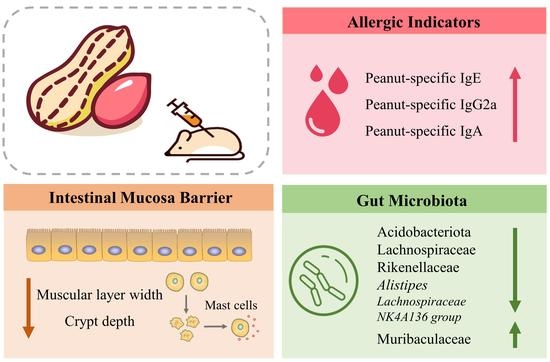
Gut Microbial Signatures Associated with Peanut Allergy in a BALB/c Mouse Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Peanut Protein Extraction
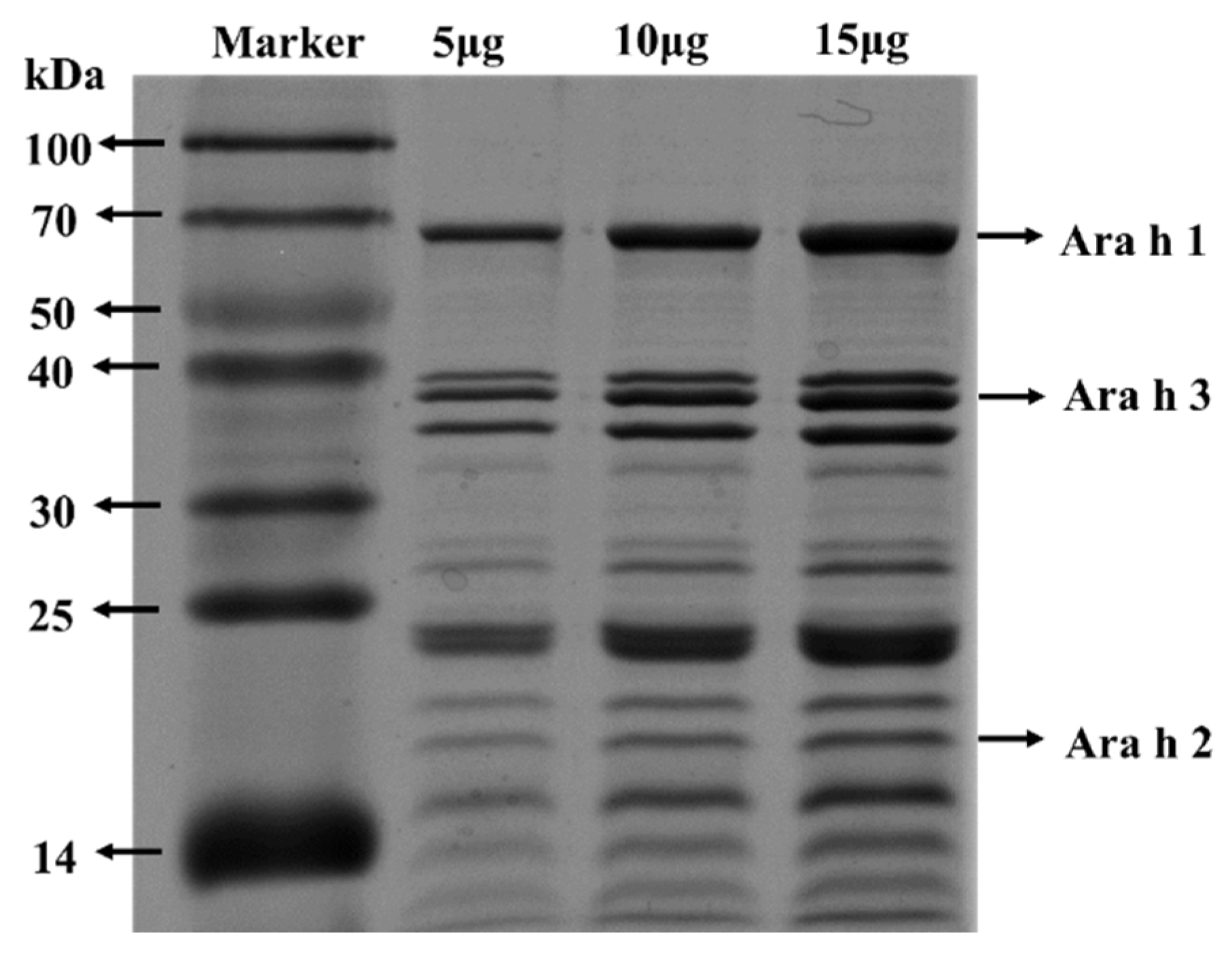
2.3. SDS-PAGE Analysis
2.4. Ethics Statement
2.5. Mice
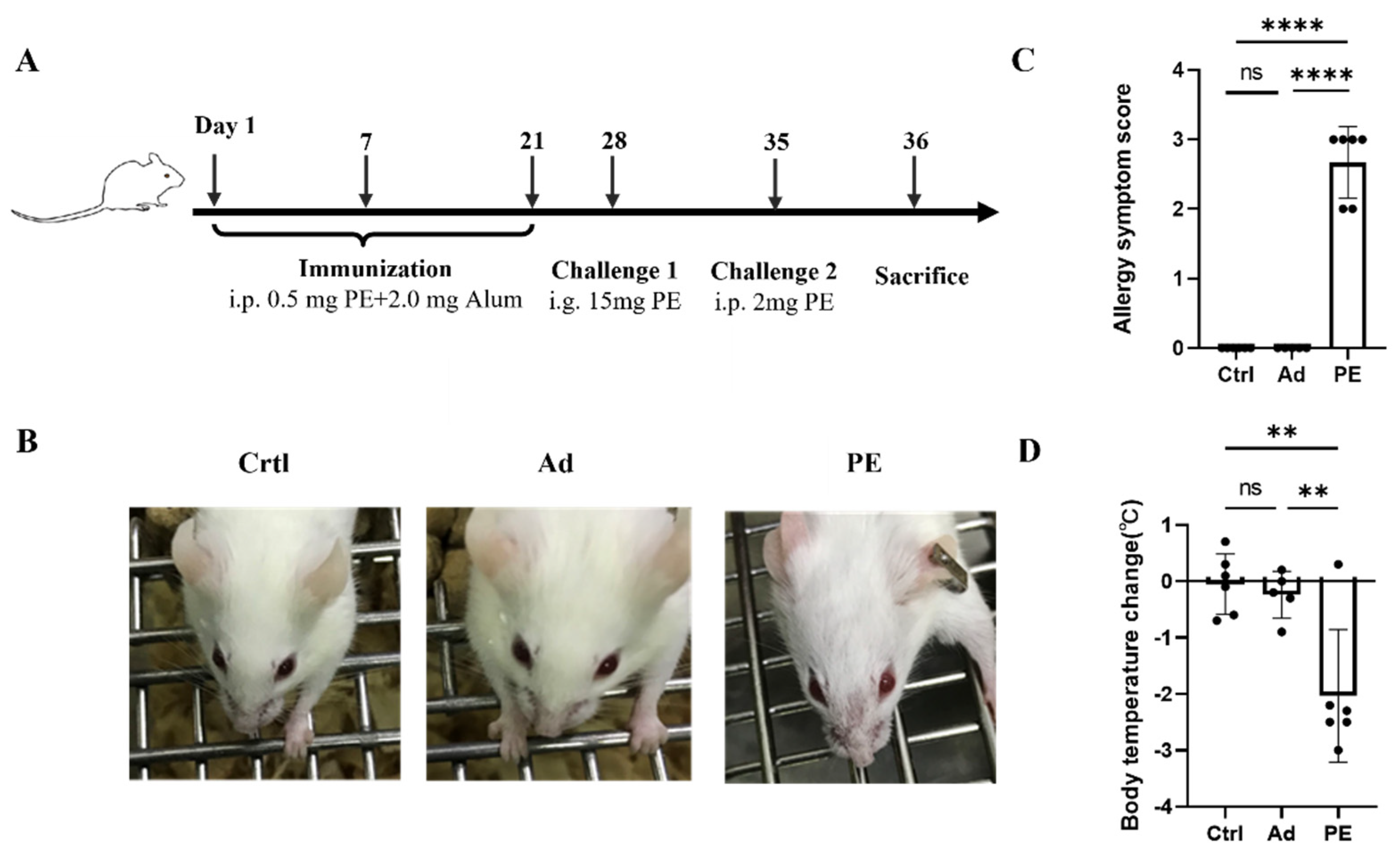
2.6. Immunization Protocols
2.7. Measurement of Levels of sIgE, sIgG2a, and sIgA in Serum
2.8. Cytokine Production in Duodenum and Spleen
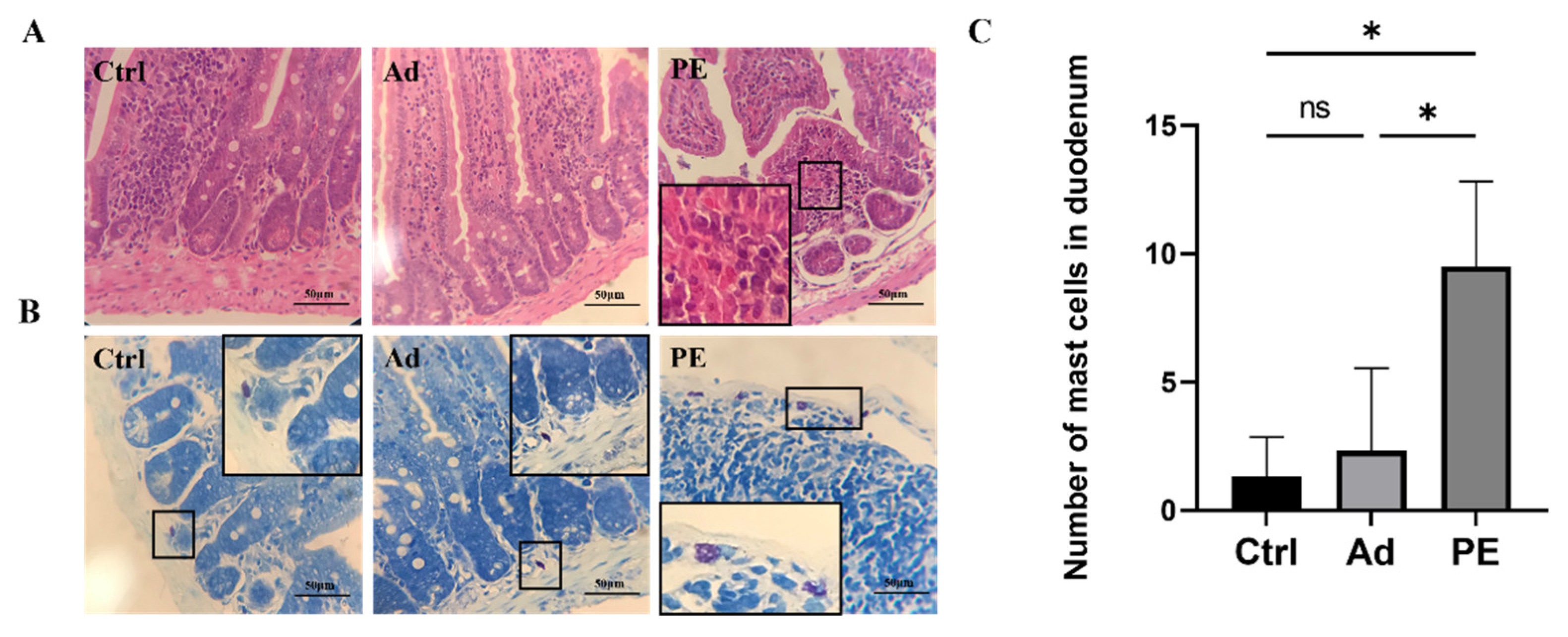
2.9. Pathological Analysis of Duodenal Tissue in Mice
2.10. 16S rRNA Gene Sequencing
2.11. Statistical Analysis
3. Results
3.1. SDS-PAGE Analysis of PE
3.2. PE Sensitization Caused Severe Allergic Responses in Mice
3.3. PE Sensitization Increased the Levels of Peanut-Specific Antibodies and IL-4
3.4. PE Sensitization Impaired the Intestinal Mucosa Barrier and Promoted Mast Cell Activation in Mice
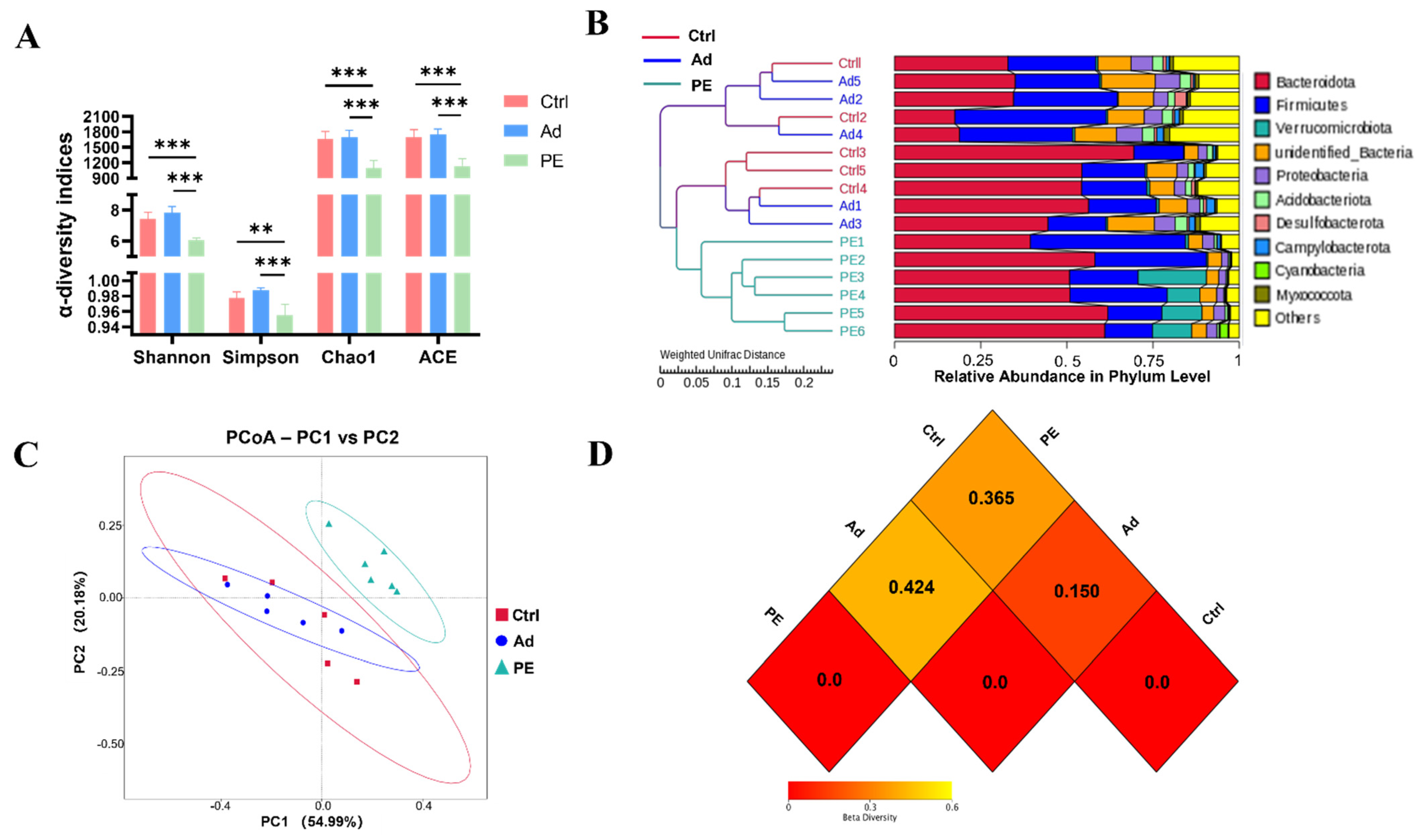
3.5. PE Sensitization Changed Microbiota Diversity and Structure
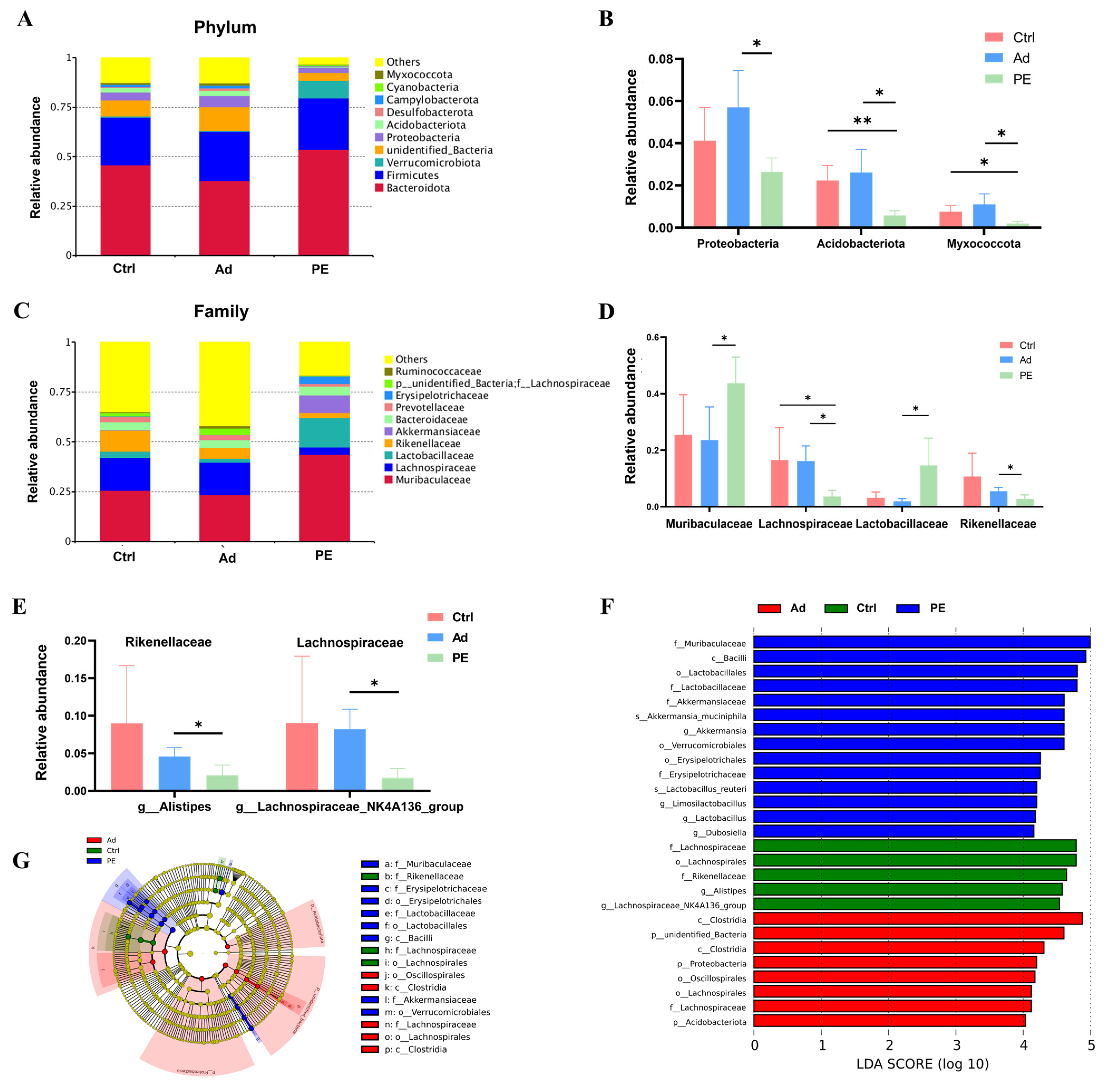
3.6. PE Sensitization Altered the Gut Microbiota Composition at Different Taxonomic Levels
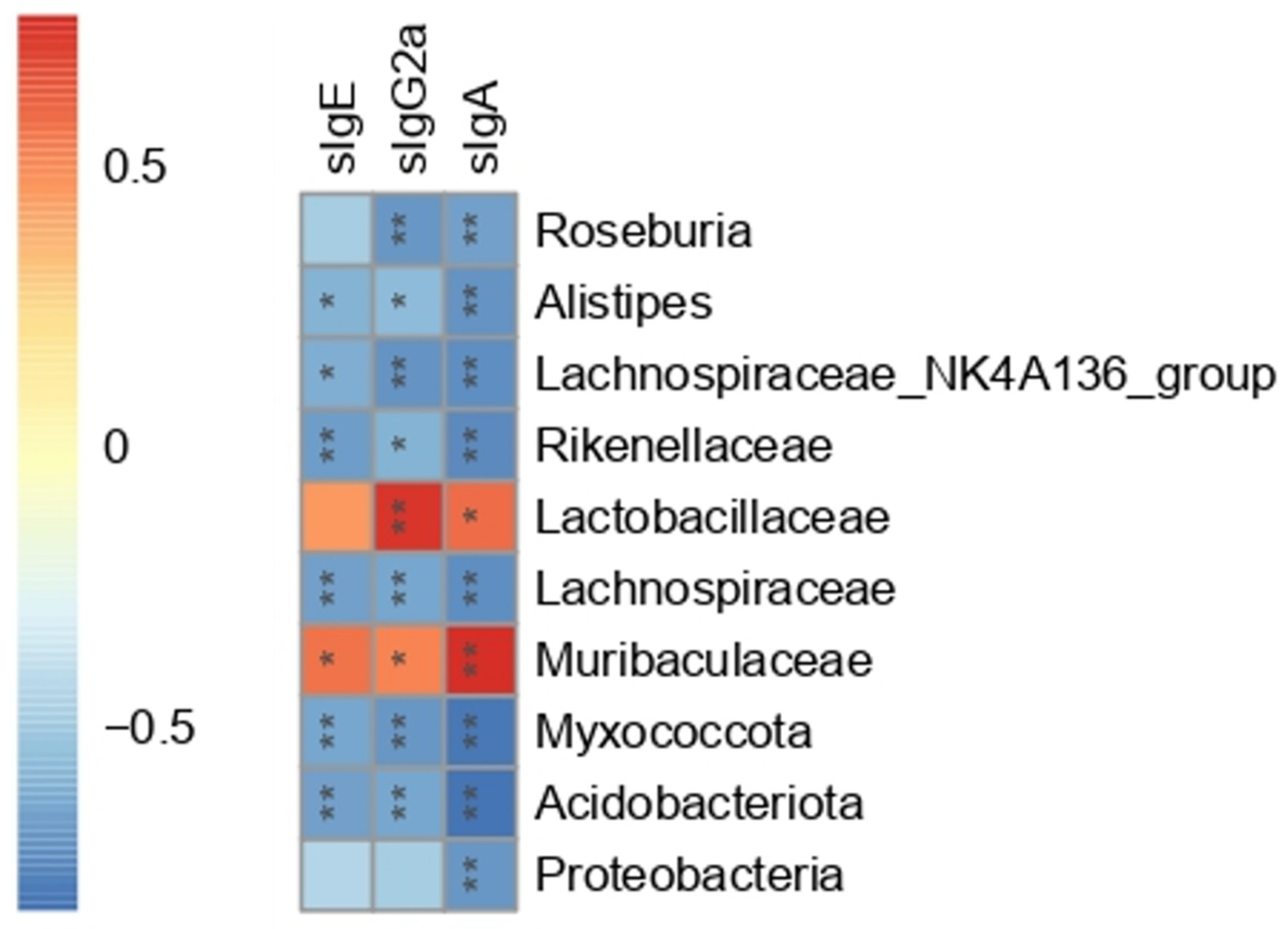
3.7. The Correlation between Gut Microbiota and Serum Peanut-Specific Antibodies
3.8. PE Sensitization Altered the Predicted Functions of Gut Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fazlollahi, M.; Chun, Y.; Grishin, A.; Wood, R.A.; Burks, A.W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.A.; Sicherer, S.H.; et al. Early-life gut microbiome and egg allergy. Allergy 2018, 73, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Boyce, J.A.; Assa’ad, A.; Burks, A.W.; Jones, S.M.; Sampson, H.A.; Wood, R.A.; Plaut, M.; Cooper, S.F.; Fenton, M.J.; Arshad, S.H.; et al. Guidelines for the Diagnosis and Management of Food Allergy in the United States: Summary of the NIAID-Sponsored Expert Panel Report. J. Allergy Clin. Immunol. 2010, 126, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Al-Muhsen, S.; Clarke, A.E.; Kagan, R.S. Peanut allergy: An overview. Can. Med. Assoc. J. 2003, 168, 1529. [Google Scholar]
- Venter, C.; Sicherer, S.H.; Greenhawt, M. Management of Peanut Allergy. J. Allergy Clin. Immunol. Pract. 2019, 7, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.R.; Mor, H.; Neriya, D.M.; Magzal, F.; Muller, E.; Appel, M.Y.; Nachshon, L.; Borenstein, E.; Tamir, S.; Louzoun, Y.; et al. Microbial signature in IgE-mediated food allergies. Genome Med. 2020, 12, 92. [Google Scholar] [CrossRef]
- Andreassen, M.; Rudi, K.; Angell, I.L.; Dirven, H.; Nygaard, U.C. Allergen Immunization Induces Major Changes in Microbiota Composition and Short-Chain Fatty Acid Production in Different Gut Segments in a Mouse Model of Lupine Food Allergy. Int. Arch. Allergy Immunol. 2018, 177, 311–323. [Google Scholar] [CrossRef]
- Bunyavanich, S.; Berin, M.C. Food allergy and the microbiome: Current understandings and future directions. J. Allergy Clin. Immunol. 2019, 144, 1468–1477. [Google Scholar] [CrossRef]
- Sharma, G.; Im, S.H. Probiotics as a Potential Immunomodulating Pharmabiotics in Allergic Diseases: Current Status and Future Prospects. Allergy Asthma Immunol. Res. 2018, 10, 575–590. [Google Scholar] [CrossRef]
- Stefka, A.T.; Feehley, T.; Tripathi, P.; Qiu, J.; McCoy, K.; Mazmanian, S.K.; Tjota, M.Y.; Seo, G.Y.; Cao, S.; Theriault, B.R.; et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA 2014, 111, 13145–13150. [Google Scholar] [CrossRef] [Green Version]
- Sivaprakasam, S.; Prasad, P.D.; Singh, N. Benefits of short-chain fatty acids and their receptors in inflammation and carcinogenesis. Pharmacol. Ther. 2016, 164, 144–151. [Google Scholar] [CrossRef] [Green Version]
- Gao, B.B.; Wang, R.J.; Peng, Y.; Li, X.B. Effects of a homogeneous polysaccharide from Sijunzi decoction on human intestinal microbes and short chain fatty acids in vitro. J. Ethnopharmacol. 2018, 224, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Org, E.; Blum, Y.; Kasela, S.; Mehrabian, M.; Kuusisto, J.; Kangas, A.J.; Soininen, P.; Wang, Z.N.; Ala-Korpela, M.; Hazen, S.L.; et al. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017, 18, 70. [Google Scholar] [CrossRef] [PubMed]
- Walker, W.A.; Iyengar, R.S. Breast milk, microbiota, and intestinal immune homeostasis. Pediatr. Res. 2015, 77, 220–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Ringel-Kulka, T.; Heikamp-de Jong, I.; Ringel, Y.; Carroll, I.; de Vos, W.M.; Salojarvi, J.; Satokari, R. Discordant temporal development of bacterial phyla and the emergence of core in the fecal microbiota of young children. ISME J. 2016, 10, 1002–1014. [Google Scholar] [CrossRef] [Green Version]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007, 5, 1556–1573. [Google Scholar] [CrossRef] [Green Version]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Koppelman, S.J.; Knol, E.F.; Vlooswijk, R.A.A.; Wensing, M.; Knulst, A.C.; Hefle, S.L.; Gruppen, H.; Piersma, S. Peanut allergen Ara h 3: Isolation from peanuts and biochemical characterization. Allergy 2003, 58, 1144–1151. [Google Scholar] [CrossRef]
- Pons, L.; Ponnappan, U.; Hall, R.A.; Simpson, P.; Cockrell, G.; West, C.M.; Sampson, H.A.; Helm, R.M.; Burks, A.W. Soy immunotherapy for peanut-allergic mice: Modulation of the peanut-allergic response. J. Allergy Clin. Immunol. 2004, 114, 915–921. [Google Scholar] [CrossRef]
- Leonard, S.A.; Martos, G.; Wang, W.; Nowak-Wegrzyn, A.; Berin, M.C. Oral immunotherapy induces local protective mechanisms in the gastrointestinal mucosa. J. Allergy Clin. Immunol. 2012, 129, 1579–1587 e1. [Google Scholar] [CrossRef] [Green Version]
- Lifrani, A.; Dubarry, M.; Rautureau, M.; Aattouri, N.; Boyaka, P.N.; Tomé, D. Peanut-lupine antibody cross-reactivity is not associated to cross-allergenicity in peanut-sensitized mouse strains. Int. Immunopharmacol. 2005, 5, 1427–1435. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, S.J.; Cui, J.L.; Guo, T.T.; Zhang, J.T.; Li, B.L. Alleviative Effects of Exopolysaccharide Produced by Lactobacillus helveticus KLDS1.8701 on Dextran Sulfate Sodium-Induced Colitis in Mice. Microorganisms 2021, 9, 2086. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, X.X.; Guo, F.; Wu, W.M.; Zhang, T. Characterization of tetracycline resistant bacterial community in saline activated sludge using batch stress incubation with high-throughput sequencing analysis. Water Res. 2013, 47, 4207–4216. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [Green Version]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef]
- Palladino, C.; Breiteneder, H. Peanut allergens. Mol. Immunol. 2018, 100, 58–70. [Google Scholar] [CrossRef]
- Md, A.; Maeda, M.; Matsui, T.; Takasato, Y.; Ito, K.; Kimura, Y. Purification and molecular characterization of a truncated-type Ara h 1, a major peanut allergen: Oligomer structure, antigenicity, and glycoform. Glycoconj. J. 2021, 38, 67–76. [Google Scholar] [CrossRef]
- Rabjohn, P.; Helm, E.M.; Stanley, J.S.; West, C.M.; Sampson, H.A.; Burks, A.W.; Bannon, G.A. Molecular cloning and epitope analysis of the peanut allergen Ara h 3. J. Clin. Investig. 1999, 103, 535–542. [Google Scholar] [CrossRef] [Green Version]
- Ratnaparkhe, M.B.; Lee, T.H.; Tan, X.; Wang, X.Y.; Li, J.P.; Kim, C.; Rainville, L.K.; Lemke, C.; Compton, R.O.; Robertson, J.; et al. Comparative and Evolutionary Analysis of Major Peanut Allergen Gene Families. Genome Biol. Evol. 2014, 6, 2468–2488. [Google Scholar] [CrossRef] [Green Version]
- Rao, H.; Tian, Y.; Fu, W.H.; Xue, W.T. In vitro digestibility and immunoreactivity of thermally processed peanut. Food Agric. Immunol. 2018, 29, 989–1001. [Google Scholar] [CrossRef] [Green Version]
- Kleber-Janke, T.; Crameri, R.; Appenzeller, U.; Schlaak, M.; Becker, W.M. Selective cloning of peanut allergens, including profilin and 2S albumins, by phage display technology. Int. Arch. Allergy Immunol. 1999, 119, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Petersen, A.; Kull, S.; Rennert, S.; Becker, W.M.; Krause, S.; Ernst, M.; Gutsmann, T.; Bauer, J.; Lindner, B.; Jappe, U. Peanut defensins: Novel allergens isolated from lipophilic peanut extract. J. Allergy Clin. Immunol. 2015, 136, 1295–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castan, L.; Bogh, K.L.; Maryniak, N.Z.; Epstein, M.M.; Kazemi, S.; O’Mahony, L.; Bodinier, M.; Smit, J.J.; van Bilsen, J.H.M.; Blanchard, C.; et al. Overview of in vivo and ex vivo endpoints in murine food allergy models: Suitable for evaluation of the sensitizing capacity of novel proteins? Allergy 2020, 75, 289–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gould, H.J.; Sutton, B.J.; Beavil, A.J.; Beavil, R.L.; McCloskey, N.; Coker, H.A.; Fear, D.; Smurthwaite, L. The biology of IgE and the basis of allergic disease. Annu. Rev. Immunol. 2003, 21, 579–628. [Google Scholar] [CrossRef]
- Shamji, M.H.; Valenta, R.; Jardetzky, T.; Verhasselt, V.; Durham, S.R.; Wurtzen, P.A.; van Neerven, R.J.J. The role of allergen-specific IgE, IgG and IgA in allergic disease. Allergy 2021, 76, 3627–3641. [Google Scholar] [CrossRef]
- Pali-Scholl, I.; Szollosi, H.; Starkl, P.; Scheicher, B.; Stremnitzer, C.; Hofmeister, A.; Roth-Walter, F.; Lukschal, A.; Diesner, S.C.; Zimmer, A.; et al. Protamine nanoparticles with CpG-oligodeoxynucleotide prevent an allergen-induced Th2-response in BALB/c mice. Eur. J. Pharm. Biopharm. 2015, 97, 290. [Google Scholar] [CrossRef]
- Swain, S.L.; Weinberg, A.D.; English, M.; Huston, G. Il-4 Directs the Development of Th2-like Helper Effectors. J. Immunol. 1990, 145, 3796–3806. [Google Scholar]
- Wang, K.; Yang, Q.Q.; Ma, Q.X.; Wang, B.; Wan, Z.R.; Chen, M.L.; Wu, L.M. Protective Effects of Salvianolic Acid A against Dextran Sodium Sulfate-Induced Acute Colitis in Rats. Nutrients 2018, 10, 791. [Google Scholar] [CrossRef] [Green Version]
- Iwaszko, M.; Bialy, S.; Bogunia-Kubik, K. Significance of Interleukin (IL)-4 and IL-13 in Inflammatory Arthritis. Cells 2021, 10, 3000. [Google Scholar] [CrossRef]
- Nakayama, J.; Kobayashi, T.; Tanaka, S.; Korenori, Y.; Tateyama, A.; Sakamoto, N.; Kiyohara, C.; Shirakawa, T.; Sonomoto, K. Aberrant structures of fecal bacterial community in allergic infants profiled by 16S rRNA gene pyrosequencing. FEMS Immunol. Med. Microbiol. 2011, 63, 397–406. [Google Scholar] [CrossRef] [Green Version]
- Di Costanzo, M.; Carucci, L.; Canani, R.B.; Biasucci, G. Gut Microbiome Modulation for Preventing and Treating Pediatric Food Allergies. Int. J. Mol. Sci. 2020, 21, 5275. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Liu, Q.B.; Wang, W. Bifidobacterium bifidum TMC3115 ameliorates milk protein allergy in by affecting gut microbiota: A randomized double-blind control trial. J. Food Biochem. 2021, 45, e13489. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ye, Y.; Ji, J.; Sun, J.; Wang, J.S.; Sun, X. Untargeted Metabolomic Profiling Reveals Changes in Gut Microbiota and Mechanisms of Its Regulation of Allergy in OVA-Sensitive BALB/c Mice. J. Agric. Food Chem. 2022, 70, 3344–3356. [Google Scholar] [CrossRef] [PubMed]
- He, Z.Y.; Vadali, V.L.G.; Szabady, R.L.; Zhang, W.M.; Norman, J.M.; Roberts, B.; Tibshirani, R.; Desai, M.; Chinthrajah, R.S.; Galli, S.J.; et al. Increased diversity of gut microbiota during active oral immunotherapy in peanut-allergic adults. Allergy 2021, 76, 927–930. [Google Scholar] [CrossRef] [PubMed]
- Kourosh, A.; Luna, R.A.; Balderas, M.; Nance, C.; Anagnostou, A.; Devaraj, S.; Davis, C.M. Fecal microbiome signatures are different in food-allergic children compared to siblings and healthy children. Pediatr. Allergy Immunol. 2018, 29, 545–554. [Google Scholar] [CrossRef]
- Rivas, M.N.; Burton, O.T.; Wise, P.; Zhang, Y.Q.; Hobson, S.A.; Lloret, M.G.; Chehoud, C.; Kuczynski, J.; DeSantis, T.; Warrington, J.; et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J. Allergy Clin. Immunol. 2013, 131, 201–212. [Google Scholar] [CrossRef]
- Vonk, M.M.; Engen, P.A.; Naqib, A.; Green, S.J.; Keshavarzian, A.; Blokhuis, B.R.J.; Garssen, J.; Knippels, L.M.J.; van Esch, B.C.A.M. Altered microbial community structure and metabolism in cow’s milk allergic mice treated with oral immunotherapy and fructo-oligosaccharides. Benef. Microbes 2020, 11, 19–32. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria with Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Zuo, K.; Li, J.; Li, K.B.; Hu, C.W.; Gao, Y.F.; Chen, M.L.; Hu, R.M.; Liu, Y.; Chi, H.J.; Wang, H.J.; et al. Disordered gut microbiota and alterations in metabolic patterns are associated with atrial fibrillation. Gigascience 2019, 8, giz058. [Google Scholar] [CrossRef] [Green Version]
- Frank, D.N.; Amand, A.L.S.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [Green Version]
- Ran, B.B.; Guo, C.E.; Li, W.D.; Li, W.S.; Wang, Q.; Qian, J.X.; Li, H.L. Sea buckthorn (Hippophae rhamnoides L.) fermentation liquid protects against alcoholic liver disease linked to regulation of liver metabolome and the abundance of gut microbiota. J. Sci. Food Agric. 2021, 101, 2846–2854. [Google Scholar] [CrossRef] [PubMed]
- Rachid, R.A.; Gerber, G.; Li, N.; Umetsu, D.T.; Bry, L.; Chatila, T.A. Food Allergy in Infancy Is Associated with Dysbiosis of the Intestinal Microbiota. J. Allergy Clin. Immunol. 2016, 137, Ab235. [Google Scholar] [CrossRef]
- Hu, S.S.; Li, S.; Liu, Y.; Sun, K.; Luo, L.Y.; Zeng, L. Aged Ripe Pu-erh Tea Reduced Oxidative Stress-Mediated Inflammation in Dextran Sulfate Sodium-Induced Colitis Mice by Regulating Intestinal Microbes. J. Agric. Food Chem. 2021, 69, 10592–10605. [Google Scholar] [CrossRef]
- Hu, S.W.; Yang, H.C.; Gao, X.; Li, S.J.; Jiang, W.; Liu, Y. Egg oil from Portunus trituberculatus alleviated obesity and regulated gut microbiota in mice. Sci. Rep. 2020, 10, 8454. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.M.; Wu, W.; Liu, Z.J.; Cong, Y.Z. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.Y.; Ni, Y.H.; Wang, Z.; Tu, W.Q.; Ni, L.Y.; Zhuge, F.; Zheng, A.Q.; Hu, L.T.; Zhao, Y.F.; Zheng, L.J.; et al. Spermidine improves gut barrier integrity and gut microbiota function in diet-induced obese mice. Gut Microbes 2020, 12, 1832857. [Google Scholar] [CrossRef]
- Kong, J.; Chalcraft, K.; Mandur, T.S.; Jimenez-Saiz, R.; Walker, T.D.; Goncharova, S.; Gordon, M.E.; Naji, L.; Flader, K.; Larche, M.; et al. Comprehensive metabolomics identifies the alarmin uric acid as a critical signal for the induction of peanut allergy. Allergy 2015, 70, 495–505. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, S.; Xie, Q.; Chen, C.; Liu, C.; Xue, W. Gut Microbial Signatures Associated with Peanut Allergy in a BALB/c Mouse Model. Foods 2022, 11, 1395. https://doi.org/10.3390/foods11101395
Gu S, Xie Q, Chen C, Liu C, Xue W. Gut Microbial Signatures Associated with Peanut Allergy in a BALB/c Mouse Model. Foods. 2022; 11(10):1395. https://doi.org/10.3390/foods11101395
Chicago/Turabian StyleGu, Shimin, Qiang Xie, Chen Chen, Chenglong Liu, and Wentong Xue. 2022. "Gut Microbial Signatures Associated with Peanut Allergy in a BALB/c Mouse Model" Foods 11, no. 10: 1395. https://doi.org/10.3390/foods11101395
APA StyleGu, S., Xie, Q., Chen, C., Liu, C., & Xue, W. (2022). Gut Microbial Signatures Associated with Peanut Allergy in a BALB/c Mouse Model. Foods, 11(10), 1395. https://doi.org/10.3390/foods11101395