Proteolytic Traits of Psychrotrophic Bacteria Potentially Causative of Sterilized Milk Instability: Genotypic, Phenotypic and Peptidomic Insight
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Phenotypic and Genotypic Traits of Bacterial Strains
2.2.1. Proteolytic Activity on Milk Agar Plate
2.2.2. Presence of the AprX Gene
2.2.3. Expression of Extracellular Heat-Resistant Proteases
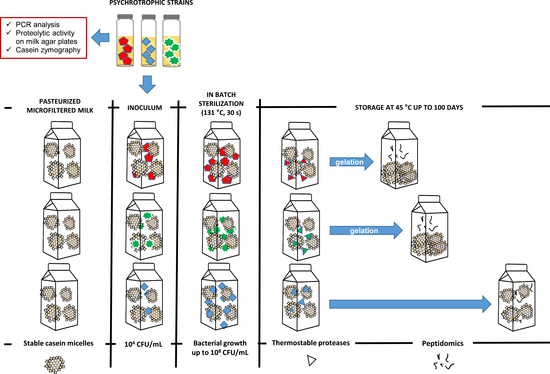
2.3. Proteolytic Activity of the Psychrotrophic Strains in Milk
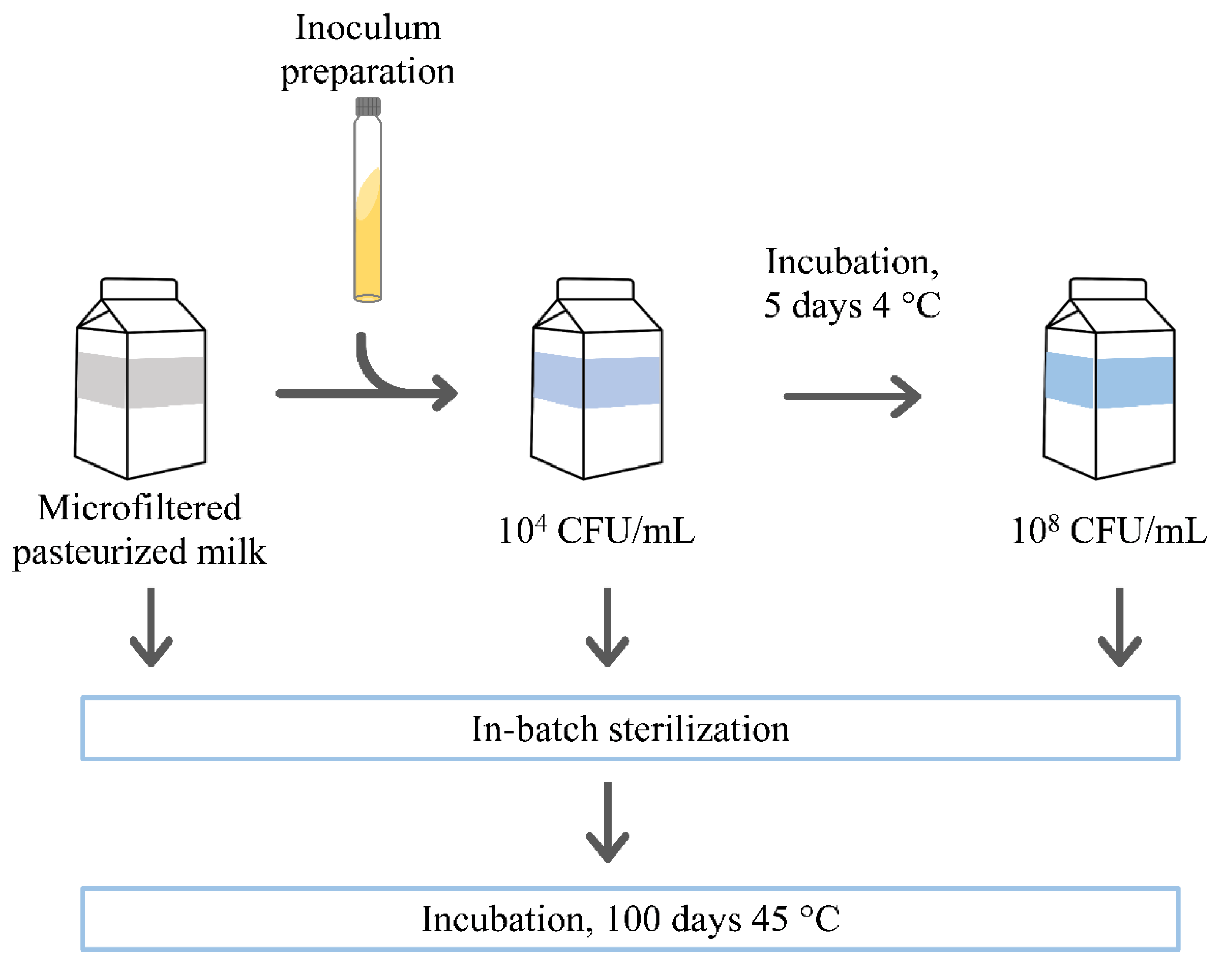
2.3.1. Preparation of Bacterial Inoculum
2.3.2. Assessment of Thermostable Protease Production in Milk
2.3.3. Inoculation and Incubation of Experimental Milk Samples
2.4. UPLC/HR-MS/MS Analysis, Peptide Identification, and Peptidomic Data Analysis
3. Results and Discussion
3.1. Phenotypic and Genotypic Characterization of Psychrotrophic Strains
3.2. Characterization of Proteolytic Activity of the Psychrotrophic Strains in Pasteurized Milk Prior to and after In-Batch Sterilization
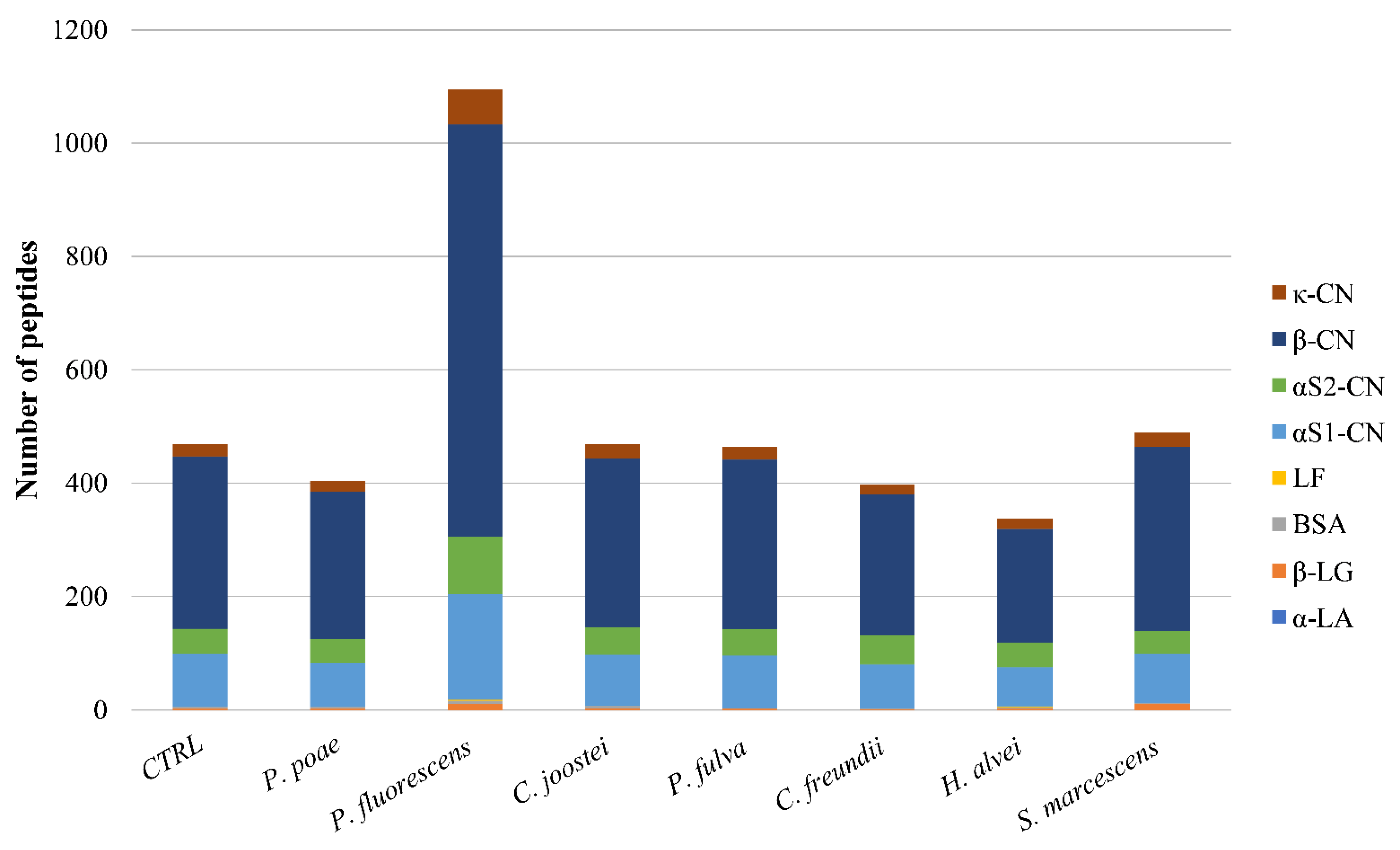
3.3. Peptidomic of Milk Samples Inoculated with the Studied Psychrotrophic Strains
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, B.; Grandison, A.S.; Lewis, M.J. Effect of Seasonal Variation on Some Physical Properties and Heat Stability of Milk Subjected to Ultra-High Temperature and in-Container Sterilisation. Food Chem. 2015, 181, 227–234. [Google Scholar] [CrossRef]
- Raynes, J.; Vincent, D.; Zawadzki, J.; Savin, K.; Mertens, D.; Logan, A.; Williams, R. Investigation of Age Gelation in UHT Milk. Beverages 2018, 4, 95. [Google Scholar] [CrossRef]
- Anema, S.G. Age Gelation, Sedimentation, and Creaming in UHT Milk: A Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 140–166. [Google Scholar] [CrossRef]
- Zhang, C.; Bijl, E.; Svensson, B.; Hettinga, K. The Extracellular Protease AprX from Pseudomonas and Its Spoilage Potential for UHT Milk: A Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 834–852. [Google Scholar] [CrossRef]
- Newstead, D.F.; Paterson, G.; Anema, S.G.; Coker, C.J.; Wewala, A.R. Plasmin Activity in Direct-Steam-Injection UHT-Processed Reconstituted Milk: Effects of Preheat Treatment. Int. Dairy J. 2006, 16, 573–579. [Google Scholar] [CrossRef]
- Ismail, B.; Nielsen, S.S. Invited Review: Plasmin Protease in Milk: Current Knowledge and Relevance to Dairy Industry. J. Dairy Sci. 2010, 93, 4999–5009. [Google Scholar] [CrossRef]
- Datta, N.; Deeth, H.C. Age Gelation of UHT Milk—A Review. Food Bioprod. Process. 2001, 79, 197–210. [Google Scholar] [CrossRef]
- Dufour, D.; Nicodème, M.; Perrin, C.; Driou, A.; Brusseaux, E.; Humbert, G.; Gaillard, J.-L.; Dary, A. Molecular Typing of Industrial Strains of Pseudomonas Spp. Isolated from Milk and Genetical and Biochemical Characterization of an Extracellular Protease Produced by One of Them. Int. J. Food Microbiol. 2008, 125, 188–196. [Google Scholar] [CrossRef]
- Matéos, A.; Guyard-Nicodème, M.; Baglinière, F.; Jardin, J.; Gaucheron, F.; Dary, A.; Humbert, G.; Gaillard, J.-L. Proteolysis of Milk Proteins by AprX, an Extracellular Protease Identified in Pseudomonas LBSA1 Isolated from Bulk Raw Milk, and Implications for the Stability of UHT Milk. Int. Dairy J. 2015, 49, 78–88. [Google Scholar] [CrossRef]
- Stuknytė, M.; Decimo, M.; Colzani, M.; Silvetti, T.; Brasca, M.; Cattaneo, S.; Aldini, G.; De Noni, I. Extracellular Thermostable Proteolytic Activity of the Milk Spoilage Bacterium Pseudomonas Fluorescens PS19 on Bovine Caseins. J. Dairy Sci. 2016, 99, 4188–4195. [Google Scholar] [CrossRef]
- Decimo, M.; Morandi, S.; Silvetti, T.; Brasca, M. Characterization of Gram-Negative Psychrotrophic Bacteria Isolated from Italian Bulk Tank Milk. J. Food Sci. 2014, 79, M2081–M2090. [Google Scholar] [CrossRef]
- Zhang, C.; Bijl, E.; Hettinga, K. Destabilization of UHT Milk by Protease AprX from Pseudomonas Fluorescens and Plasmin. Food Chem. 2018, 263, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Machado, S.G.; da Silva, F.L.; Bazzolli, D.M.S.; Heyndrickx, M.; Costa, P.M. de A.; Vanetti, M.C.D. Pseudomonas Spp. and Serratia Liquefaciens as Predominant Spoilers in Cold Raw Milk. J. Food Sci. 2015, 80, M1842–M1849. [Google Scholar] [CrossRef]
- Machado, S.G.; Baglinière, F.; Marchand, S.; Van Coillie, E.; Vanetti, M.C.D.; De Block, J.; Heyndrickx, M. The Biodiversity of the Microbiota Producing Heat-Resistant Enzymes Responsible for Spoilage in Processed Bovine Milk and Dairy Products. Front. Microbiol. 2017, 8, 302. [Google Scholar] [CrossRef]
- Vithanage, N.R. Mapping the Thermo-Tolerant Proteases in Ultra High Temperature (UHT) Treated Milk Using Molecular Approaches. Ph.D. Thesis, Victoria University, Melbourne, Australia, 2017. [Google Scholar]
- Kaibara, F.; Iiyama, K.; Chieda, Y.; Lee, J.M.; Kusakabe, T.; Yasunaga-Aoki, C.; Shimizu, S. Construction of Serralysin-like Metalloprotease-Deficient Mutants of Serratia Liquefaciens and Their Virulence in the Silkworm, Bombyx mori. J. Insect Biotechnol. Sericology 2012, 81, 55–61. [Google Scholar] [CrossRef]
- Anema, S.G. Storage Stability and Age Gelation of Reconstituted Ultra-High Temperature Skim Milk. Int. Dairy J. 2017, 75, 56–67. [Google Scholar] [CrossRef]
- Stoeckel, M.; Lidolt, M.; Achberger, V.; Glück, C.; Krewinkel, M.; Stressler, T.; von Neubeck, M.; Wenning, M.; Scherer, S.; Fischer, L.; et al. Growth of Pseudomonas Weihenstephanensis, Pseudomonas Proteolytica and Pseudomonas Sp. in Raw Milk: Impact of Residual Heat-Stable Enzyme Activity on Stability of UHT Milk during Shelf-Life. Int. Dairy J. 2016, 59, 20–28. [Google Scholar] [CrossRef]
- Zhang, D.; Li, S.; Palmer, J.; Teh, K.H.; Leow, S.; Flint, S. The Relationship between Numbers of Pseudomonas Bacteria in Milk Used to Manufacture UHT Milk and the Effect on Product Quality. Int. Dairy J. 2020, 105, 104687. [Google Scholar] [CrossRef]
- Marchand, S.; Vandriesche, G.; Coorevits, A.; Coudijzer, K.; De Jonghe, V.; Dewettinck, K.; De Vos, P.; Devreese, B.; Heyndrickx, M.; De Block, J. Heterogeneity of Heat-Resistant Proteases from Milk Pseudomonas Species. Int. J. Food Microbiol. 2009, 133, 68–77. [Google Scholar] [CrossRef] [PubMed]
- ISO 4833-1:2013. Microbiology of the Food Chain–Horizontal Method for the Enumeration of Microorganisms; Part 1: Colony Count at 30 C by the Pour Plate Technique; International Organization for Standardization: Geneva, Switzerland, 2013. [Google Scholar]
- Cattaneo, S.; Pica, V.; Stuknytė, M.; Masotti, F.; Mallardi, D.; Tabasso, C.; Roggero, P.; Noni, I.D. Effect of Protein Fortification on Heat Damage and Occurrence of β-Casomorphins in (Un)Digested Donor Human Milk Intended for Nutrition of Preterm Infants. Food Chem. 2020, 314, 126176. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D.; Russo, F.; Ferrocino, I.; Villani, F. Molecular Identification of Mesophilic and Psychrotrophic Bacteria from Raw Cow’s Milk. Food Microbiol. 2009, 26, 228–231. [Google Scholar] [CrossRef]
- Baglinière, F.; Jardin, J.; Gaucheron, F.; de Carvalho, A.F.; Vanetti, M.C.D. Proteolysis of Casein Micelles by Heat-Stable Protease Secreted by Serratia Liquefaciens Leads to the Destabilisation of UHT Milk during Its Storage. Int. Dairy J. 2017, 68, 38–45. [Google Scholar] [CrossRef]
- Zarei, M.; Yousefvand, A.; Maktabi, S.; Pourmahdi Borujeni, M.; Mohammadpour, H. Identification, Phylogenetic Characterisation and Proteolytic Activity Quantification of High Biofilm-Forming Pseudomonas Fluorescens Group Bacterial Strains Isolated from Cold Raw Milk. Int. Dairy J. 2020, 109, 104787. [Google Scholar] [CrossRef]
- Machado, S.G.; Heyndrickx, M.; De Block, J.; Devreese, B.; Vandenberghe, I.; Vanetti, M.C.D.; Van Coillie, E. Identification and Characterization of a Heat-Resistant Protease from Serratia Liquefaciens Isolated from Brazilian Cold Raw Milk. Int. J. Food Microbiol. 2016, 222, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Brasca, M.; Decimo, M.; Morandi, S.; Machado, S.G.; Bagliniére, F.; Vanetti, M.C.D. Psychrotrophic bacteria. In Microbiology in Dairy Processing; Poltronieri, P., Ed.; John Wiley & Sons Ltd and the Institute of Food Technologists: Chichester, UK, 2017; pp. 37–61. [Google Scholar]
- Koka, R.; Weimer, B.C. Isolation and Characterization of a Protease from Pseudomonas Fluorescens RO98. J. Appl. Microbiol. 2000, 89, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Nicodème, M.; Grill, J.-P.; Humbert, G.; Gaillard, J.-L. Extracellular Protease Activity of Different Pseudomonas Strains: Dependence of Proteolytic Activity on Culture Conditions. J. Appl. Microbiol. 2005, 99, 641–648. [Google Scholar] [CrossRef]
- Hellio, F.C.; Orange, N.; Guespin-Michel, J.F. Growth Temperature Controls the Production of a Single Extracellular Protease by Pseudomonas Fluorescens MF0, in the Presence of Various Inducers. Res. Microbiol. 1993, 144, 617–625. [Google Scholar] [CrossRef]
- Rajmohan, S.; Dodd, C.E.R.; Waites, W.M. Enzymes from Isolates of Pseudomonas Fluorescens Involved in Food Spoilage. J. Appl. Microbiol. 2002, 93, 205–213. [Google Scholar] [CrossRef]
- Meng, L.; Zhang, Y.; Liu, H.; Zhao, S.; Wang, J.; Zheng, N. Characterization of Pseudomonas Spp. and Associated Proteolytic Properties in Raw Milk Stored at Low Temperatures. Front. Microbiol. 2017, 8, 2158. [Google Scholar] [CrossRef]
- Wang, Y.; Han, X.; Chen, X.; Deng, Y. Potential Harmful of Extracellular Proteases Secreted by Pseudomonas Fluorescens W3 on Milk Quality. J. Food Process. Preserv. 2021, 45, e15192. [Google Scholar] [CrossRef]
- Bylund, G. Dairy Processing Handbook; Tetra Pak Processing Systems AB: Lund, Sweden, 1995; pp. 215–232. [Google Scholar]
- Zhang, S.; Li, H.; Uluko, H.; Liu, L.; Pang, X.; Lv, J. Investigation of Protease Production by Pseudomonas Fluorescens BJ-10 and Degradation on Milk Proteins. J. Food Process. Preserv. 2015, 39, 2466–2472. [Google Scholar] [CrossRef]
- D’Incecco, P.; Brasca, M.; Rosi, V.; Morandi, S.; Ferranti, P.; Picariello, G.; Pellegrino, L. Bacterial Proteolysis of Casein Leading to UHT Milk Gelation: An Applicative Study. Food Chem. 2019, 292, 217–226. [Google Scholar] [CrossRef]
- Liu, M.; Wang, H.; Griffiths, M.W. Regulation of Alkaline Metalloprotease Promoter by N-Acyl Homoserine Lactone Quorum Sensing in Pseudomonas Fluorescens: Protease Promoter Regulation by AHLs in P. Fluorescens. J. Appl. Microbiol. 2007, 103, 2174–2184. [Google Scholar] [CrossRef]
- Alves, M.P.; Salgado, R.L.; Eller, M.R.; Dias, R.S.; Oliveira de Paula, S.; Fernandes de Carvalho, A. Temperature Modulates the Production and Activity of a Metalloprotease from Pseudomonas Fluorescens 07A in Milk. J. Dairy Sci. 2018, 101, 992–999. [Google Scholar] [CrossRef]
- Baglinière, F.; Tanguy, G.; Jardin, J.; Matéos, A.; Briard, V.; Rousseau, F.; Robert, B.; Beaucher, E.; Humbert, G.; Dary, A.; et al. Quantitative and Qualitative Variability of the Caseinolytic Potential of Different Strains of Pseudomonas Fluorescens: Implications for the Stability of Casein Micelles of UHT Milks during Their Storage. Food Chem. 2012, 135, 2593–2603. [Google Scholar] [CrossRef] [PubMed]
- Baglinière, F.; Matéos, A.; Tanguy, G.; Jardin, J.; Briard-Bion, V.; Rousseau, F.; Robert, B.; Beaucher, E.; Gaillard, J.L.; Amiel, C.; et al. Proteolysis of Ultra High Temperature-Treated Casein Micelles by AprX Enzyme from Pseudomonas Fluorescens F Induces Their Destabilisation. Int. Dairy J. 2013, 31, 55–61. [Google Scholar] [CrossRef]
- Costa, M.; Gómez, M.F.; Molina, L.H.; Simpson, R.; Romero, A. Purification and characterization of proteases from Pseudomonas fluorescens and their effects on milk proteins. Arch. Latinoam. Nutr. 2002, 52, 160–166. [Google Scholar]
- De Noni, I.; Resmini, P. Identification of Rennet-Whey Solids in “Traditional Butter” by Means of HPLC/ESI-MS of Non-Glycosylated Caseinomacropeptide A. Food Chem. 2005, 93, 65–72. [Google Scholar] [CrossRef]


| Genus | Pseudomonas | Pseudomonas | Chryseobacterium | Pseudomonas | Citrobacter | Hafnia | Serratia |
|---|---|---|---|---|---|---|---|
| Species | poae | fluorescens | joostei | fulva | freundii | alvei | marcescens |
| Strain | LP5 | LPF3 | LPR1 | PS1 | PS37 | PS46 | PS92 |
| Proteolytic activity on milk agar 1 | + | + | + | + | + | - | + |
| aprX gene 2 | + | + | + | - | + | - | - |
| Extracellular thermostable proteases | AprX | AprX | AprX | - | AprX | - | + (Ser1) |
| Genus | Pseudomonas | Pseudomonas | Chryseobacterium | Pseudomonas | Citrobacter | Hafnia | Serratia | |
|---|---|---|---|---|---|---|---|---|
| Species | poae | fluorescens | joostei | fulva | freundii | alvei | marcescens | |
| Strain | LP5 | LPF3 | LPR1 | PS1 | PS37 | PS46 | PS92 | |
| Pasteurized milk 1 | Sedimentation 3 | no | no | no | no | no | no | no |
| Gelation 3 | no | no | no | no | no | no | no | |
| In-batch sterilized milk 2 | Sedimentation 3 | no | no | yes (104 CFU/mL) | no | yes (104 CFU/mL) | no | no |
| Gelation 3 | yes 4 (108 CFU/mL) | yes 4 (108 CFU/mL) | yes 4 (108 CFU/mL) | no | yes 4 (108 CFU/mL) | no | yes 4 (108 CFU/mL) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morandi, S.; Pica, V.; Masotti, F.; Cattaneo, S.; Brasca, M.; De Noni, I.; Silvetti, T. Proteolytic Traits of Psychrotrophic Bacteria Potentially Causative of Sterilized Milk Instability: Genotypic, Phenotypic and Peptidomic Insight. Foods 2021, 10, 934. https://doi.org/10.3390/foods10050934
Morandi S, Pica V, Masotti F, Cattaneo S, Brasca M, De Noni I, Silvetti T. Proteolytic Traits of Psychrotrophic Bacteria Potentially Causative of Sterilized Milk Instability: Genotypic, Phenotypic and Peptidomic Insight. Foods. 2021; 10(5):934. https://doi.org/10.3390/foods10050934
Chicago/Turabian StyleMorandi, Stefano, Valentina Pica, Fabio Masotti, Stefano Cattaneo, Milena Brasca, Ivano De Noni, and Tiziana Silvetti. 2021. "Proteolytic Traits of Psychrotrophic Bacteria Potentially Causative of Sterilized Milk Instability: Genotypic, Phenotypic and Peptidomic Insight" Foods 10, no. 5: 934. https://doi.org/10.3390/foods10050934
APA StyleMorandi, S., Pica, V., Masotti, F., Cattaneo, S., Brasca, M., De Noni, I., & Silvetti, T. (2021). Proteolytic Traits of Psychrotrophic Bacteria Potentially Causative of Sterilized Milk Instability: Genotypic, Phenotypic and Peptidomic Insight. Foods, 10(5), 934. https://doi.org/10.3390/foods10050934