Salivary Biomarkers as Prognostic Tools in Oral Squamous Cell Carcinoma: A Systematic Review of Survival and Progression Outcomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Focused Question
2.2. Eligibility Criteria
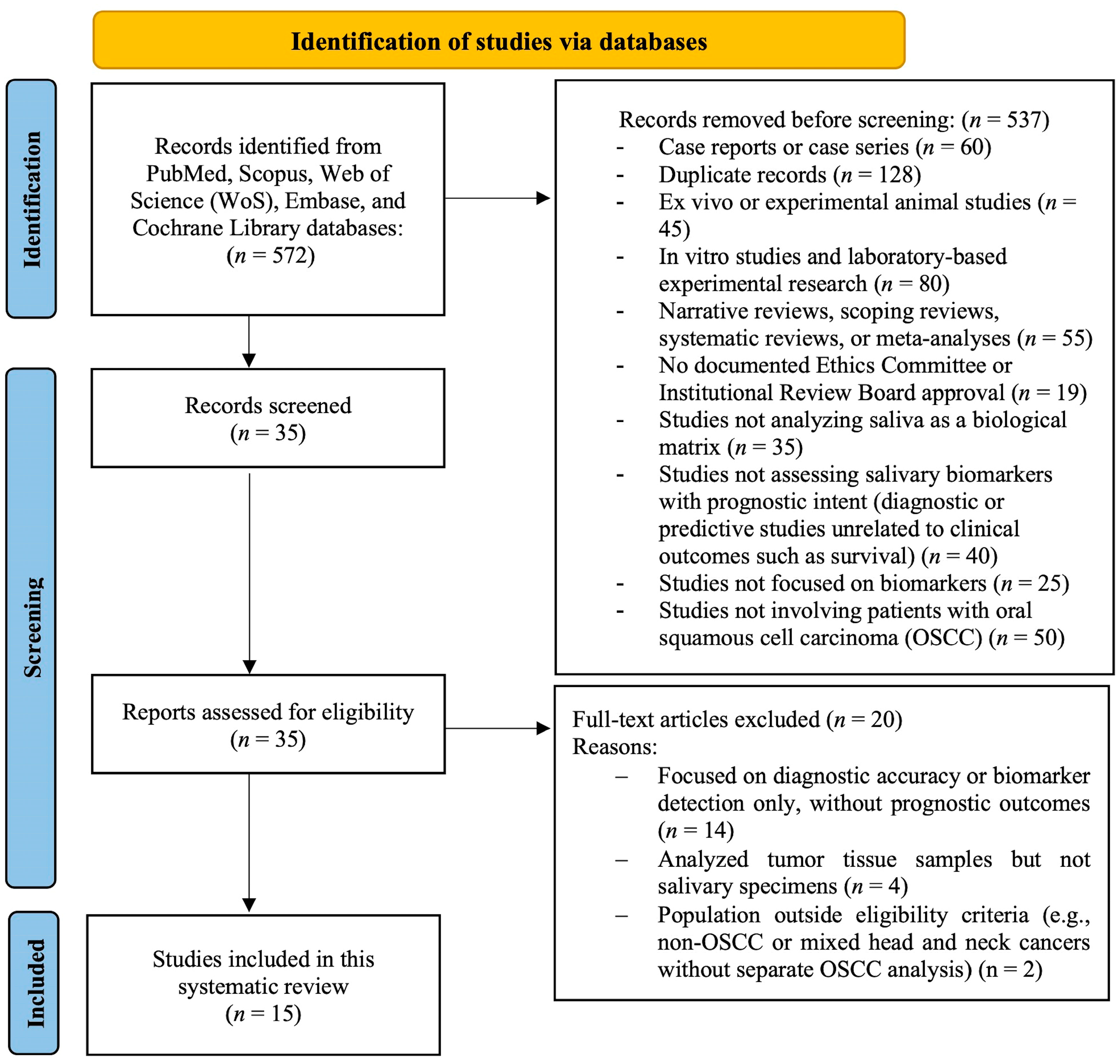
2.3. Search Strategy and Study Selection
2.4. Data Extraction
2.5. Quality Assessment of Included Studies
3. Results
3.1. Risk of Bias Assessment
3.2. Results of Syntheses
3.2.1. Proteomic Biomarkers
3.2.2. Transcriptomic Biomarkers
3.2.3. Metabolomic Biomarkers
3.2.4. Comparative Trends
3.2.5. Prognostic Endpoints Versus Surrogate Measures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AKR1B10 | Aldo-Keto Reductase Family 1 Member B10 |
| CE-TOFMS | Capillary Electrophoresis Time-Of-Flight Mass Spectrometry |
| CK19 | Cytokeratin 19 |
| CST1 | Cystatin SN |
| DFS | Disease-Free Survival |
| ECLIA | Electrochemiluminescence Immunoassay |
| ELISA | Enzyme-Linked Immunosorbent Assay |
| EMT | Epithelial–Mesenchymal Transition |
| ENEPT | Extranodal Extension-Positive Tumors |
| ENsT | Extranodal Stage Tumor |
| ETNMsT | Early Tumor With Nodal And Metastatic Spread |
| ETsT | Early Tumor Stage Tumor |
| HC | Healthy Controls |
| HR | Hazard Ratio |
| IL-10 | Interleukin-10 |
| IL-13 | Interleukin-13 |
| IL-1RA | Interleukin-1 Receptor Antagonist |
| KPNA2 | Karyopherin Subunit Alpha 2 |
| LC-MS/MS | Liquid Chromatography Tandem Mass Spectrometry |
| LTsT | Low Tumor Stage Tumor |
| Mac-2 | Galectin-3 Binding Protein |
| MALDI-TOF-MS | Matrix-Assisted Laser Desorption Ionization Time-Of-Flight Mass Spectrometry |
| miR | Microrna |
| N.R. | Not Reported |
| OS | Overall Survival |
| OSCC | Oral Squamous Cell Carcinoma |
| PDT | Poorly Differentiated Tumor |
| qRT-PCR | Quantitative Reverse Transcription Polymerase Chain Reaction |
| RNA-Seq | RNA Sequencing |
| RT-qPCR | Reverse Transcription Quantitative Polymerase Chain Reaction |
| S100A7 | S100 Calcium-Binding Protein A7 |
| S100A9 | S100 Calcium-Binding Protein A9 |
| S100P | S100 Calcium-Binding Protein P |
| SMVs | Salivary Microvesicles |
| TNF-α | Tumor Necrosis Factor Alpha |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Wang, Z.; Xu, M.; Li, B.; Huang, Z.; Qin, S.; Nice, E.C.; Tang, J.; Huang, C. Oral squamous cell carcinomas: State of the field and emerging directions. Int. J. Oral Sci. 2023, 15, 44. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Yu, H.; Yuan, W.; Wu, J.; Xu, Q.; Mei, N.; Wang, X.; Wang, C. Alveolar bone loss, tooth loss and oral cancer mortality in older patients: A retrospective cohort study. Clin. Interv. Aging 2020, 15, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Menditti, D.; Santagata, M.; Imola, G.; Staglianò, S.; Vitagliano, R.; Boschetti, C.E.; Inchingolo, A.M. Personalized medicine in oral oncology: Imaging methods and biological markers to support diagnosis of oral squamous cell carcinoma (OSCC): A narrative literature review. J. Pers. Med. 2023, 13, 1397. [Google Scholar] [CrossRef]
- Santosh, A.B.; Jones, T.; Harvey, J. A review on oral cancer biomarkers: Understanding the past and learning from the present. J. Cancer Res. Ther. 2016, 12, 486–492. [Google Scholar] [CrossRef]
- Cristaldi, M.; Mauceri, R.; Di Fede, O.; Giuliana, G.; Campisi, G.; Panzarella, V. Salivary biomarkers for oral squamous cell carcinoma diagnosis and follow-up: Current status and perspectives. Front. Physiol. 2019, 10, 1476. [Google Scholar] [CrossRef]
- Khurshid, Z.; Zafar, M.S.; Khan, R.S.; Najeeb, S.; Slowey, P.D.; Rehman, I.U. Role of salivary biomarkers in oral cancer detection. Adv. Clin. Chem. 2018, 86, 23–70. [Google Scholar]
- Khijmatgar, S.; Yong, J.; Rübsamen, N.; Lorusso, F.; Rai, P.; Cenzato, N.; Gaffuri, F.; Del Fabbro, M.; Tartaglia, G.M. Salivary biomarkers for early detection of oral squamous cell carcinoma (OSCC) and head/neck squamous cell carcinoma (HNSCC): A systematic review and network meta-analysis. Jpn. Dent. Sci. Rev. 2024, 60, 32–39. [Google Scholar] [CrossRef]
- Liu, D.; Xin, Z.; Guo, S.; Li, S.; Cheng, J.; Jiang, H. Blood and salivary microRNAs for diagnosis of oral squamous cell carcinoma: A systematic review and meta-analysis. J. Oral Maxillofac. Surg. 2021, 79, 1082.e1–1082.e13. [Google Scholar] [CrossRef]
- Benito-Ramal, E.; Egido-Moreno, S.; González-Navarro, B.; Jané-Salas, E.; Roselló-Llabrés, X.; López-López, J. Role of selected salivary inflammatory cytokines in the diagnosis and prognosis of oral squamous cell carcinoma: A systematic review and meta-analysis. Med. Oral Patol. Oral Cir. Bucal 2023, 28, e474–e486. [Google Scholar] [CrossRef]
- Nguyen, T.T.H.; Sodnom-Ish, B.; Choi, S.W.; Jung, H.I.; Cho, J.; Hwang, I.; Kim, S.M. Salivary biomarkers in oral squamous cell carcinoma. J. Korean Assoc. Oral Maxillofac. Surg. 2020, 46, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.C.; Wang, S.G.; Gao, Z.; Qing, M.F.; Pan, S.; Liu, Y.Y.; Li, F. Emerging salivary biomarkers for early detection of oral squamous cell carcinoma. World J. Clin. Oncol. 2025, 16, 103803. [Google Scholar] [CrossRef] [PubMed]
- Bastías, D.; Maturana, A.; Marín, C.; Martínez, R.; Niklander, S.E. Salivary biomarkers for oral cancer detection: An exploratory systematic review. Int. J. Mol. Sci. 2024, 25, 2634. [Google Scholar] [CrossRef] [PubMed]
- Panneerselvam, K.; Rajkumar, K.; Kumar, S.; Mohan, A.M.; Arockiam, A.S.; Sugimoto, M. Salivary metabolomics in early detection of oral squamous cell carcinoma—A meta-analysis. Expert Rev. Proteom. 2024, 21, 317–332. [Google Scholar] [CrossRef]
- Rengasamy, G.; Kasirajan, H.S.; Veeraraghavan, V.P.; Ramani, P.; Cervino, G.; Minervini, G. Salivary cytokines as a biomarker for diagnosis, prognosis and treatment of oral squamous cell carcinoma: A systematic review. Dent. Med. Probl. 2025, 62, 351–359. [Google Scholar] [CrossRef]
- Ravindran, S.; Ranganathan, S.; R, K.; J, N.; A, S.; Kannan, S.K.; K, D.P.; Marri, J.; K, R. The role of molecular biomarkers in the diagnosis, prognosis, and treatment stratification of oral squamous cell carcinoma: A comprehensive review. J. Liq. Biopsy 2025, 7, 100285. [Google Scholar] [CrossRef]
- Qin, Y.; Dong, X.; Li, B. Salivary miRNAs and cytokines associated with diagnosis and prognosis of oral squamous cell carcinoma. Front. Cell Dev. Biol. 2025, 13, 1531016. [Google Scholar] [CrossRef]
- Aromataris, E.; Lockwood, C.; Porritt, K.; Pilla, B.; Jordan, Z. Ed. JBI Manual for Evidence Synthesis. JBI. 2024. Available online: https://synthesismanual.jbi.global (accessed on 31 August 2025).
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Hayden, J.A.; van der Windt, D.A.; Cartwright, J.L.; Côté, P.; Bombardier, C. Assessing bias in studies of prognostic factors. Ann. Intern. Med. 2013, 158, 280–286. [Google Scholar] [CrossRef]
- Brundha, M.P.; Raveendran, S.R.; Rajeshkar, N. Salivary tumour necrosis factor-alpha and receptor for advanced glycation end products as prognostic and predictive markers for recurrence in oral squamous cell carcinoma—A pilot study. Eur. J. Clin. Exp. Med. 2023, 21, 36–43. [Google Scholar] [CrossRef]
- Gai, C.; Camussi, F.; Broccoletti, R.; Gambino, A.; Cabras, M.; Molinaro, L.; Carossa, S.; Camussi, G.; Arduino, P.G. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer 2018, 18, 439. [Google Scholar] [CrossRef]
- Gallo, C.; Ciavarella, D.; Santarelli, A.; Ranieri, E.; Colella, G.; Lo Muzio, L.; Lo Russo, L. Potential salivary proteomic markers of oral squamous cell carcinoma. Cancer Genom. Proteom. 2016, 13, 55–61. [Google Scholar]
- Gonçalves, A.S.; Arantes, D.A.; Bernardes, V.F.; Jaeger, F.; Silva, J.M.; Silva, T.A.; Aguiar, M.C.F.; Batista, A.C. Immunosuppressive mediators of oral squamous cell carcinoma in tumour samples and saliva. Hum. Immunol. 2015, 76, 52–58. [Google Scholar] [CrossRef]
- Honarmand, M.H.; Farhad-Mollashahi, L.; Nakhaee, A.; Nehi, M. Salivary levels of ErbB2 and CEA in oral squamous cell carcinoma patients. Asian Pac. J. Cancer Prev. 2016, 17 (Suppl. S3), 77–80. [Google Scholar] [CrossRef]
- Ishikawa, S.; Ishizawa, K.; Tanaka, A.; Kimura, H.; Kitabatake, K.; Sugano, A.; Edamatsu, K.; Ueda, S.; Iino, M. Identification of salivary proteomic biomarkers for oral cancer screening. Vivo 2021, 35, 541–547. [Google Scholar] [CrossRef]
- Javaraiah, R.K.; David, C.M.; Namitha, J.; Tiwari, R.; Benakanal, P. Evaluation of salivary lactate dehydrogenase as a prognostic biomarker in tobacco users with and without potentially malignant disorders of the oral cavity. South Asian J. Cancer 2020, 9, 93–98. [Google Scholar] [CrossRef]
- Jayarajkumar, S.; Ramamoorthi, R.; Muniapillai, S.; Gopalakrishnan, S.; Jayaseelan, V.P. Assessment of salivary levels of ErbB2 in oral squamous cell carcinoma. J. Oral Maxillofac. Pathol. 2023, 27, 777. [Google Scholar] [CrossRef]
- Kravets, O.; Burtyn, O.; Borikun, T.; Rossylna, O. The study of prognostic value of microRNAs (miR-10b and -155) and CDKN2A/P16INK4A in oral squamous cell carcinoma. Exp. Oncol. 2023, 45, 187–194. [Google Scholar] [CrossRef]
- Kumari, P.; Syed, S.A.; Wahid, M.; Qureshi, M.A.; Kumar, R. Expression of miR-31 in saliva-liquid biopsy in patients with oral squamous cell carcinoma. J. Taibah Univ. Med. Sci. 2021, 16, 733–739. [Google Scholar] [CrossRef]
- Mohamed, N.; Litlekalsøy, J.; Ahmed, I.A.; Martinsen, E.M.H.; Furriol, J.; Javier-Lopez, R.; Elsheikh, M.; Gaafar, N.M.; Morgado, L.; Mundra, S.; et al. Analysis of salivary mycobiome in a cohort of oral squamous cell carcinoma patients from Sudan identifies higher salivary carriage of Malassezia as an independent and favorable predictor of overall survival. Front. Cell. Infect. Microbiol. 2021, 11, 673465. [Google Scholar] [CrossRef]
- Nandakumar, A.; Nataraj, P.; James, A.; Krishnan, R.; M, K.M. Estimation of salivary 8-hydroxydeoxyguanosine (8-OHdG) as a potential biomarker in assessing progression towards malignancy: A case-control study. Asian Pac. J. Cancer Prev. 2020, 21, 2325–2329. [Google Scholar] [CrossRef]
- Oka, R.; Nakashiro, K.; Goda, H.; Iwamoto, K.; Tokuzen, N.; Hamakawa, H. Annexin A8 is a novel molecular marker for detecting lymph node metastasis in oral squamous cell carcinoma. Oncotarget 2016, 7, 4882–4889. [Google Scholar] [CrossRef]
- Sasahira, T.; Kurihara, M.; Nishiguchi, Y.; Nakashima, C.; Kirita, T.; Kuniyasu, H. Pancreatic adenocarcinoma up-regulated factor has oncogenic functions in oral squamous cell carcinoma. Histopathology 2017, 70, 539–548. [Google Scholar] [CrossRef]
- Sawant, S.; Gokulan, R.; Dongre, H.; Vaidya, M.; Chaukar, D.; Prabhash, K.; Ingle, A.; Joshi, S.; Dange, P.; Joshi, S.; et al. Prognostic role of Oct4, CD44 and c-Myc in radio-chemo-resistant oral cancer patients and their tumourigenic potential in immunodeficient mice. Clin. Oral Investig. 2016, 20, 43–56. [Google Scholar] [CrossRef]
- Scholtz, B.; Horváth, J.; Tar, I.; Kiss, C.; Márton, I.J. Salivary miR-31-5p, miR-345-3p, and miR-424-3p are reliable biomarkers in patients with oral squamous cell carcinoma. Pathogens 2022, 11, 229. [Google Scholar] [CrossRef]
- Tavakoli, F.; Ghavimi, M.A.; Fakhrzadeh, V.; Abdolzadeh, D.; Afshari, A.; Eslami, H. Evaluation of salivary transferrin in patients with oral squamous cell carcinoma. Clin. Exp. Dent. Res. 2024, 10, e809. [Google Scholar] [CrossRef]
- Ueda, S.; Goto, M.; Hashimoto, K.; Imazawa, M.; Takahashi, M.; Oh-Iwa, I.; Shimozato, K.; Nagao, T.; Nomoto, S. Salivary CPLANE1 levels as a biomarker of oral squamous cell carcinoma. Anticancer Res. 2021, 41, 765–772. [Google Scholar] [CrossRef]
- Vimal, J.; George, N.A.; Kumar, R.R.; Kattoor, J.; Kannan, S. Identification of salivary metabolic biomarker signatures for oral tongue squamous cell carcinoma. Arch. Oral Biol. 2023, 155, 105780. [Google Scholar] [CrossRef]
- Wang, K.; Shen, Y.; Xu, J.; Li, Z.; Liu, Y.; Yu, C.; Peng, L.; Zheng, J.; Zeng, Y. Evaluation of synuclein-γ levels by novel monoclonal antibody in saliva and cancer tissues from oral squamous cell carcinoma patients. Neoplasma 2020, 67, 707–713. [Google Scholar] [CrossRef]
- Aziz, S.; Ahmed, S.S.; Ali, A.; Khan, F.A.; Zulfiqar, G.; Iqbal, J.; Khan, A.A.; Shoaib, M. Salivary immunosuppressive cytokines IL-10 and IL-13 are significantly elevated in oral squamous cell carcinoma patients. Cancer Investig. 2015, 33, 318–328. [Google Scholar] [CrossRef]
- Bu, J.; Bu, X.; Liu, B.; Chen, F.; Chen, P. Increased expression of tissue/salivary transgelin mRNA predicts poor prognosis in patients with oral squamous cell carcinoma (OSCC). Med. Sci. Monit. 2015, 21, 2275–2281. [Google Scholar] [PubMed]
- Winck, F.V.; Prado Ribeiro, A.C.; Ramos Domingues, R.; Ling, L.Y.; Riaño-Pachón, D.M.; Rivera, C.; Brandão, T.B.; Gouvea, A.F.; Santos-Silva, A.R.; Coletta, R.D.; et al. Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. Sci. Rep. 2015, 5, 16305. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, R.; Urs, A.B.; Chakravarti, A.; Kumar, S.; Gupta, V.K.; Mahajan, B. Correlation of Cyfra 21-1 levels in saliva and serum with CK19 mRNA expression in oral squamous cell carcinoma. Tumour Biol. 2016, 37, 9263–9271. [Google Scholar] [CrossRef]
- Pathiyil, V.; D’Cruz, A.M. Salivary lactate dehydrogenase as a prognostic marker in oral squamous cell carcinoma patients following surgical therapy. J. Exp. Ther. Oncol. 2017, 11, 133–137. [Google Scholar]
- Ko, H.H.; Peng, H.H.; Cheng, S.J.; Kuo, M.Y. Increased salivary AKR1B10 level: Association with progression and poor prognosis of oral squamous cell carcinoma. Head Neck 2018, 40, 2642–2647. [Google Scholar] [CrossRef]
- Patel, A.; Patel, S.; Patel, P.; Mandlik, D.; Patel, K.; Tanavde, V. Salivary exosomal miRNA-1307-5p predicts disease aggressiveness and poor prognosis in oral squamous cell carcinoma patients. Int. J. Mol. Sci. 2022, 23, 10639. [Google Scholar] [CrossRef]
- Shabbir, A.; Waheed, H.; Ahmed, S.; Shaikh, S.S.; Farooqui, W.A. Association of salivary Cathepsin B in different histological grades among patients presenting with oral squamous cell carcinoma. BMC Oral Health 2022, 22, 63. [Google Scholar] [CrossRef]
- Patel, A.; Patel, P.; Mandlik, D.; Patel, K.; Malaviya, P.; Johar, K.; Swamy, K.B.; Patel, S.; Tanavde, V. A novel 3-miRNA network regulates tumour progression in oral squamous cell carcinoma. Biomark. Res. 2023, 11, 64. [Google Scholar] [CrossRef]
- Premkumar, A.; Walia, C.; Roy, S. The comparative evaluation of salivary survivin levels between different grades of oral squamous cell carcinoma. J. Biomed. Biotechnol. Res. 2023, 7, 293–297. [Google Scholar]
- Wang, C.I.; Yu, C.J.; Huang, Y.; Yi, J.S.; Cheng, H.W.; Kao, H.K.; Lao, W.W.; Chang, K. Association of overexpressed karyopherin alpha 2 with poor survival and its contribution to interleukin-1β-induced matrix metalloproteinase expression in oral cancer. Head Neck 2018, 40, 1719–1733. [Google Scholar] [CrossRef]
- Zhong, W.Q.; Ren, J.G.; Xiong, X.P.; Man, Q.W.; Zhang, W.; Gao, L.; Li, C.; Liu, B.; Sun, Z.; Jia, J.; et al. Increased salivary microvesicles are associated with the prognosis of patients with oral squamous cell carcinoma. J. Cell. Mol. Med. 2019, 23, 4054–4062. [Google Scholar] [CrossRef]
- Romani, C.; Salviato, E.; Paderno, A.; Zanotti, L.; Ravaggi, A.; Deganello, A.; Berretti, G.; Gualtieri, T.; Marchini, S.; D’Incalci, M.; et al. Genome-wide study of salivary miRNAs identifies miR-423-5p as promising diagnostic and prognostic biomarker in oral squamous cell carcinoma. Theranostics 2021, 11, 2987–2999. [Google Scholar] [CrossRef]
- Ishikawa, S.; Sugimoto, M.; Konta, T.; Kitabatake, K.; Ueda, S.; Edamatsu, K.; Okuyama, N.; Yusa, K.; Iino, M. Salivary metabolomics for prognosis of oral squamous cell carcinoma. Front. Oncol. 2022, 11, 789248. [Google Scholar] [CrossRef]
- Hema Shree, K.; Gayathri, R.; Veeraraghavan, V.P.; Ramani, P.; Ramadoss, R.; Yuwanati, M. Gold nanoparticle enhanced TNFα antibody interface using saliva for predicting prognosis in OSCC. Arch. Oral Biol. 2025, 173, 106196. [Google Scholar] [CrossRef] [PubMed]
| Domain | Description |
|---|---|
| P—Population | Adults diagnosed with OSCC, regardless of tumor site, stage, or histologic grading |
| I—Index Prognostic Factors | Salivary biomarkers of proteomic, transcriptomic, and metabolomic nature, assessed for prognostic significance |
| C—Comparator/Covariates | Healthy individuals or OSCC patients with early-stage disease (stage I–II), low-grade tumors (grade I), or absence of lymph node and distant metastasis. Reported covariates included age, gender, tobacco or alcohol use, TNM stage, and histological grade |
| O—Outcomes of Interest | Prognostic outcomes included tumor stage at diagnosis, histological grade, presence of lymph node involvement, presence of distant metastases, disease progression, local recurrence, OS, and DFS |
| T—Timing (Prognostication Time and Follow-up Period) | At the time of OSCC diagnosis or before initiation of treatment. When available, outcome data were reported during treatment, immediately after treatment completion, at 6 months, 1 year, or at the longest follow-up time specified by the study authors |
| S—Setting | Clinical studies conducted in hospital-based or university-based research settings, without restriction by geographic location |
| Authors, Year, and Country of Publication | N° of Patients and Women (%) | N° of Controls and Women (%) | Mean Age ± SD and/or Range (Years) of Patients | TNM Staging and/or Grading (N° of Patients) | Inclusion (IC) and Exclusion Criteria (EC) |
|---|---|---|---|---|---|
| Aziz et al., 2015, Pakistan [41] | 30; 6 (20%) | 33; 6 (18.2%) | 50.33 ± 11.77 | Grade I: 20 Grade II: 5 Grade III: 5 | IC: histopathologically confirmed untreated OSCC; healthy, age- and gender-matched controls; EC: any oral or systemic illness (including periodontal disease). |
| Bu et al., 2015, Cina [42] | 78; 30 (38.5%) | N.R. | N.R. | Stage I-II: 46 Stage III-IV: 32 | IC: pathologically confirmed OSCC, no prior treatment before sample collection; EC: N.R. |
| Winck et al., 2015, Brasil [43] | 24; 3 (12.5%) | 10; N.R. | N.R. | Grade I: 5 Grade II: 8 Grade III: 5 Grade N.R. for 6 patients | IC: N.R. EC: N.R. |
| Malhotra et al., 2016, India [44] | 50; N.R. | 50; N.R. | N.R. | Grade I: 27 Grade II: 23 | IC: histologically diagnosed OSCC and healthy controls; EC: history of drugs with anticholinergic effect. |
| Pathiyil et al., 2017, India [45] | 20; N.R. | 20; N.R. | N.R. | N.R. | IC: untreated cases of OSCC, healthy controls; EC: patients with previous history of malignancy and treatments and with systemic diseases. |
| Ko et al., 2018, Taiwan [46] | 86; 10 (11.6%) | 35; N.R. | N.R. | Stage I-II: 70 Stage III-IV: 16 | IC: untreated OSCC patients and healthy controls; EC: patients with prior malignancy, metastasis, recurrence, or preoperative treatment. |
| Patel et al., 2022, India [47] | 21; 0 (0%) | 15; 0 (%) | 48 ± 8.3 | Stage I/II: 8 Stage III/IV: 13 | IC: diagnosis of OSCC, history of smokeless tobacco, healthy controls; EC: benign leukoplakia, HIV, HBsAg, HPV, COVID-19 positivity, pediatric patients, samples needed for further histopathological diagnosis, disease-oriented complications |
| Shabbir et al., 2022, Pakistan [48] | 60; 17 (28.3%) | 20; 8 (40%) | 18–65 | Grade I: 20 Grade II: 20 Grade III: 20 | IC: OSCC cases and healthy controls; EC: patients with underlying systemic illness, controls with a history of smoking. |
| Patel et al., 2023, India [49] | 23; N.R. | 21; N.R. | N.R. | N.R. | IC: untreated cases of OSCC, healthy controls; EC: leucoplakia, HIV, HBsAg, HPV, COVID-19 infected patients, pediatric patients. |
| Premkumar et al., 2023, India [50] | 38; N.R. | 38; N.R. | 25–80 | Grade I: 18 Grade II: 10 Grade III: 10 | IC: 25–80 years aged patients diagnosed with OSCC and healthy controls; EC: patients with previous treatments and malignancy history, patients with autoimmune diseases. |
| Wang et al., 2018, Taiwan [51] | 116; N.R. | 65; N.R. | N.R. | Early stage: 35 Late stage: 81 | IC: OSCC patients who did not receive radiation or chemotherapy and healthy controls; EC: patients with occurrence of synchronous or metachronous primary cancer and who failed to receive the adjuvant therapy when indicated. |
| Zhong et al., 2019, China [52] | 65; N.R. | 42; N.R. | N.R. | N.R. | IC: OSCC patients free from any other systemic illness and healthy controls; EC: N.R. |
| Romani et al., 2021, Italy [53] | 89; 27 (30.3%) | 58; 20 (34.5%) | 24–91 | Grade I: 19 Grade II: 49 Grade III: 20 Grade N.R. for 1 patient. | IC: primary OSCC cases and healthy controls; EC: N.R. |
| Ishikawa et al., 2022, Japan [54] | 72; 34 (47.2%) | N.R. | 23–94 | Stage 0: 3 (4.2%) Stage I: 24 (33.3%) Stage II: 14 (19.4%) Stage III: 13 (18.1%) Stage IV: 18 (25.0%) | IC: diagnosed OSCC, treated with curative intent (surgery or chemoradiotherapy); informed opt-out consent; EC: Non-curative treatment (palliative/symptomatic); declined participation (none did). |
| Hema Shree et al., 2025, India [55] | 40; 16 (40%) | 10; 5 (50%) | 50–60 | N.R. | IC: OSCC patients undergoing longitudinal salivary sampling for TNF-α analysis; EC: patients with recent food intake, oral hygiene procedures, or interfering oral conditions. |
| Authors, Year, and Country of Publication | Study Design and Aim | Sample Analysis | Biomarkers | OSCC vs. Controls | Prognostic Association | Disease-Free Survival | Overall Survival |
|---|---|---|---|---|---|---|---|
| Aziz et al., 2015, Pakistan [41] | Cross-sectional study aimed to evaluate the importance of immunosuppressive cytokines as prospective salivary biomarkers of OSCC | xMAP | IL-4, IL-10, IL-13, IL-1RA (proteomic) | IL-10 ↑ (p = 0.004), IL-13 ↑ (p = 0.010), IL-1RA ↑ in PDT vs. MDT, WDT, and controls (p < 0.002) | Elevated salivary levels of IL-4, IL-10, IL-13, and IL-1RA in OSCC patients vs. controls; IL-1RA significantly higher in PDT vs. MDT and WDT, suggesting involvement of these cytokines in tumor progression and immune evasion | N.R. | N.R. |
| Bu et al., 2015, Cina [42] | Cross-sectional study to assess salivary transgelin mRNA in OSCC | Real-time PCR | Transgelin mRNA (transcriptomic) | ↑ in OSCC vs. HC (p < 0.01); ↑ in ETsT, ENsT, ETNMsT, PDT, ENEPT (p < 0.05) | Higher salivary transgelin mRNA levels associated with advanced tumor stage (ETsT), nodal stage (ENsT), metastatic stage (ETNMsT), and PDT, indicating correlation with aggressive disease phenotype | N.R. | N.R. |
| Winck et al., 2015, Brasil [43] | Cross-sectional comparative proteomic profiling of whole saliva from OSCC patients and healthy controls to identify diagnostic and prognostic markers | Shotgun proteomics (LC-MS/MS) | 14-3-3 protein zeta/delta, Annexin A1, S100A9 (proteomic) | 14-3-3 protein ↑ in OSCC (p < 0.05); Annexin A1 and S100A9 ↓ in OSCC (p < 0.01) | 14-3-3 protein associated with tumor cell proliferation and survival; Annexin A1 downregulation linked to loss of anti-inflammatory and pro-apoptotic control; S100A9 reduction may indicate impaired immune response—together suggesting potential markers of poor prognosis | N.R. | N.R. |
| Malhotra et al., 2016, India [44] | Cross-sectional study to evaluate Cyfra 21-1 levels and their correlation with tissue CK19 mRNA | ECLIA | Cyfra 21-1 (proteomic) | ↑ in OSCC vs. HC (p < 0.001); ↑ in grade II vs. grade I; ↑ in recurrence cases (p < 0.005) | Higher salivary Cyfra 21-1 levels correlated with increased tumor grade (grade II > I), recurrence, and CK19 mRNA tissue expression, supporting its role in predicting recurrence and histological aggressiveness | N.R. | N.R. |
| Pathiyil et al., 2017, India [45] | Cross-sectional study investigating the utility of salivary LDH as a prognostic marker in OSCC | Spectrophotometry at 340 nm | LDH (metabolomic) | LDH significantly ↑ in OSCC vs. HC (457.06 ± 88.93 IU/L vs. 178.35 ± 120.54 IU/L; p < 0.001) | Salivary LDH significantly ↑ one month post-surgery vs. preoperative levels (p < 0.001); potential marker of disease burden and recurrence | N.R. | N.R. |
| Ko et al., 2018, Taiwan [46] | Cross-sectional study to assess salivary AKR1B10 as screening and prognostic marker | ELISA | AKR1B10 (proteomic) | ↑ in OSCC vs. HC (p < 0.001); ↑ in ETsT and ETNMsT (p < 0.05) | AKR1B10 levels increased in OSCC vs. controls and in advanced tumor (ETsT) and nodal stages (ETNMsT); levels > 646 pg/mL in early-stage tumors (LTsT) predicted shorter survival, supporting its prognostic significance | N.R. | Lower OS associated with AKR1B10 levels > 646 pg/mL in early-stage OSCC |
| Patel et al., 2022, India [47] | Cross-sectional study to identify OSCC-specific salivary exosomal miRNAs | TEM, RNA extraction, miRNA sequencing | miR-1307-5p (transcriptomic) | ↑ in OSCC vs. HC (493 vs. 23 reads); ↑ in ETNMsT, ENsT, chemoresistant cases (p < 0.05) | Overexpression of miR-1307-5p in OSCC saliva correlated with advanced nodal stage (ETNMsT), extranodal spread (ENsT), and chemoresistance, indicating prognostic value in aggressive and treatment-resistant OSCC | N.R. | N.R. |
| Shabbir et al., 2022, Pakistan [48] | Cross-sectional study evaluating salivary Cathepsin B levels among patients with different OSCC histological grades | ELISA | Cathepsin B (proteomic) | Salivary Cathepsin B significantly ↑ in OSCC vs. HC (detected in 70% OSCC vs. 15% controls; p < 0.001); ↑ with increasing tumor burden | Cathepsin B levels reflect histological grade and tumor progression; potential for prognostic stratification | N.R. | N.R. |
| Patel et al., 2023, India [49] | Cross-sectional study aimed to identify differentially expressed miRNAs in OSCC and validate their diagnostic/prognostic potential in saliva | Small RNA-Seq, qRT-PCR, transcriptome analysis | miR-140-5p, miR-143-5p, miR-145-5p (transcriptomic) | miRNAs differentially expressed in OSCC vs. HC (p ≤ 0.05); miR-140-5p, miR-143-5p, miR-145-5p significantly ↓ in OSCC (validated via qPCR; p < 0.01). | Downregulation of the 3-miRNA panel associated with increased tumor aggressiveness, EMT, and poor prognosis | N.R. | N.R. |
| Premkumar et al., 2023, India [50] | Comparative cross-sectional study investigating survivin expression in salivary secretions of OSCC patients with different histological grades | ELISA | Survivin (proteomic) | Mean salivary survivin significantly ↑ in OSCC vs. HC (9.51 ± 2.9 vs. 2.69 ± 1.2 pg/mL; p < 0.001); ↑ with increasing tumor grade (p < 0.001). | Elevated survivin levels strongly correlated with tumor grade; higher in poorly differentiated OSCC, suggesting worse prognosis | N.R. | N.R. |
| Wang et al., 2018, Taiwan [51] | Cohort proteomic study assessing salivary protein biomarkers for early OSCC detection using 2D-DIGE and MS | 2D-DIGE, MALDI-TOF-MS, Western blot | Mac-2 binding protein, S100A7, S100P, CST1, KPNA2 (proteomic) | Multiple proteins significantly ↑ in OSCC vs. HC (fold change > 2; p < 0.05); KPNA2 significantly ↑ in OSCC (p < 0.01) | CST1, S100P, and KPNA2 overexpression associated with proliferation, nuclear transport dysregulation, and poor prognosis | N.R. | N.R. |
| Zhong et al., 2019, China [52] | Cohort study evaluating the association between salivary microvesicles (SMVs) and clinicopathological features and prognosis in OSCC. | Flow cytometry, TEM, immunohistochemistry | Salivary SMVs (proteomic) | SMVs significantly ↑ in OSCC vs. HC and oral ulcer (p < 0.001); ↑ in OSCC with lymph node metastasis and advanced clinical stage (p < 0.01) | Lower apoptotic/non-apoptotic SMV ratio associated with higher pathological grade and reduced overall survival (p < 0.01) | N.R. | Overall survival significantly reduced in patients with a low apoptotic/non-apoptotic SMV ratio (p = 0.001). |
| Romani et al., 2021, Italy [53] | Cohort genome-wide salivary miRNA study profiling to assess diagnostic and prognostic potential in OSCC patients | Microarray, RT-qPCR | miR-423-5p, miR-106b-5p, miR-193b-3p (transcriptomic) | Multiple miRNAs differentially expressed in OSCC vs. HC (p ≤ 0.05); miR-423-5p ↑ in OSCC, ↓ post-surgery (p < 0.001). | High salivary miR-423-5p is an independent predictor of shorter DFS in multivariate analysis (p < 0.05) | High miR-423-5p was an independent predictor of shorter DFS in multivariate analysis (HR = 2.58; 95% CI: 1.42–4.71; p = 0.002). | N.R. |
| Ishikawa et al., 2022, Japan [54] | Prospective observational study aiming to identify salivary metabolomic biomarkers with prognostic value in OSCC | CE-TOFMS | 3-methylhistidine, 5-hydroxylysine, N-acetylglucosamine, proline, creatinine (metabolomic) | Not evaluated (no healthy control group included) | 3-methylhistidine and 5-hydroxylysine associated with OS in training set; only 3-methylhistidine retained significance in validation set | N-acetylglucosamine significant for DFS in training set, not confirmed in validation set | 3-methylhistidine associated with reduced OS in both training and validation sets |
| Hema Shree et al., 2025, India [55] | Cohort study aimed at assessing the efficacy of a gold nanoparticle-enhanced ELISA for the detection of TNF-α in saliva of OSCC patients, and evaluating its prognostic value for survival | ELISA and gold nanoparticle-enhanced ELISA | TNF-α (proteomic) | Mean salivary Tumor Necrosis Factor alpha (TNF-α) was significantly ↑ in OSCC patients compared to healthy controls, both with conventional ELISA (47.52 ± 20.23 vs. 10.13 ± 3.07 pg/mL; p < 0.001) and gold nano-enhanced ELISA (57.63 ± 24.99 vs. 12.07 ± 3.66 pg/mL; p < 0.001). TNF-α levels were also found to ↑ progressively with increasing tumor grade and clinical stage (p < 0.001). | Although TNF-α levels were higher in more advanced stages and tumor grades, the Cox proportional hazards model showed no significant association between TNF-α levels and overall survival (p = 0.653). Therefore, its prognostic value remains uncertain in this dataset. | N.R. | Kaplan-Meier analysis showed variable survival trends depending on the method and TNF-α levels. The gold nano-enhanced method suggested better survival in patients with TNF-α above the cutoff after 9 months, but no statistically significant differences were confirmed (log-rank p = 0.78). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pellegrini, M.; Pascadopoli, M.; Faretta, M.R.; Nobili, A.; Martínez, C.P.-A.; Spadari, F.; Scribante, A. Salivary Biomarkers as Prognostic Tools in Oral Squamous Cell Carcinoma: A Systematic Review of Survival and Progression Outcomes. Dent. J. 2025, 13, 479. https://doi.org/10.3390/dj13100479
Pellegrini M, Pascadopoli M, Faretta MR, Nobili A, Martínez CP-A, Spadari F, Scribante A. Salivary Biomarkers as Prognostic Tools in Oral Squamous Cell Carcinoma: A Systematic Review of Survival and Progression Outcomes. Dentistry Journal. 2025; 13(10):479. https://doi.org/10.3390/dj13100479
Chicago/Turabian StylePellegrini, Matteo, Maurizio Pascadopoli, Mario Romolo Faretta, Alessandro Nobili, Carlos Pérez-Albacete Martínez, Francesco Spadari, and Andrea Scribante. 2025. "Salivary Biomarkers as Prognostic Tools in Oral Squamous Cell Carcinoma: A Systematic Review of Survival and Progression Outcomes" Dentistry Journal 13, no. 10: 479. https://doi.org/10.3390/dj13100479
APA StylePellegrini, M., Pascadopoli, M., Faretta, M. R., Nobili, A., Martínez, C. P.-A., Spadari, F., & Scribante, A. (2025). Salivary Biomarkers as Prognostic Tools in Oral Squamous Cell Carcinoma: A Systematic Review of Survival and Progression Outcomes. Dentistry Journal, 13(10), 479. https://doi.org/10.3390/dj13100479









