Effects of Microbial Communities on Volatile Profiles and Biogenic Amines in Beef Jerky from Inner Mongolian Districts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. High-Throughput Sequencing
2.2.1. DNA Extraction and PCR Amplification
2.2.2. Sequencing and Data Analysis
2.3. Determination of Volatile Flavor Compounds
2.4. Biogenic Amine Analysis
2.5. Statistical Analysis
3. Results and Discussion
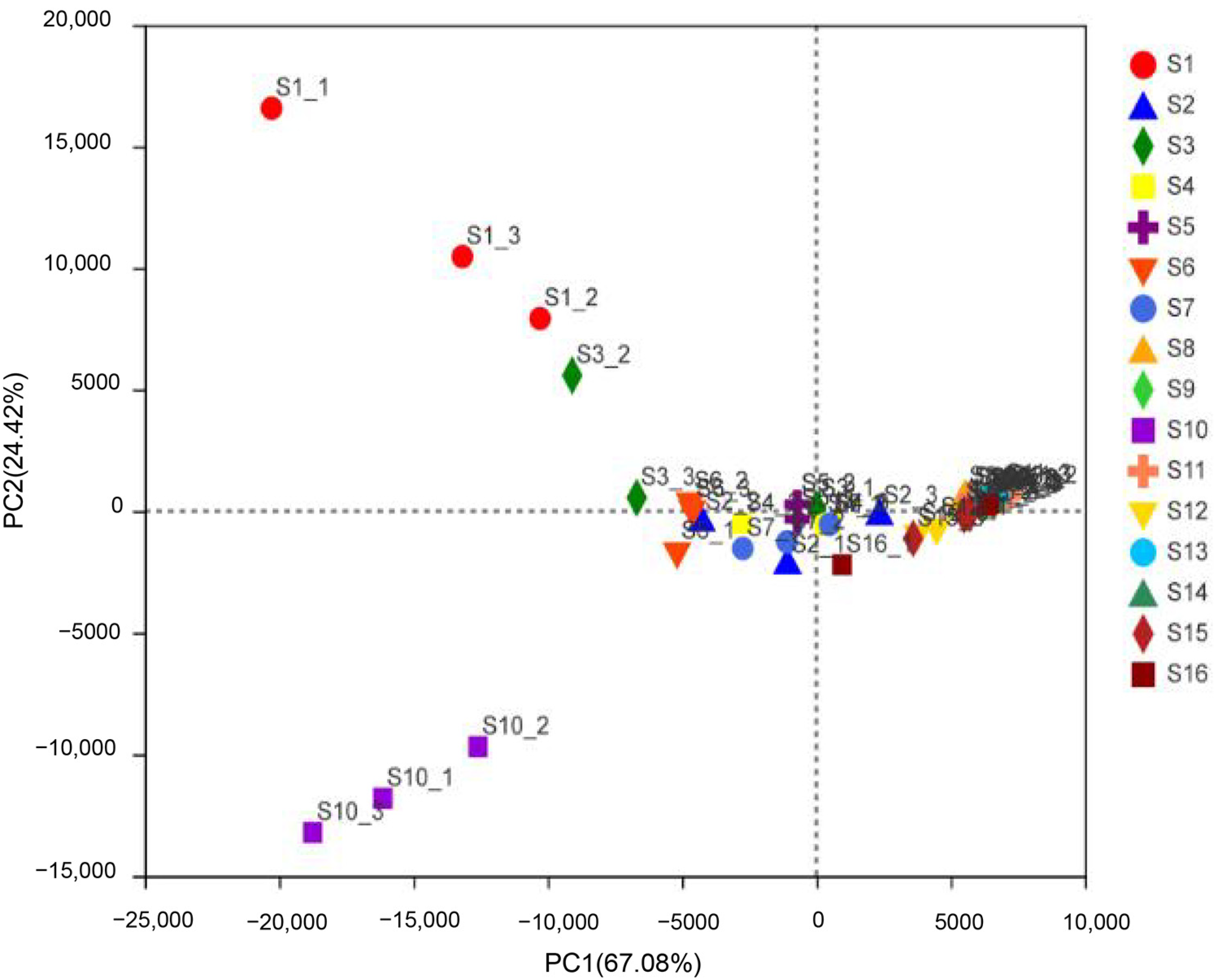
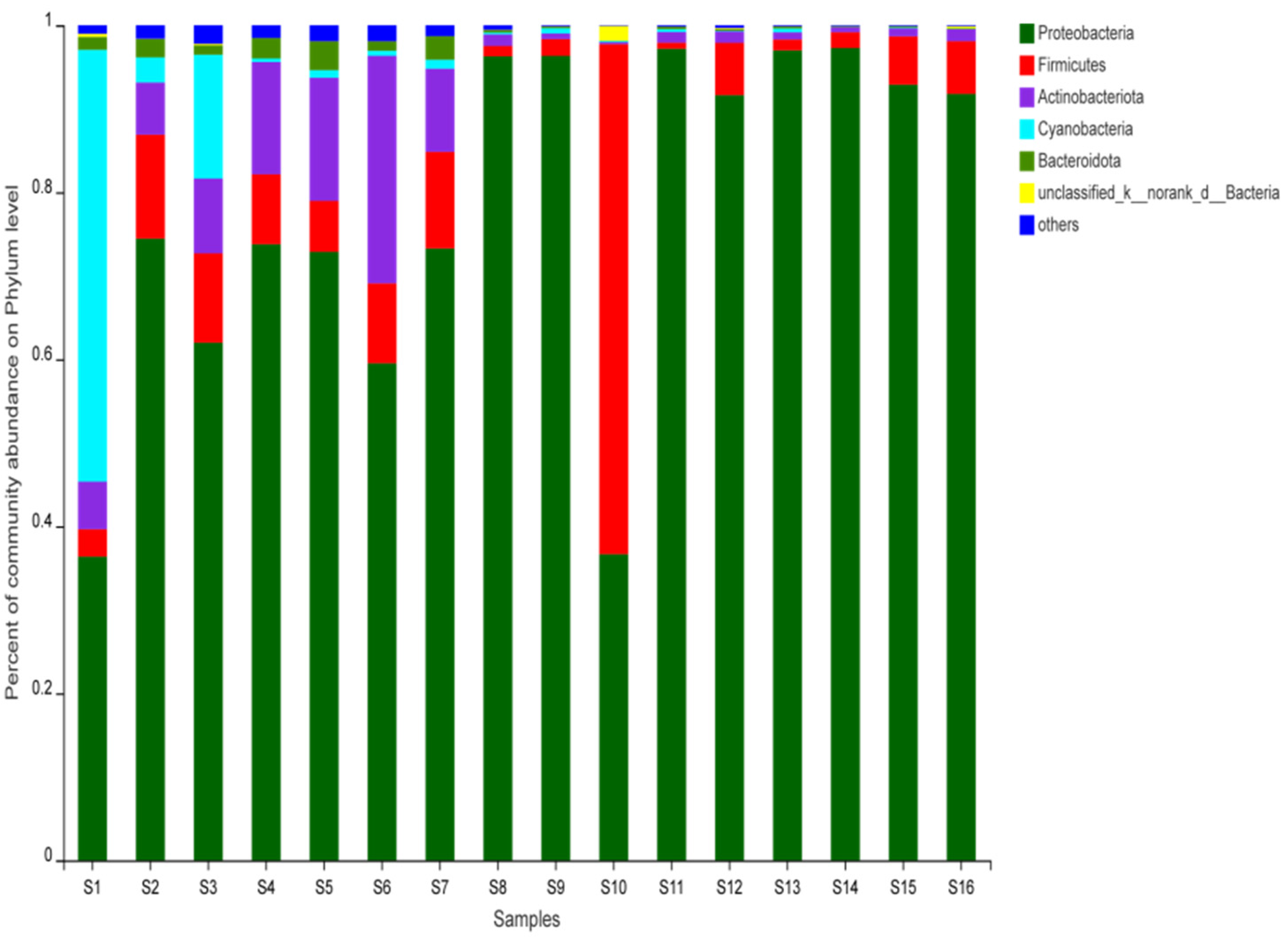
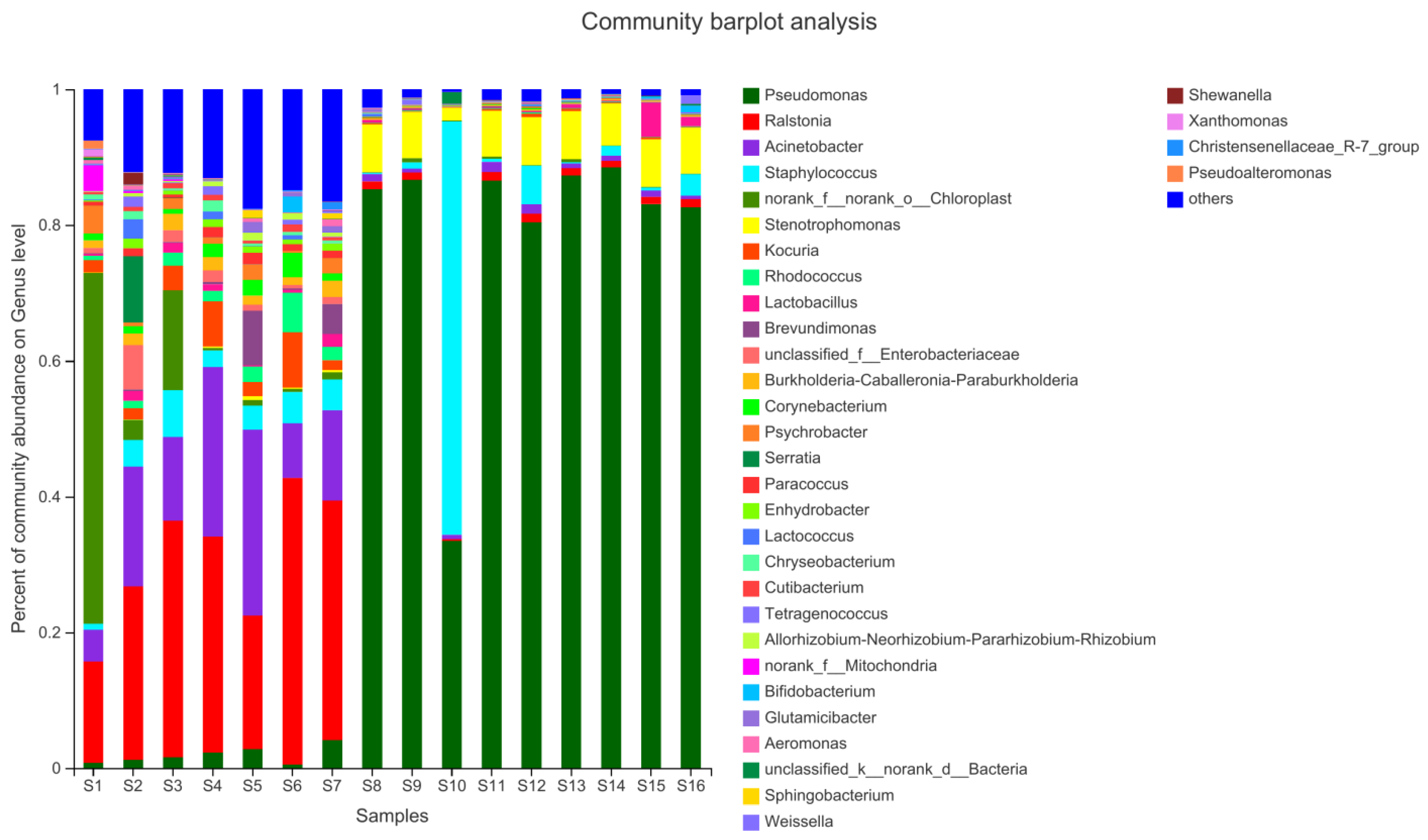
3.1. Bacterial Community Composition of Beef Jerky from Different Areas of Inner Mongolia
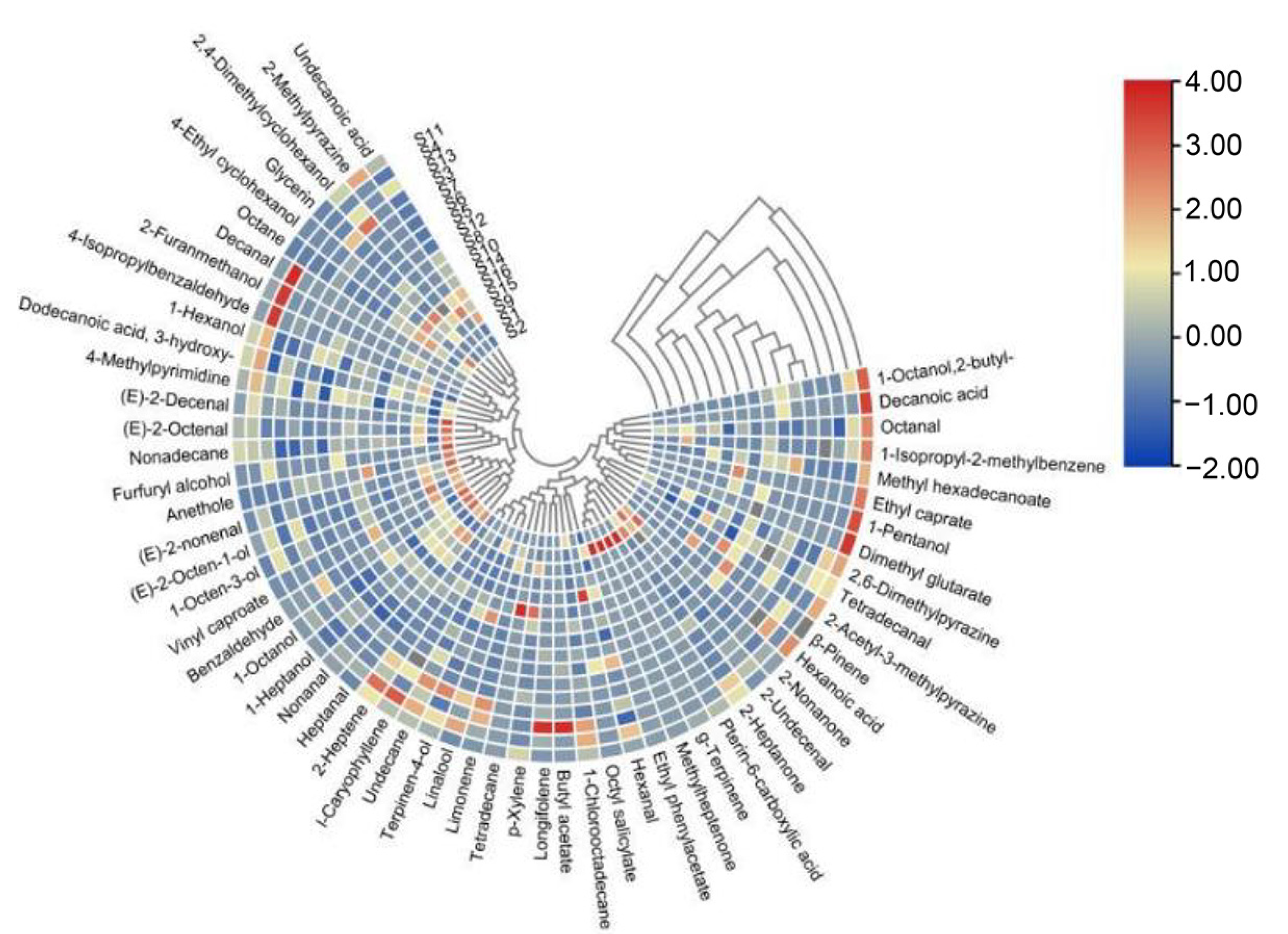
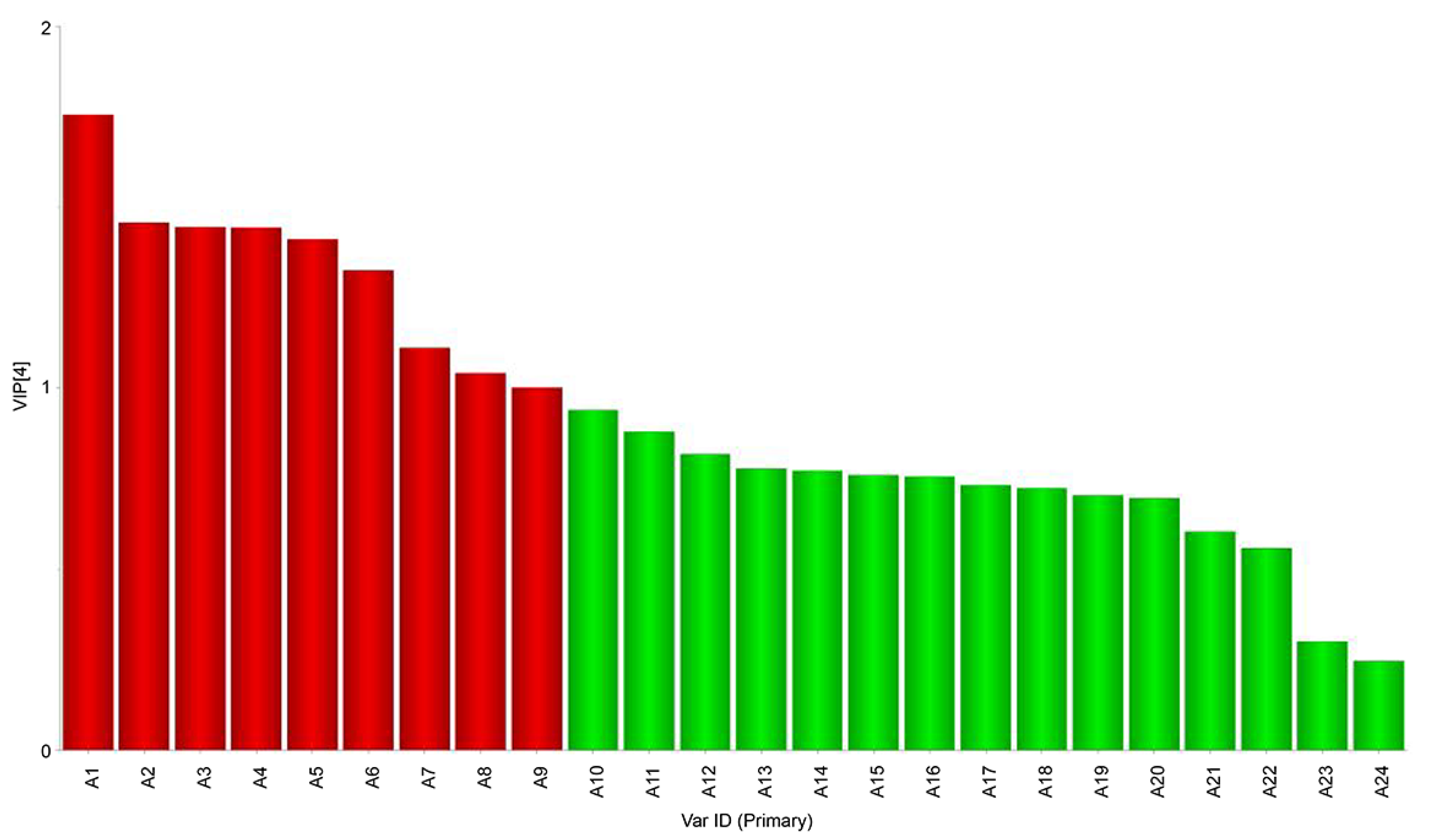
3.2. Composition of Volatile Flavor Compounds of Beef Jerky from Different Areas of Inner Mongolia
3.3. Biogenic Amine Concentrations of Beef Jerky from Different Areas of Inner Mongolia
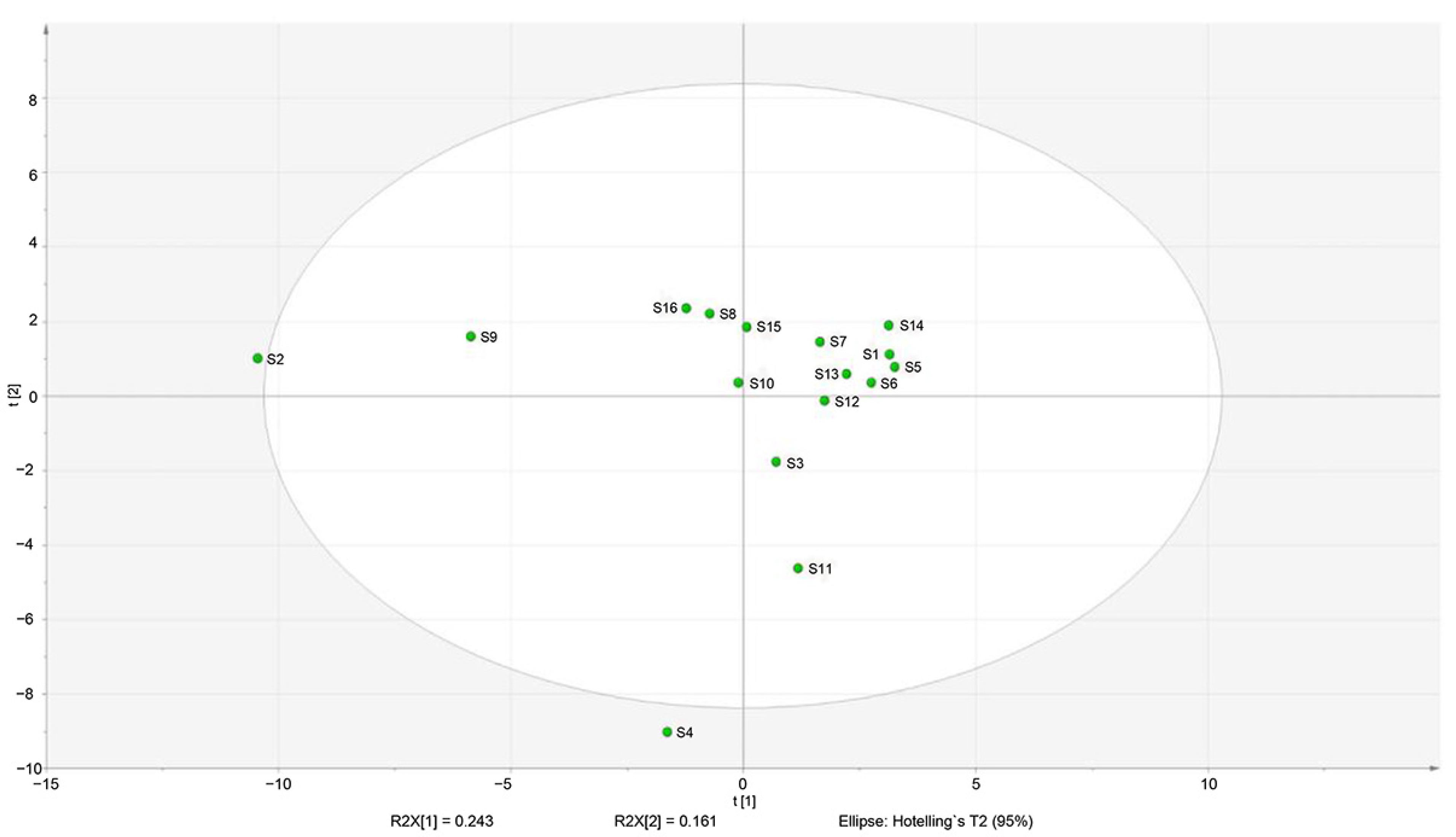
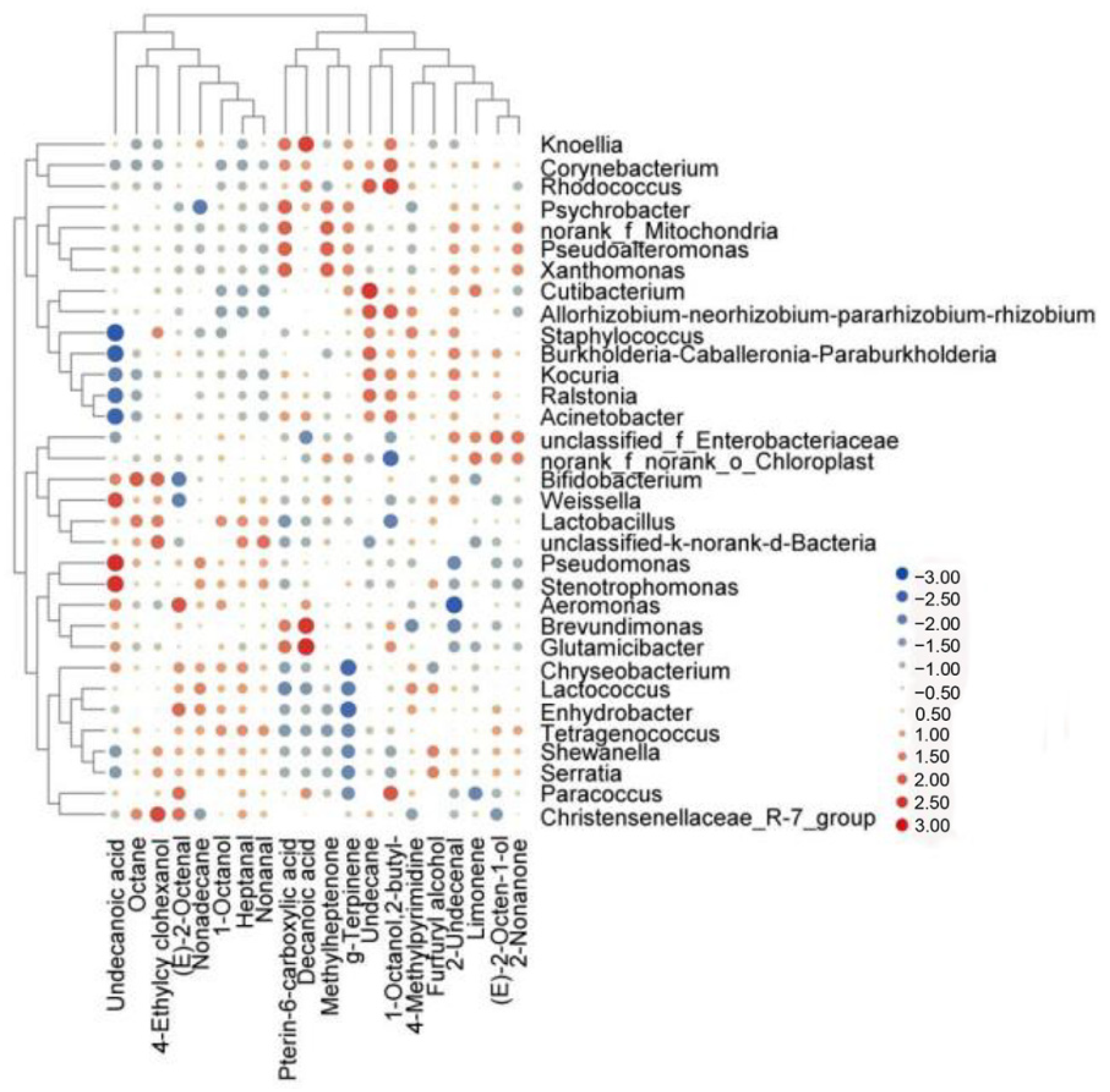
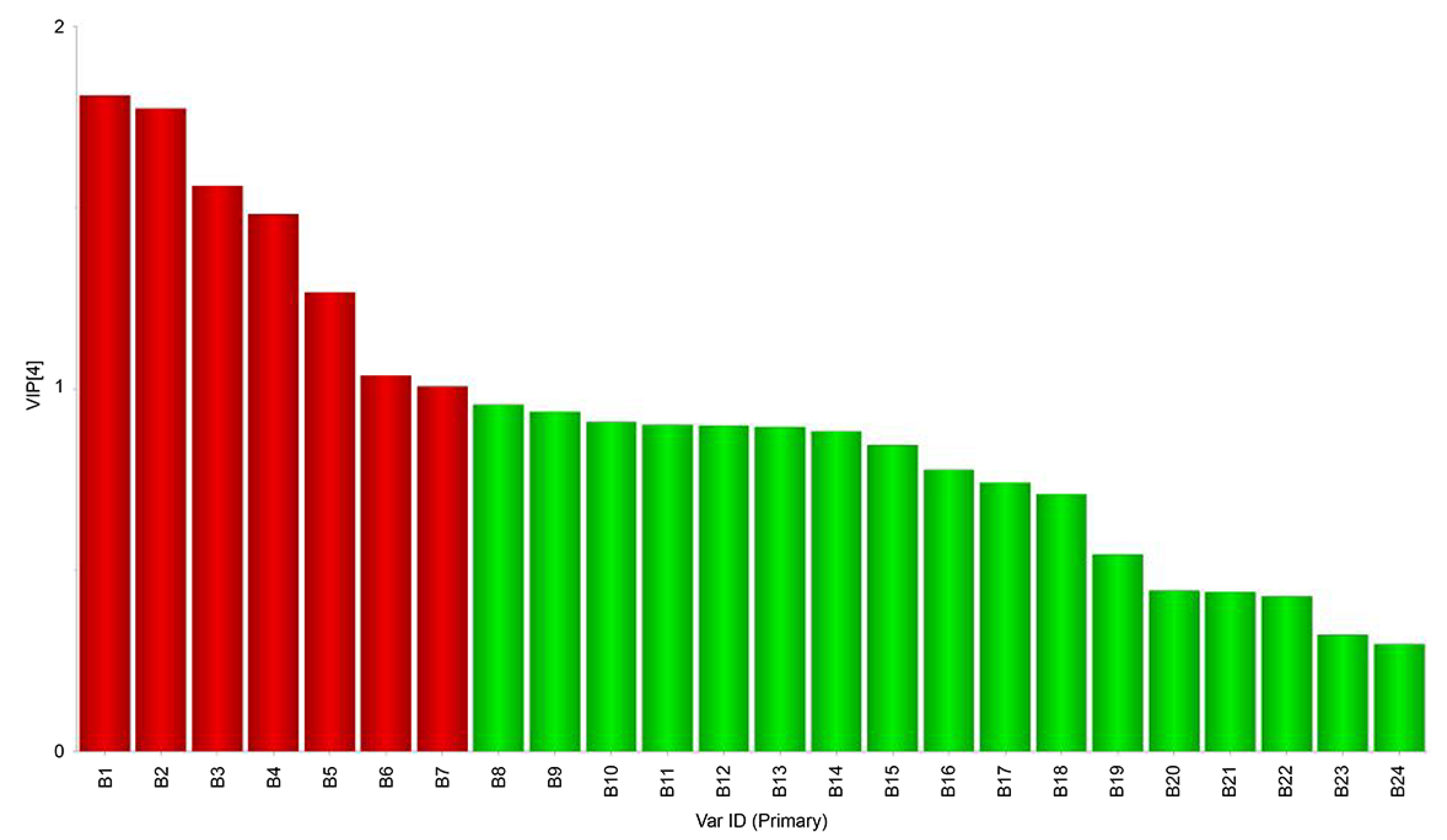
3.4. Correlation of Volatile Compound Composition with Microbial Community
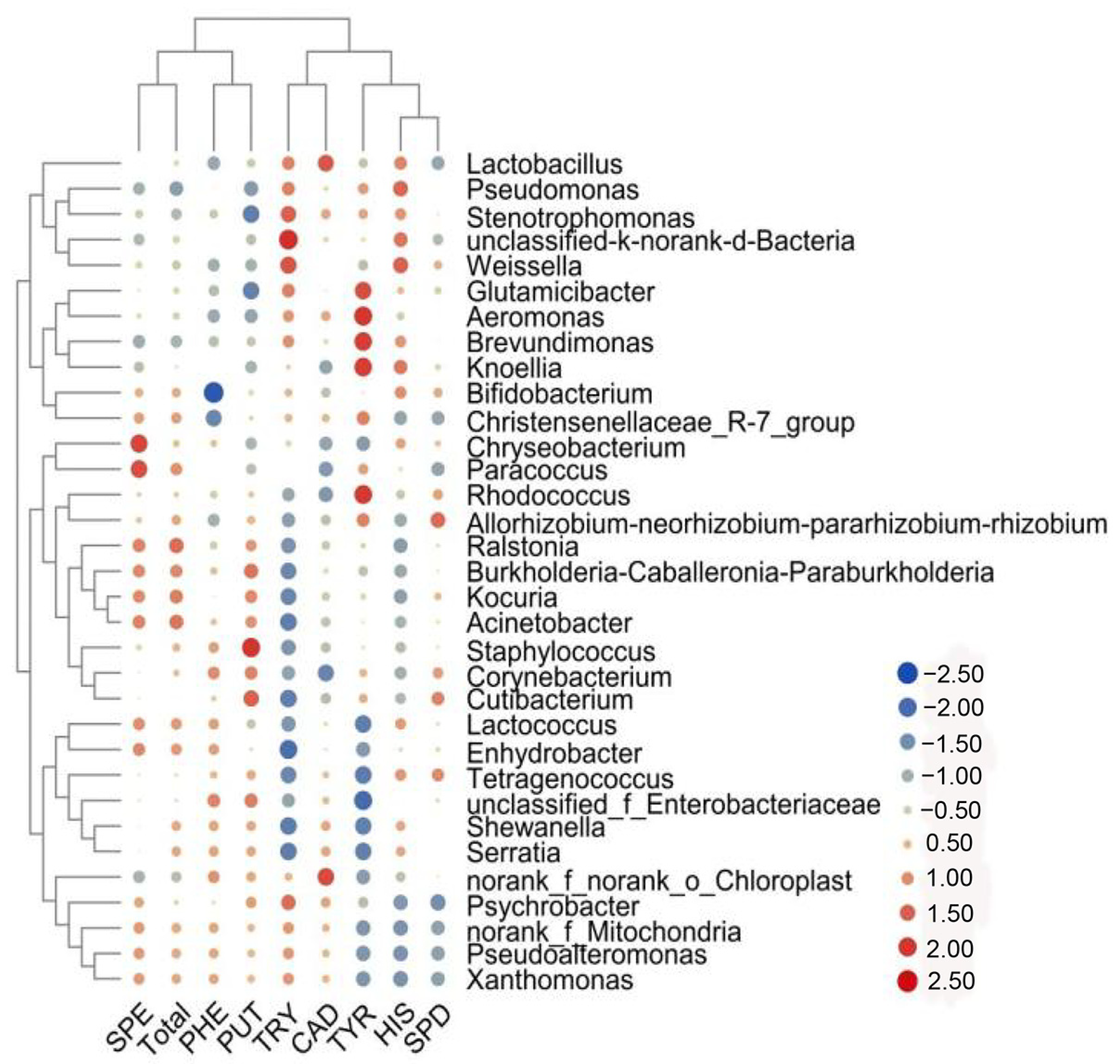
3.5. Correlation of Biogenic Amine Concentration with Microbial Community
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wen, R.; Sun, F.; Wang, Y.; Chen, Q.; Kong, B. Evaluation the potential of lactic acid bacteria isolates from traditional beef jerky as starter cultures and their effects on flavor formation during fermentation. LWT 2021, 142, 110982. [Google Scholar] [CrossRef]
- Leroy, F.; Verluyten, J.; De Vuyst, L. Functional meat starter cultures for improved sausage fermentation. Int. J. Food Microbiol. 2006, 106, 270–285. [Google Scholar] [CrossRef]
- Inguglia, E.; Oliveira, M.; Burgess, C.; Kerry, J.; Tiwari, B. Plasma activated water as an alternative nitrite source for the curing of beef jerky: Influence on quality and inactivation of listeria innocua. Innov. Food Sci. Emerg. 2020, 59, 102276. [Google Scholar] [CrossRef]
- Shi, S.; Kong, B.; Wang, Y.; Liu, Q.; Xia, X. Comparison of the quality of beef jerky processed by traditional and modern drying methods from different districts in inner mongolia. Meat Sci. 2020, 163, 108080. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Ren, H. Comparison of bacterial diversity profiles and microbial safety assessment of salami, chinese dry-cured sausage and chinese smoked-cured sausage by high-throughput sequencing. LWT 2018, 90, 108–115. [Google Scholar] [CrossRef]
- Quijada, N.; Filippis, F.; Sanz, J.; García-Fernández, M.; Rodríguez-Lázaro, D.; Ercolini, D.; Hernández, M. Different lactobacillus populations dominate in chorizo de leon manufacturing performed in different production plants. Food Microbiol. 2018, 70, 94–102. [Google Scholar] [CrossRef]
- Fontana, C.; Bassi, D.; López, C.; Pisacane, V.; Otero, M.; Puglisi, E.; Rebecchi, A.; Cocconcelli, P.; Vignolo, G. Microbial ecology involved in the ripening of naturally fermented llama meat sausages. a focus on lactobacilli diversity. Int. J. Food Microbiol. 2016, 236, 17–25. [Google Scholar] [CrossRef]
- Flores, M.; Toldrá, F. Microbial enzymatic activities for improved fermented meats. Trends Food Sci. Technol. 2011, 22, 81–90. [Google Scholar] [CrossRef]
- Corral, S.; Leitner, E.; Siegmund, B.; Flores, M. Determination of sulphur and nitrogen compounds during the processing of dry fermented sausages and their relation to amino acid generation. Food Chem. 2016, 190, 657–664. [Google Scholar] [CrossRef]
- Flores, M. Understanding the implications of current health trends on the aroma of wet and dry cured meat products. Meat Sci. 2018, 144, 53–61. [Google Scholar] [CrossRef]
- Huang, L.; Xiong, Y.; Kong, B.; Huang, X.; Li, J. Influence of storage temperature and duration on lipid and protein oxidation and flavour changes in frozen pork dumpling filler. Meat Sci. 2013, 95, 295–301. [Google Scholar] [CrossRef]
- Kim, B.; Bo, Y.; Man, J. Biogenic amine formation and bacterial contribution in natto products. Food Chem. 2012, 135, 2005–2011. [Google Scholar] [CrossRef]
- Kalac, P. Health effects and occurrence of dietary polyamines: A review for the period 2005–mid 2013. Food Chem. 2014, 161, 27–39. [Google Scholar] [CrossRef]
- Tittarelli, F.; Perpetuini, G.; Gianvito, P.; Tofalo, R. Biogenic amines producing and degrading bacteria: A snapshot from raw ewes’ cheese. LWT 2019, 101, 1–9. [Google Scholar] [CrossRef]
- Öz, M. Effects of black cumin (nigella sativa) oil on ammonia and biogenic amine production in rainbow trout. Trends Food Sci. Technol. 2017, 52, 265–269. [Google Scholar] [CrossRef]
- Linsalata, M.; Russo, F. Nutritional factors and polyamine metabolism in colorectal cancer. Nutrition 2008, 24, 382–389. [Google Scholar] [CrossRef]
- Vidal-Carou, M.; Bover-Cid, S.; Comas-Baste, O.; Latorre-Moratalla, L. Tyramine and histamine risk assessment related to consumption of dry fermented sausages by the spanish population. Food Chem. Toxicol. 2017, 99, 78–85. [Google Scholar]
- Xie, C.; Wang, H.; Deng, S.; Xu, X. The inhibition of cell-free supernatant of lactobacillus plantarum on production of putrescine and cadaverine by four amine-positive bacteria in vitro. LWT 2016, 67, 106–111. [Google Scholar] [CrossRef]
- Papavergou; Ekaterini, J. Biogenic amine levels in dry fermented sausages produced and sold in greece. Procedia Food Sci. 2011, 1, 1126–1131. [Google Scholar] [CrossRef]
- Liang, J.; Li, D.; Shi, R.; Wang, J.; Guo, S.; Ma, Y.; Xiong, K. Effects of microbial community succession on volatile profiles and biogenic amine during sufu fermentation. LWT 2019, 114, 108379. [Google Scholar] [CrossRef]
- Wang, B.; Luo, Y.; Wang, Y.; Wang, D.; Hou, Y.; Yao, D.; Tian, J.; Jin, Y. Rumen bacteria and meat fatty acid composition of sunit sheep reared under different feeding regimens in china. J. Sci. Food Agric. 2021, 101, 1110. [Google Scholar] [CrossRef]
- Yang, Y.; Term, C.; Shan, W.; Zheng, F.; Li, Q. Physicochemical, flavor and microbial dynamic changes during low-salt doubanjiang (broad bean paste) fermentation. Food Chem. 2021, 351, 128454. [Google Scholar] [CrossRef]
- Yang, P.; Zhong, G.; Yang, J.; Zhao, L.; Sun, D.; Tian, Y. Metagenomic and metabolomic profiling reveals the correlation between the microbiota and flavor compounds and nutrients in fermented sausages. Food Chem. 2021, 375, 131645. [Google Scholar] [CrossRef]
- Lu, S.; Ji, H.; Wang, Q.; Li, B.; Li, K.; Xu, C. The effects of starter cultures and plant extracts on the biogenic amine accumulation in traditional chinese smoked horsemeat sausages. Food Control 2015, 50, 869–875. [Google Scholar] [CrossRef]
- Lemos, L.; Fulthorpe, R.; Triplett, E.; Roesch, L. Rethinking microbial diversity analysis in the high throughput sequencing era. J. Microbiol. Meth. 2011, 86, 42–51. [Google Scholar] [CrossRef]
- Yao, D.; Xu, L.; Wu, M.; Wang, X.; Wang, C. Effects of microbial community succession on flavor compounds and physicochemical properties during cs sufu fermentation. LWT 2021, 152, 112313. [Google Scholar] [CrossRef]
- Jeong, S.-J.; Ryu, M.-S.; Yang, H.-J.; Wu, X.-H.; Jeong, D.-Y.; Park, S.-M. Bacterial Distribution, Biogenic Amine Contents, and Functionalities of Traditionally Made Doenjang, a Long-Term Fermented Soybean Food, from Different Areas of Korea. Microorganisms 2021, 9, 1348. [Google Scholar] [CrossRef]
- Zhong, A.; Chen, W.; Duan, Y.; Li, K.; Wang, C. The potential correlation between microbial communities and flavors in traditional fermented sour meat. LWT 2021, 149, 111873. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, Q.; Wen, R.; Wang, Y.; Kong, B. Quality characteristics and flavor profile of harbin dry sausages inoculated with lactic acid bacteria and staphylococcus xylosus. LWT 2019, 114, 108392. [Google Scholar] [CrossRef]
- Jin, Y.; Li, D.; Ai, M.; Tang, Q.; Huang, J.; Ding, X. Correlation between volatile profiles and microbial communities: A metabonomic approach to study jiang-flavor liquor daqu. Food Res. Int. 2019, 121, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Hu, J.; Chen, W. Analysis of the relationship between microorganisms and flavour development in dry-cured grass carp by high-throughput sequencing, volatile flavour analysis and metabolomics. Food Chem. 2022, 368, 130889. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Ge, Z.; Lin, K.; Zhang, D.; Shi, X. Dynamic changes in bacterial microbiota succession and flavour development during milk fermentation of kazak artisanal cheese. Int. Dairy J. 2021, 113, 104878. [Google Scholar] [CrossRef]
- Dabadé, D.S.; Jacxsens, L.; Miclotte, L.; Abatih, E.; Devlieghere, F.; de Meulenaer, B. Survey of multiple biogenic amines and correlation to microbiological quality and free amino acids in foods. Food Control 2021, 120, 107497. [Google Scholar] [CrossRef]
| Sample | Total Reads | OTUs | Shannon | Simpson | ACE | Chao1 | Goods Coverage |
|---|---|---|---|---|---|---|---|
| S1 | 55,861 | 1068 | 2.39 | 0.32 | 696.01 | 639.2 | 99.52 |
| S2 | 49,546 | 1133 | 3.43 | 0.10 | 634.08 | 639.09 | 99.62 |
| S3 | 53,181 | 1051 | 3.10 | 0.16 | 538.74 | 541.76 | 99.75 |
| S4 | 51,515 | 1039 | 3.16 | 0.14 | 601.34 | 608.36 | 99.64 |
| S5 | 55,592 | 730 | 3.45 | 0.12 | 413.35 | 414.46 | 99.83 |
| S6 | 75,682 | 1083 | 3.02 | 0.20 | 561.98 | 563.54 | 99.70 |
| S7 | 46,200 | 928 | 3.45 | 0.14 | 536.29 | 539.23 | 99.75 |
| S8 | 65,063 | 790 | 0.85 | 0.73 | 579.19 | 478.76 | 99.55 |
| S9 | 60,658 | 518 | 0.74 | 0.75 | 546.15 | 462.00 | 99.60 |
| S10 | 73,426 | 281 | 1.06 | 0.47 | 290.19 | 257.00 | 99.83 |
| S11 | 46,579 | 399 | 0.73 | 0.75 | 373.79 | 311.64 | 99.71 |
| S12 | 57,201 | 584 | 1.00 | 0.65 | 437.79 | 356.04 | 99.66 |
| S13 | 57,823 | 477 | 0.69 | 0.76 | 388.62 | 335.63 | 99.66 |
| S14 | 38,659 | 230 | 0.62 | 0.78 | 251.29 | 201.42 | 99.81 |
| S15 | 57,355 | 416 | 0.83 | 0.70 | 477.39 | 340.92 | 99.68 |
| S16 | 56,819 | 467 | 0.91 | 0.67 | 490.42 | 341.02 | 99.68 |
| mg/kg | TRY | PHE | PUT | CAD | HIS | TYR | SPD | SPE | Total |
|---|---|---|---|---|---|---|---|---|---|
| S1 | 62.48 ± 3.93 a | 33.55 ± 10.16 b | 68.99 ± 8.23 b | 87.64 ± 8.6 c | 47 ± 1.88 bcd | - | - | 351.75 ± 97.75 a | 651.41 ± 96.89 b |
| S2 | 40.65 ± 0.53 fg | 63.31 ± 12.29 a | 269.66 ± 65.73 a | 867.61 ± 163.53 a | 157.65 ± 27.05 a | - | 34.11 ± 0.54 | 258.75 ± 81.46 bc | 1691.73 ± 350.04 a |
| S3 | 41.24 ± 3.26 fg | 16.21 ± 0.67 de | 59.19 ± 9.64 b | 63.86 ± 6.05 c | 43.46 ± 2.78 d | - | - | 132.27 ± 43.66 f | 350.82 ± 61.21 def |
| S4 | 31.66 ± 0.31 h | 24.36 ± 2.78 bcd | 65.25 ± 18.02 b | 59.32 ± 12.34 c | 61.14 ± 24.72 bcd | - | 52.62 ± 8.5 ab | 268.73 ± 61.36 bc | 563.08 ± 78.79 bc |
| S5 | 42.07 ± 2.4 fg | 20.32 ± 3.43 cde | 56.26 ± 3.15 b | 59.07 ± 13.19 c | 55.93 ± 8.6 bcd | 17.85 ± 2.24 e | - | 172.14 ± 23.88 def | 423.65 ± 25.01 cdef |
| S6 | 31.85 ± 0.47 h | - | 59.5 ± 0.46 b | 59.42 ± 0.57 c | 46.58 ± 1.24 cd | 58.19 ± 5.83 b | 72.2 ± 33.4 a | 243.04 ± 37.37 cd | 527.32 ± 58.06 bcd |
| S7 | 50.71 ± 4.34 cde | - | 57.78 ± 1.45 b | 77.93 ± 3.96 c | 51.44 ± 3.33 bcd | 38.96 ± 0.39 cd | - | 252.3 ± 48.84 bcd | 516.14 ± 72.26 bcde |
| S8 | 56.03 ± 9.73 abc | - | - | 179.48 ± 83.53 b | - | 44.56 ± 13.5 bc | - | 327.6 ± 43.64 ab | 498.46 ± 180.96 bcde |
| S9 | 47.08 ± 3.84 def | 23.26 ± 2.35 cd | 57.5 ± 0.59 b | 80.71 ± 1.83 c | 58.04 ± 2.12 bcd | 18.09 ± 1.48 e | 35.4 ± 0.74 bc | - | 308.29 ± 11.9 f |
| S10 | 57.18 ± 6.32 abc | 28.1 ± 1.87 bc | 70.76 ± 10.46 b | 79.7 ± 1.86 c | 63.49 ± 1.21 bcd | 28.83 ± 12.91 cde | 34.15 ± 14.29 bc | 173.55 ± 9.04 def | 477.92 ± 78.59 bcdef |
| S11 | 46.02 ± 0.05 def | 11.22 ± 0.35 e | 47.16 ± 0.13 b | 52.33 ± 0.38 c | 71.04 ± 0.45 bc | 86.19 ± 6.04 a | 0 ± 0 | 128.34 ± 0.1 f | 442.29 ± 5.28 cdef |
| S12 | 50.43 ± 6.1 cde | 23.73 ± 1.44 cd | 55.14 ± 4.46 b | 69.9 ± 6.81 c | 59.49 ± 5.13 bcd | 27.74 ± 4.98 de | 0 ± 0 | 223.24 ± 9.23 cde | 509.67 ± 12.8 bced |
| S13 | 43.11 ± 2.19 efg | - | 50.07 ± 2.95 b | 63.68 ± 3.12 c | 52.74 ± 3.26 bcd | - | 12.43 ± 1.41 c | 115.19 ± 6.81 f | 333.08 ± 11.99 ef |
| S14 | 37.55 ± 0.6 gh | 28 ± 3.33 bc | 57.25 ± 11.61 b | 59.13 ± 3.79 c | 70.63 ± 27.66 bc | - | - | 153.03 ± 25.04 ef | 405.58 ± 35.08 cdef |
| S15 | 52.9 ± 1.11 bcd | - | 56.19 ± 1.96 b | 74.14 ± 1 c | 71.44 ± 3.21 bc | - | - | 220.5 ± 9.89 cde | 475.17 ± 11.54 bcdef |
| S16 | 60.48 ± 1.39 ab | - | 52.6 ± 1.2 b | 62.38 ± 2.66 c | 71.97 ± 9 b | - | - | 219.07 ± 13.66 cde | 466.5 ± 7.02 bcdef |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Sun, L.; Su, L.; Wang, H.; Wang, D.; Liu, J.; Sun, E.; Hu, G.; Liu, C.; Gao, A.; et al. Effects of Microbial Communities on Volatile Profiles and Biogenic Amines in Beef Jerky from Inner Mongolian Districts. Foods 2022, 11, 2659. https://doi.org/10.3390/foods11172659
Sun X, Sun L, Su L, Wang H, Wang D, Liu J, Sun E, Hu G, Liu C, Gao A, et al. Effects of Microbial Communities on Volatile Profiles and Biogenic Amines in Beef Jerky from Inner Mongolian Districts. Foods. 2022; 11(17):2659. https://doi.org/10.3390/foods11172659
Chicago/Turabian StyleSun, Xueying, Lina Sun, Lin Su, Huiting Wang, Dan Wang, Jianlin Liu, Erke Sun, Guanhua Hu, Chang Liu, Aiwu Gao, and et al. 2022. "Effects of Microbial Communities on Volatile Profiles and Biogenic Amines in Beef Jerky from Inner Mongolian Districts" Foods 11, no. 17: 2659. https://doi.org/10.3390/foods11172659
APA StyleSun, X., Sun, L., Su, L., Wang, H., Wang, D., Liu, J., Sun, E., Hu, G., Liu, C., Gao, A., Jin, Y., & Zhao, L. (2022). Effects of Microbial Communities on Volatile Profiles and Biogenic Amines in Beef Jerky from Inner Mongolian Districts. Foods, 11(17), 2659. https://doi.org/10.3390/foods11172659






