Investigation by Synchrotron Radiation Circular Dichroism of the Secondary Structure Evolution of Pepsin under Oxidative Environment
Abstract
1. Introduction
2. Materials and Methods
2.1. Oxidation Induction of Pepsin
2.2. Creatine Kinase Hydrolysis
2.3. Protein Oxidation Determination
2.4. Synchrotron-Radiation Circular Dichroism
2.5. MALDI-TOF Mass Spectrometry
2.6. Statistical Analysis
3. Results and Discussion
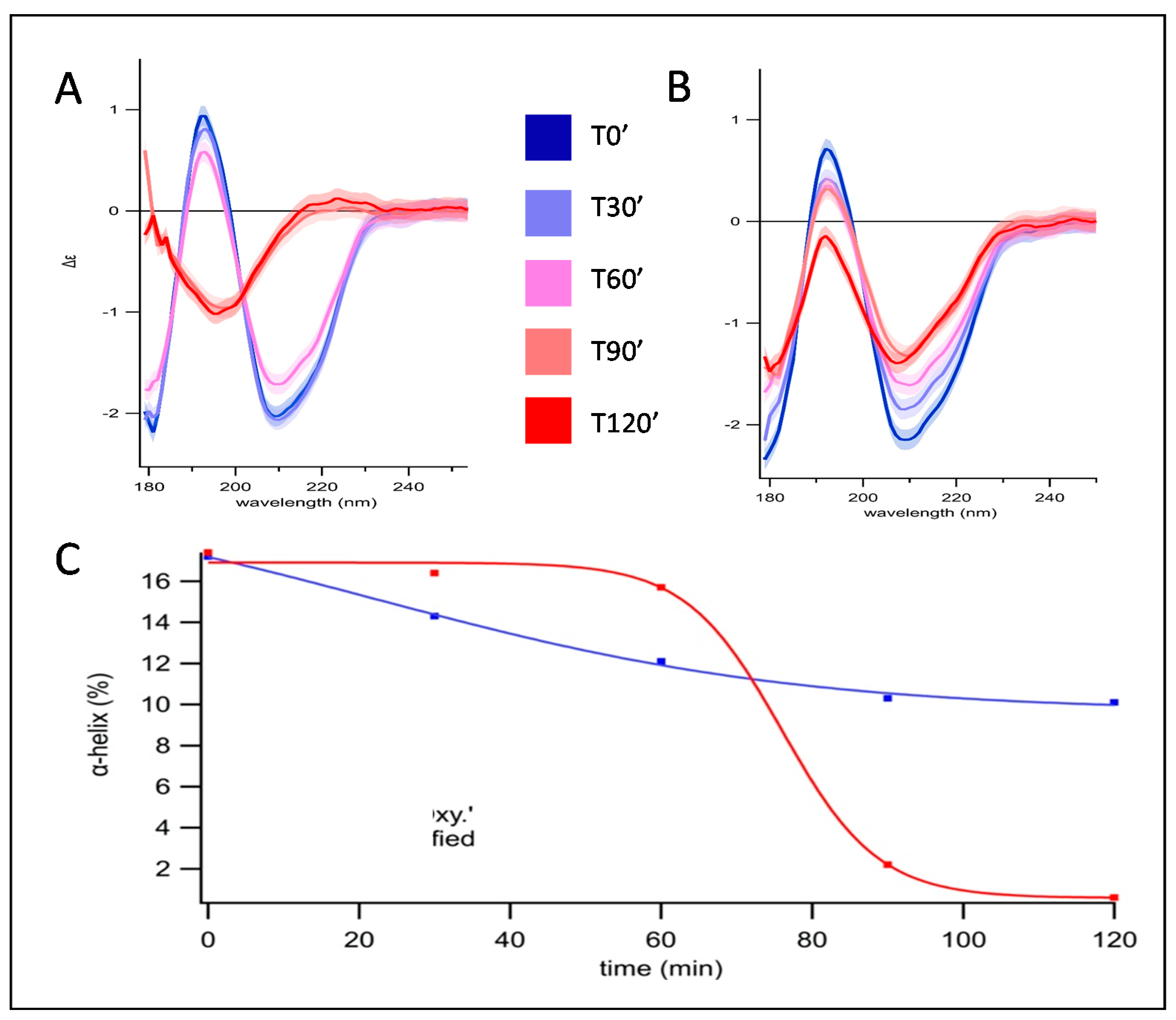
3.1. Secondary Structure Evolution of Oxidized Pepsin Investigated by SRCD
3.2. Proteolytic Activity of Oxidized Pepsin Investigated by MALDI-TOF Mass Spectrometry
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hellwig, M. The Chemistry of Protein Oxidation in Food. Angew. Chem. 2019, 58, 16742–16763. [Google Scholar] [CrossRef] [PubMed]
- De La Pomélie, D.; Santé-Lhoutellier, V.; Sayd, T.; Gatellier, P. Oxidation and nitrosation of meat proteins under gastro-intestinal conditions: Consequences in terms of nutritional and health values of meat. Food Chem. 2018, 243, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Soladoye, O.P.; Juarez, M.L.; Aalhus, J.L.; Shand, P.; Estévez, M. Protein Oxidation in Processed Meat: Mechanisms and Potential Implications on Human Health. Compr. Rev. Food Sci. Food Saf. 2015, 14, 106–122. [Google Scholar] [CrossRef]
- Estévez, M. Protein carbonyls in meat systems: A review. Meat Sci. 2011, 89, 259–279. [Google Scholar] [CrossRef]
- Santé-Lhoutellier, V.; Aubry, L.; Gatellier, P. Effect of oxidation on in vitro digestibility of skeletal muscle myofibrillar proteins. J. Agric. Food Chem. 2007, 55, 5343–5348. [Google Scholar] [CrossRef] [PubMed]
- Rutherfurd, S.M.; Montoya, C.A.; Moughan, P.J. Effect of Oxidation of Dietary Proteins with Performic Acid on True Ileal Amino Acid Digestibility as Determined in the Growing Rat. J. Agric. Food Chem. 2014, 62, 699–707. [Google Scholar] [CrossRef]
- Rogers, D.M.; Jasim, S.B.; Dyer, N.T.; Auvray, F.; Réfrégiers, M.; Hirst, J.D. Electronic Circular Dichroism Spectroscopy of Proteins. Chem 2019, 5, 2751–2774. [Google Scholar] [CrossRef]
- Duconseille, A.; Wien, F.; Audonnet, F.; Traore, A.; Refregiers, M.; Astruc, T.; Santé-Lhoutellier, V. The effect of origin of the gelatine and ageing on the secondary structure and water dissolution. Food Hydrocoll. 2017, 66, 378–388. [Google Scholar] [CrossRef]
- Auvray, F.; Dennetiere, D.; Giuliani, A.; Jamme, F.; Wien, F.; Nay, B.; Zirah, S.; Polack, F.; Menneglier, C.; Lagarde, B.; et al. Time resolved transient circular dichroism spectroscopy using synchrotron natural polarization. Struct. Dyn. 2019, 6, 054307. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef]
- Oliver, C.N.; Ahn, B.; Moermant, E.J.; Goldstein, S.; Stadtman, E.R. Age-related Changes in Oxidized Proteins. J. Biol. Chem. 1987, 262, 5488–5491. [Google Scholar] [CrossRef]
- Mercier, Y.; Gatellier, P.; Viau, M.; Remignon, H.; Renerre, M. Effect of dietary fat and vitamin E on colour stability and on lipid and protein oxidation in turkey meat during storage. Meat Sci. 1998, 48, 301–318. [Google Scholar] [CrossRef]
- Réfrégiers, M.; Wien, F.; Ta, H.-P.; Premvardhan, L.; Bac, S.; Jamme, F.; Rouam, V.; Lagarde, B.; Polack, F.; Giorgetta, J.-L.; et al. DISCO synchrotron-radiation circular-dichroism endstation at SOLEIL. J. Synchrotron Radiat. 2012, 19, 831–835. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, A.; Giorgetta, J.-L.; Ricaud, J.-P.; Jamme, F.; Rouam, V.; Wien, F.; Laprevote, O.; Refregiers, M. Atmospheric pressure photoionization using tunable VUV synchrotron radiation. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 2012, 279, 114–117. [Google Scholar] [CrossRef]
- Miles, A.J.; Janes, R.W.; Brown, A.; Clarke, D.T.; Sutherland, J.C.; Tao, Y.; Wallace, B.A.; Hoffmann, S.V. Light flux density threshold at which protein denaturation is induced by synchrotron radiation circular dichroism beamlines. J. Synchrotron Radiat. 2008, 15, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef]
- Lopes, J.L.S.; Miles, A.J.; Whitmore, L.; Wallace, B.A. Distinct circular dichroism spectroscopic signatures of polyproline II and unordered secondary structures: Applications in secondary structure analyses. Protein Sci. 2014, 23, 1765–1772. [Google Scholar] [CrossRef]
- Malagelada, J.R.; Longstreth, G.F.; Summerskill, W.H.J.; Go, V.L.W. Measurement of gastric functions during digestion of ordinary solid meals in man. Gastroenterology 1976, 70, 203–210. [Google Scholar] [CrossRef]
- Dressman, J.B.; Berardi, R.R.; Dermentzoglou, L.C.; Russell, T.L.; Schmaltz, S.P.; Barnett, J.L.; Jarvenpaa, K.M. Upper Gastrointestinal (GI) pH in Young, Healthy Men and Women. Pharm. Res. 1990, 7, 756–761. [Google Scholar] [CrossRef]
- Gardner, J.D. Meal-stimulated gastric acid secretion and integrated gastric acidity in gastro-oesophageal reflux disease. Aliment. Pharmacol. Ther. 2003, 17, 945–953. [Google Scholar] [CrossRef]
- Sicard, J.; Mirade, P.-S.; Portanguen, S.; Clerjon, S.; Kondjoyan, A. Simulation of the gastric digestion of proteins of meat bolus using a reaction-diffusion model. Food Funct. 2018, 9, 6455–6469. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, K.; Tashiro, T. The Oxidation of Trypsin and Pepsin by Periodate. J. Agric. Chem. Soc. Jpn. 1956, 30, 183–187. [Google Scholar] [CrossRef]
- Sielecki, A.R.; Fedorov, A.A.; Boodhoo, A.; Andreeva, N.S.; James, M.N. Molecular and crystal structures of monoclinic porcine pepsin refined at 1.8A°resolution. J. Mol. Biol. 1990, 214, 143–170. [Google Scholar] [CrossRef]
- Zhu, R.-R.; Wang, W.-R.; Sun, X.-Y.; Liu, H.; Wang, S.-L. Enzyme activity inhibition and secondary structure disruption of nano-TiO2 on pepsin. Toxicol. Vitr. 2010, 24, 1639–1647. [Google Scholar] [CrossRef] [PubMed]

| Kinetic, in Minutes | 0 | 30 | 60 | 90 | 120 | |
|---|---|---|---|---|---|---|
| pH | 5 | 4.5 | 3.5 | 2.5 | 2 | |
| Carbonyl group (nmoles DTNP/mg of protein) | Control | 3.20 ± 1.03 a | 2.56 ± 0.79 a | 2.57 ± 0.74 a | 1.99 ± 0.21 a | 2.04 ± 0.22 a |
| Oxidized | 15.43 ± 0.54 b | 14.95 ± 0.66 b | 14.95 ± 0.89 b | 14.75 ± 0.34 b | 13.98 ± 0.44 b | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Théron, L.; Bonifacie, A.; Delabre, J.; Sayd, T.; Aubry, L.; Gatellier, P.; Ravel, C.; Chambon, C.; Astruc, T.; Rouel, J.; et al. Investigation by Synchrotron Radiation Circular Dichroism of the Secondary Structure Evolution of Pepsin under Oxidative Environment. Foods 2021, 10, 998. https://doi.org/10.3390/foods10050998
Théron L, Bonifacie A, Delabre J, Sayd T, Aubry L, Gatellier P, Ravel C, Chambon C, Astruc T, Rouel J, et al. Investigation by Synchrotron Radiation Circular Dichroism of the Secondary Structure Evolution of Pepsin under Oxidative Environment. Foods. 2021; 10(5):998. https://doi.org/10.3390/foods10050998
Chicago/Turabian StyleThéron, Laetitia, Aline Bonifacie, Jérémy Delabre, Thierry Sayd, Laurent Aubry, Philippe Gatellier, Christine Ravel, Christophe Chambon, Thierry Astruc, Jacques Rouel, and et al. 2021. "Investigation by Synchrotron Radiation Circular Dichroism of the Secondary Structure Evolution of Pepsin under Oxidative Environment" Foods 10, no. 5: 998. https://doi.org/10.3390/foods10050998
APA StyleThéron, L., Bonifacie, A., Delabre, J., Sayd, T., Aubry, L., Gatellier, P., Ravel, C., Chambon, C., Astruc, T., Rouel, J., Santé-Lhoutellier, V., Réfrégiers, M., & Wien, F. (2021). Investigation by Synchrotron Radiation Circular Dichroism of the Secondary Structure Evolution of Pepsin under Oxidative Environment. Foods, 10(5), 998. https://doi.org/10.3390/foods10050998







