I.; Bourassa, P.; Tarantilis, P.A.; Polissiou, M.G.; Tajmir-Riahi, H.-A. Milk β-lactoglobulin complexes with tea polyphenols. Food Chem. 2011, 127, 1046–1055. [Google Scholar] [CrossRef] [PubMed]
- Ozdal, T.; Capanoglu, E.; Altay, F. A review on protein–phenolic interactions and associated changes. Food Res. Int. 2013, 51, 954–970. [Google Scholar] [CrossRef]
- Buitimea-Cantúa, N.E.; Gutiérrez-Uribe, J.A.; Serna-Saldívar, S.O. Phenolic–protein interactions: Effects on food properties and health benefits. J. Med. Food 2018, 21, 188–198. [Google Scholar] [CrossRef]
- Yildirim-Elikoglu, S.; Erdem, Y.K. Interactions between milk proteins and polyphenols: Binding mechanisms, related changes, and the future trends in the dairy industry. Food Rev. Int. 2018, 34, 665–697. [Google Scholar] [CrossRef]
- Yang, Y.L.; Guo, S.J.; Su, H.; Cai, R.M.; Zhai, F.L.; Chen, M.J. Changes in the aroma components of Yashixiang Dancong black tea during processing. Food Res. Dev. 2019, 40, 82–90. [Google Scholar] [CrossRef]
- Anderson, T.J.; Lamsal, B.P. Zein extraction from corn, corn products, and coproducts and modifications for various applications: A review. Cereal Chem. 2011, 88, 159–173. [Google Scholar] [CrossRef] [Green Version]
- Paliwal, R.; Palakurthi, S. Zein in controlled drug delivery and tissue engineering. J. Control. Release 2014, 189, 108–122. [Google Scholar] [CrossRef]
- Torres-Giner, S.; Gimenez, E.; Lagaron, J.M. Characterization of the morphology and thermal properties of Zein Prolamine nanostructures obtained by electrospinning. Food Hydrocoll. 2008, 22, 601–614. [Google Scholar] [CrossRef]
- Patel, A.R.; Heussen, P.C.M.; Hazekamp, J.; Drost, E.; Velikov, K.P. Quercetin loaded biopolymeric colloidal particles prepared by simultaneous precipitation of quercetin with hydrophobic protein in aqueous medium. Food Chem. 2012, 133, 423–429. [Google Scholar] [CrossRef]
- Patel, A.R.; Heussen, P.C.M.; Dorst, E.; Hazekamp, J.; Velikov, K.P. Colloidal approach to prepare colour blends from colourants with different solubility profiles. Food Chem. 2013, 141, 1466–1471. [Google Scholar] [CrossRef]
- Liu, F.G.; Ma, C.C.; McClements, D.J.; Gao, Y.X. A comparative study of covalent and non-covalent interactions between zein and polyphenols in ethanolwater solution. Food Hydrocoll. 2017, 63, 625–634. [Google Scholar] [CrossRef]
- Chen, S.; Li, Q.; McClements, D.J.; Han, Y.H.; Dai, L.; Mao, L.; Gao, Y.X. Codelivery of curcumin and piperine in zein-carrageenan core-shell nanoparticles: Formation, structure, stability and in vitro gastrointestinal digestion. Food Hydrocoll. 2019, 99, 105334. [Google Scholar] [CrossRef]
- Xue, P.; Zhang, G.; Zhao, H.; Wang, W.; Zhang, J.; Ren, L. Serum albumin complexed with ellagic acid from pomegranate peel and its metabolite urolithin B. Food Biosci. 2022, 46, 101618. [Google Scholar] [CrossRef]
- Jin, N.; Zhang, S.; Sun, S.; Wu, M.; Yang, X.; Xu, J.; Ma, K.; Guan, S.; Xu, W. An Organic Solvent-Free Method for the Extraction of Ellagic Acid Compounds from Raspberry Wine Pomace with Assistance of Sodium Bicarbonate. Molecules 2022, 27, 2145. [Google Scholar] [CrossRef]
- An, J.Y.; Wang, L.T.; Lv, M.J.; Wang, J.D.; Cai, Z.H.; Wang, Y.Q.; Zhang, S.; Yang, Q.; Fu, Y.J. An efficiency strategy for extraction and recovery of ellagic acid from waste chestnut shell and its biological activity evaluation. Microchem. J. 2021, 160, 105616. [Google Scholar] [CrossRef]
- Soong, Y.Y.; Barlow, P.J. Isolation and structure elucidation of phenolic compounds from longan (Dimocarpus longan Lour.) seed by high-performance liquid chromatography–electrospray ionization mass spectrometry. J. Chromatogr. A 2005, 1085, 270–277. [Google Scholar] [CrossRef]
- Pavlova, E.L.; Zografov, N.N.; Simeonova, L.S. Comparative study on the antioxidant capacities of synthetic influenza inhibitors and ellagic acid in model systems. Biomed. Pharmacother. 2016, 83, 755–762. [Google Scholar] [CrossRef]
- Smart, R.C.; Huang, M.T.; Chang, R.L.; Sayer, J.M.; Jerina, D.M.; Conney, A.H. Disposition of the naturally occurring antimutagenic plant phenol, ellagic acid, and its synthetic derivatives, 3-O-decylellagic acid and 3,3′-di-Omethylellagic acid in mice. Carcinogenesis 1986, 7, 1663–1667. [Google Scholar] [CrossRef]
- Gil, T.Y.; Hong, C.H.; An, H.J. Anti-inflammatory effects of ellagic acid on keratinocytes via MAPK and STAT pathways. Int. J. Mol. Sci. 2021, 22, 1277. [Google Scholar] [CrossRef]
- Ding, Y.; Zhang, B.; Zhou, K.Y.; Chen, M.C.; Wang, M.M.; Jia, Y.Y.; Song, Y.; Li, Y.W.; Wen, A.D. Dietary ellagic acid improves oxidant-induced endothelial dysfunction and atherosclerosis: Role of Nrf2 activation. Int. J. Cardiol. 2014, 175, 508–514. [Google Scholar] [CrossRef]
- Fatima, N.; Hafizur, R.M.; Hameed, A.; Ahmed, S.; Nisar, M.; Kabir, N. Ellagic acid in Emblica officinalis exerts antidiabetic activity through the action on β-cells of pancreas. Eur. J. Nutr. 2017, 56, 591–601. [Google Scholar] [CrossRef]
- Kiasalari, Z.; Heydarifard, R.; Khalili, M. Ellagic acid ameliorates learning and memory deficits in a rat model of Alzheimer’s disease: An exploration of underlying mechanisms. Psychopharmacology 2017, 234, 1841–1852. [Google Scholar] [CrossRef]
- Rossi, M.; Erlebacher, J.; Zacharias, D.E.; Carrell, H.L.; Iannucci, B. The crystal and molecule-structure of ellagic acid dihydrate—A dietary anticancer agent. Carcinogenesis 1991, 12, 2227–2232. [Google Scholar] [CrossRef] [PubMed]
- Clifford, M.N.; Scalbert, A. Ellagitannins-nature, occurrence and dietary burden. J. Sci. Food Agric. 2000, 80, 1118–1125. [Google Scholar] [CrossRef]
- Zhang, L.L.; Liu, Y.C.; Hu, X.Y.; Xu, M.; Wang, Y.M. Studies on interactions of pentagalloyl glucose, ellagic acid and gallic acid with bovine serum albumin: A spectroscopic analysis. Food Chem. 2020, 324, 126872. [Google Scholar] [CrossRef]
- Pattanayak, R.; Basak, P.; Sen, S.; Bhattacharyya, M. An insight to the binding of ellagic acid with human serum albumin using spectroscopic and isothermal calorimetry studies. Biochem. Biophys. Rep. 2017, 10, 88–93. [Google Scholar] [CrossRef]
- Krekora, M.; Nawrocka, A. Effect of a polyphenol molecular size on the gluten proteins–polyphenols interactions studied with FT-Raman spectroscopy. Food Biophys. 2022. [Google Scholar] [CrossRef]
- Yan, C.J.; Zhou, Z. Ellagic acid can act as a chaperone and suppress the heat-induced amyloid-like aggregation of ovalbumin. Food Hydrocoll. 2020, 100, 105408. [Google Scholar] [CrossRef]
- Diao, M.; Liang, Y.; Zhao, J.; Zhang, J.; Zhang, T. Complexation of ellagic acid with α-lactalbumin and its antioxidant property. Food Chem. 2022, 372, 131307. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Bao, Y.H.; Sun, K.F.; Chang, C.; Liu, W.F. Effects of covalent interactions and gel characteristics on soy protein-tannic acid conjugates prepared under alkaline conditions. Food Hydrocoll. 2021, 112, 106293. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Weber, G. Quenching of fluorescence by oxygen-probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions-forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Li, D.J.; Zhu, J.F.; Jin, J.; Yao, X.J. Studies on the binding of nevadensin to human serum albumin by molecular spectroscopy and modeling. J. Mol. Struct. 2007, 846, 34–41. [Google Scholar] [CrossRef]
- Zhang, W.; Han, B.; Zhao, S.; Ge, F.; Xiong, X.; Chen, D.; Chen, C. Study on the interaction between theasinesin and bovine serum albumin by fluorescence method. Anal. Lett. 2010, 43, 289–299. [Google Scholar] [CrossRef]
- Keppler, J.K.; Stuhldreier, M.C.; Temps, F.; Schwarz, K. Influence of mathematical models and correction factors on binding results of polyphenols and retinol with beta-lactoglobulin measured with fluorescence quenching. Food Biophys. 2014, 9, 158–168. [Google Scholar] [CrossRef]
- Bose, A. Interaction of tea polyphenols with serum albumins: A fluorescence spectroscopic analysis. J. Lumin. 2016, 169, 220–226. [Google Scholar] [CrossRef]
- Joye, I.J.; Davidov-Pardo, G.; Ludescher, R.D.; McClements, D.J. Fluorescence quenching study of resveratrol binding to zein and gliadin: Towards a more rational approach to resveratrol encapsulation using water-insoluble proteins. Food Chem. 2015, 185, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.Z.; Xu, J.W.; Zhang, X.Y.; Xie, F.Y.; Zhang, S.; Jiang, L.Z.; Li, Y. Effect of pH-shifting treatment on the structural and functional properties of soybean protein isolate and its interactions with (-)-epigallocatechin-3-gallate. Process Biochem. 2021, 101, 190–198. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2000. [Google Scholar]
- Wang, Q.; Tang, Y.; Yang, Y.; Lei, L.; Lei, X.; Zhao, J.; Ming, J. Interactions and structural properties of zein/ferulic acid: The effect of calcium chloride. Food Chem. 2022, 373, 131489. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.F.; Zhang, X.; Liang, Q.F.; Hou, T.; Zhou, H.J. Effects of Different Working Modes of Ultrasound on Structural Characteristics of Zein and ACE Inhibitory Activity of Hydrolysates. J. Food Quqlity 2017, 2017, 7896037. [Google Scholar] [CrossRef]
- Dai, T.T.; Yan, X.Y.; Li, Q.; Li, T.; Liu, C.M.; McClements, D.J.; Chen, J. Characterization of binding interaction between rice glutelin and gallic acid: Multi-spectroscopic analyses and computational docking simulation. Food Res. Int. 2017, 102, 274–281. [Google Scholar] [CrossRef]
- Acharya, D.P.; Sanguansri, L.; Augustin, M.A. Binding of resveratrol with sodium caseinate in aqueous solutions. Food Chem. 2013, 141, 1050–1054. [Google Scholar] [CrossRef]
- Hemar, Y.; Gerbeaud, M.; Oliver, C.M.; Augustin, M.A. Investigation into the interaction between resveratrol and whey proteins using fluorescence spectroscopy. Int. J. Food Sci. Technol. 2011, 46, 2137–2144. [Google Scholar] [CrossRef]
- Liu, C.Z.; Lv, N.; Ren, G.R.; Wu, R.B.; Wang, B.J.; Cao, Z.X.; Xie, H.J. Explore the interaction mechanism between zein and EGCG using multi-spectroscopy and molecular dynamics simulation methods. Food Hydrocoll. 2021, 120, 106906. [Google Scholar] [CrossRef]
- Zembyla, M.; Murray, B.S.; Radford, S.J.; Sarkar, A. Water-in-oil Pickering emulsions stabilized by an interfacial complex of water-insoluble polyphenol crystals and protein. J. Colloid Interface Sci. 2019, 548, 88–99. [Google Scholar] [CrossRef]
- Shukla, R.; Cheryan, M. Zein: The industrial protein from corn. Ind. Crops Prod. 2001, 13, 171–192. [Google Scholar] [CrossRef]
- Zhan, F.C.; Yang, J.C.; Li, J.; Wang, Y.T.; Li, B. Characteristics of the interaction mechanism between tannic acid and sodium caseinate using multispectroscopic and thermodynamics methods. Food Hydrocoll. 2018, 75, 81–87. [Google Scholar] [CrossRef]
- Xie, W.; Huang, Y.; Xiang, Y.; Xiong, S.; Manyande, A.; Du, H. Insights into the binding mechanism of polyphenols and fish myofibrillar proteins explored using multi-spectroscopic methods. Food Bioprocess Technol. 2020, 13, 797–806. [Google Scholar] [CrossRef]
- Ren, C.; Xiong, W.; Li, J.; Li, B. Comparison of binding interactions of cyanidin-3-O-glucoside to β-conglycinin and glycinin using multi-spectroscopic and thermodynamic methods. Food Hydrocoll. 2019, 92, 155–162. [Google Scholar] [CrossRef]
- Li, Y.Q.; Li, J.; Xia, Q.Y.; Zhang, B.; Wang, Q.; Huang, Q.R. Understanding the Dissolution of alpha-Zein in Aqueous Ethanol and Acetic Acid Solutions. J. Phys. Chem. B 2012, 116, 12057–12064. [Google Scholar] [CrossRef]
- Tan, H.X.; Zhou, H.Y.; Guo, T.; Zhang, Y.H. Integrated multi-spectroscopic and molecular modeling techniques to study the formation mechanism of hidden zearalenone in maize. Food Chem. 2021, 351, 129286. [Google Scholar] [CrossRef]
- Tiwari, P.; Ali, R.; Ishrat, R.; Arfin, N. Study of interaction between zein and curcumin using spectroscopic and in silico techniques. J. Mol. Struct. 2021, 1230, 129637. [Google Scholar] [CrossRef]
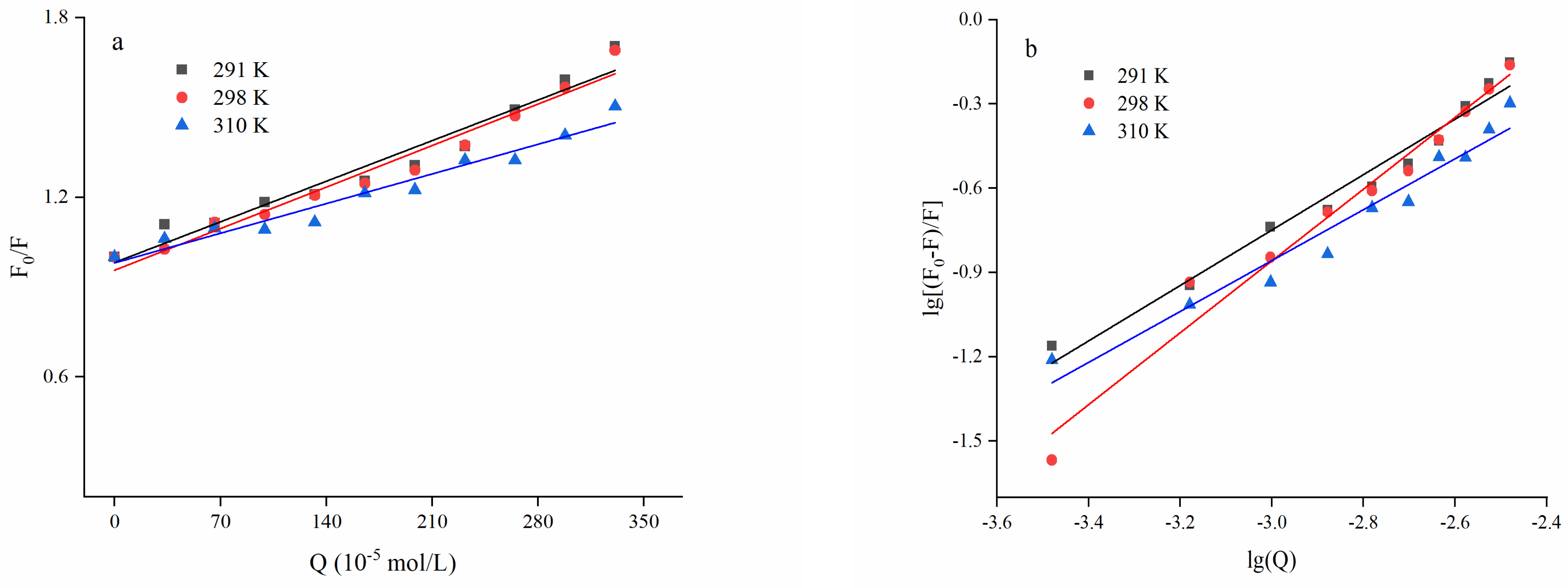
| T (K) | Ksv (L/mol) | Kq (L/(mol·s)) | R2 |
|---|---|---|---|
| 291 | 1.90 × 103 | 1.90 × 1011 | 0.9529 |
| 298 | 2.00 × 103 | 2.00 × 1011 | 0.9666 |
| 310 | 1.40 × 103 | 1.40 × 1011 | 0.9557 |
| T (K) | n | KA (L/mol) | R2 |
|---|---|---|---|
| 291 | 0.9871 | 1.63 × 102 | 0.9713 |
| 298 | 1.2786 | 9.47 × 102 | 0.9740 |
| 310 | 0.9065 | 1.27 × 103 | 0.9560 |
| T (K) | ΔH (kJ/mol) | ΔS (J/(mol•K)) | ΔG (kJ/mol) |
|---|---|---|---|
| 291 | 181.28 | 0.67 | −12.32 |
| 298 | 181.28 | 0.67 | −16.98 |
| 310 | 181.28 | 0.67 | −26.42 |
| Sample | α-helix | β-sheet | β-turn | Random Coil |
|---|---|---|---|---|
| Zein | 51.12 ± 0.83 b | 12.51 ± 0.98 c | 16.91 ± 0.77 a | 19.46 ± 0.58 a |
| Zein-0.1 mg/mL EA | 53.55 ± 0.50 a | 16.35 ± 0.39 b | 14.82 ± 0.96 b | 15.28 ± 0.19 b |
| Zein-1 mg/mL EA | 55.82 ± 0.90 a | 19.52 ± 0.36 a | 13.36 ± 0.73 b | 11.30 ± 0.65 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Deng, Y.; Yan, T.; Yang, X.; Xu, W.; Liu, D.; Wang, W. Explore the Interaction between Ellagic Acid and Zein Using Multi-Spectroscopy Analysis and Molecular Docking. Foods 2022, 11, 2764. https://doi.org/10.3390/foods11182764
Zhao S, Deng Y, Yan T, Yang X, Xu W, Liu D, Wang W. Explore the Interaction between Ellagic Acid and Zein Using Multi-Spectroscopy Analysis and Molecular Docking. Foods. 2022; 11(18):2764. https://doi.org/10.3390/foods11182764
Chicago/Turabian StyleZhao, Shunan, Yong Deng, Tianyi Yan, Xiaoling Yang, Weidong Xu, Donghong Liu, and Wenjun Wang. 2022. "Explore the Interaction between Ellagic Acid and Zein Using Multi-Spectroscopy Analysis and Molecular Docking" Foods 11, no. 18: 2764. https://doi.org/10.3390/foods11182764
APA StyleZhao, S., Deng, Y., Yan, T., Yang, X., Xu, W., Liu, D., & Wang, W. (2022). Explore the Interaction between Ellagic Acid and Zein Using Multi-Spectroscopy Analysis and Molecular Docking. Foods, 11(18), 2764. https://doi.org/10.3390/foods11182764






