Brazilian Multiethnic Association Study of Genetic Variant Interactions among FOS, CASP8, MMP2 and CRISPLD2 in the Risk of Nonsyndromic Cleft Lip with or without Cleft Palate
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Samples
2.3. Genotyping and Assessment of Genomic Ancestry
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dixon, M.J.; Marazita, M.L.; Beaty, T.H.; Murray, J.C. Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 2011, 12, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Mossey, P.A.; Little, J.; Munger, R.G.; Dixon, M.J.; Shaw, W.C. Cleft lip and palate. Lancet 2009, 374, 1773–1785. [Google Scholar] [CrossRef] [PubMed]
- Beaty, T.H.; Marazita, M.L.; Leslie, E.J. Genetic factors influencing risk to orofacial clefts: Today’s challenges and tomorrow’s opportunities. F1000Research 2016, 5, 2800. [Google Scholar] [CrossRef] [PubMed]
- Martelli-Junior, H.; Porto, L.V.; Martelli, D.R.; Bonan, P.R.; Freitas, A.B.; Della Coletta, R. Prevalence of nonsyndromic oral clefts in a reference hospital in the state of Minas Gerais, Brazil, between 2000–2005. Braz. Oral Res. 2007, 21, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, K.; Sena, M.F.; Roncalli, A.G.; Ferreira, M.A. Prevalence of orofacial clefts and social factors in Brazil. Braz. Oral Res. 2009, 23, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Alade, A.; Awotoye, W.; Butali, A. Genetic and epigenetic studies in non-syndromic oral clefts. Oral Dis. 2022, 28, 1339–1350. [Google Scholar] [CrossRef]
- Chiquet, B.T.; Lidral, A.C.; Stal, S.; Mulliken, J.B.; Moreno, L.M.; Arcos-Burgos, M.; Valencia-Ramirez, C.; Blanton, S.H.; Hecht, J.T. CRISPLD2: A novel NSCLP candidate gene. Hum. Mol. Genet. 2007, 16, 2241–2248. [Google Scholar] [CrossRef]
- Shen, X.; Liu, R.M.; Yang, L.; Wu, H.; Li, P.Q.; Liang, Y.L.; Xie, X.D.; Yao, T.; Zhang, T.T.; Yu, M. The CRISPLD2 gene is involved in cleft lip and/or cleft palate in a Chinese population. Birth Defects Res. A Clin. Mol. Teratol. 2011, 91, 918–924. [Google Scholar] [CrossRef]
- Ge, X.; Shi, Q.M.; Ding, Z.; Ju, Q.; Wang, H.; Wang, Q.; Li, M.X.; Chen, G.; Wang, H.X.; Xu, L.C. Association Between CRISPLD2 Polymorphisms and the Risk of Nonsyndromic Clefts of the Lip and/or Palate: A Meta-analysis. Cleft Palate Craniofac. J. 2018, 55, 328–334. [Google Scholar] [CrossRef]
- Yuan, Q.; Chiquet, B.T.; Devault, L.; Warman, M.L.; Nakamura, Y.; Swindell, E.C.; Hecht, J.T. Craniofacial abnormalities result from knock down of nonsyndromic clefting gene, crispld2, in zebrafish. Genesis 2012, 50, 871–881. [Google Scholar] [CrossRef]
- Chiquet, B.T.; Yuan, Q.; Swindell, E.C.; Maili, L.; Plant, R.; Dyke, J.; Boyer, R.; Teichgraeber, J.F.; Greives, M.R.; Mulliken, J.B.; et al. Knockdown of Crispld2 in zebrafish identifies a novel network for nonsyndromic cleft lip with or without cleft palate candidate genes. Eur. J. Hum. Genet. 2018, 26, 1441–1450. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.L.; Yarram, S.J.; Mansell, J.P.; Sandy, J.R. Matrix metalloproteinases have a role in palatogenesis. J. Dent. Res. 2002, 81, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Messetti, A.C.; Machado, R.A.; de Oliveira, C.E.; Martelli-Júnior, H.; de Almeida Reis, S.R.; Moreira, H.S.; Persuhn, D.C.; Wu, T.; Coletta, R.D. Brazilian multicenter study of association between polymorphisms in CRISPLD2 and JARID2 and non-syndromic oral clefts. J. Oral Pathol. Med. 2017, 46, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Bagordakis, E.; Paranaiba, L.M.; Brito, L.A.; de Aquino, S.N.; Messetti, A.C.; Martelli-Junior, H.; Swerts, M.S.; Graner, E.; Passos-Bueno, M.R.; Coletta, R.D. Polymorphisms at regions 1p22.1 (rs560426) and 8q24 (rs1530300) are risk markers for nonsyndromic cleft lip and/or palate in the Brazilian population. Am. J. Med. Genet. A 2013, 161, 1177–1180. [Google Scholar] [CrossRef]
- de Aquino, S.N.; Messetti, A.C.; Hoshi, R.; Borges, A.; Viena, C.S.; Reis, S.R.; Oliveira Swerts, M.S.; Graner, E.; Martelli-Júnior, H.; Coletta, R.D. Analysis of susceptibility polymorphisms for nonsyndromic cleft lip with or without cleft palate in the Brazilian population. Birth Defects Res. A Clin. Mol. Teratol. 2014, 100, 36–42. [Google Scholar] [CrossRef]
- do Rego Borges, A.; Sá, J.; Hoshi, R.; Viena, C.S.; Mariano, L.C.; de Castro Veiga, P.; Medrado, A.P.; Machado, R.A.; de Aquino, S.N.; Messetti, A.C.; et al. Genetic risk factors for nonsyndromic cleft lip with or without cleft palate in a Brazilian population with high African ancestry. Am. J. Med. Genet. A 2015, 167, 2344–2349. [Google Scholar] [CrossRef]
- Aidar, M.; Line, S.R. A simple and cost-effective protocol for DNA isolation from buccal epithelial cells. Braz. Dent. J. 2007, 18, 148–152. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Calle, M.L.; Urrea, V.; Malats, N.; Van Steen, K. mbmdr: An R package for exploring gene-gene interactions associated with binary or quantitative traits. Bioinformatics 2010, 26, 2198–2199. [Google Scholar] [CrossRef][Green Version]
- Shi, J.; Jiao, X.; Song, T.; Zhang, B.; Qin, C.; Cao, F. CRISPLD2 polymorphisms are associated with non-syndromic cleft lip with or without cleft palate in a northern Chinese population. Eur. J. Oral Sci. 2010, 118, 430–433. [Google Scholar] [CrossRef]
- Gowans, L.J.; Adeyemo, W.L.; Eshete, M.; Mossey, P.A.; Busch, T.; Aregbesola, B.; Donkor, P.; Arthur, F.K.; Bello, S.A.; Martinez, A.; et al. Association Studies and Direct DNA Sequencing Implicate Genetic Susceptibility Loci in the Etiology of Nonsyndromic Orofacial Clefts in Sub-Saharan African Populations. J. Dent. Res. 2016, 95, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Neela, P.K.; Gosla, S.R.; Husain, A.; Mohan, V. CRISPLD2 Gene Polymorphisms with Nonsyndromic Cleft Lip Palate in Indian Population. Glob. Med. Genet. 2020, 7, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Girardi, A.; Martinelli, M.; Carinci, F.; Morselli, P.G.; Caramelli, E.; Scapoli, L. No evidence for a role of CRISPLD2 in non-syndromic cleft lip with or without cleft palate in an Italian population. Eur. J. Oral Sci. 2011, 119, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Alfonso-Gonzalez, C.; Riesgo-Escovar, J.R. Fos metamorphoses: Lessons from mutants in model organisms. Mech. Dev. 2018, 154, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Brennan, A.; Leech, J.T.; Kad, N.M.; Mason, J.M. Selective antagonism of cJun for cancer therapy. J. Exp. Clin. Cancer Res. 2020, 39, 184. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016, 11, 1–9. [Google Scholar] [CrossRef]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- Mosig, R.A.; Dowling, O.; DiFeo, A.; Ramirez, M.C.; Parker, I.C.; Abe, E.; Diouri, J.; Aqeel, A.A.; Wylie, J.D.; Oblander, S.A.; et al. Loss of MMP-2 disrupts skeletal and craniofacial development and results in decreased bone mineralization, joint erosion and defects in osteoblast and osteoclast growth. Hum. Mol. Genet. 2007, 16, 1113–1123. [Google Scholar] [CrossRef]
- Blavier, L.; Lazaryev, A.; Groffen, J.; Heisterkamp, N.; DeClerck, Y.A.; Kaartinen, V. TGF-beta3-induced palatogenesis requires matrix metalloproteinases. Mol. Biol. Cell 2001, 12, 1457–1466. [Google Scholar] [CrossRef]
- Smane, L.; Pilmane, M.; Akota, I. Apoptosis and MMP-2, TIMP-2 expression in cleft lip and palate. Stomatologija 2013, 15, 129–134. [Google Scholar]
- Bergman, M.R.; Cheng, S.; Honbo, N.; Piacentini, L.; Karliner, J.S.; Lovett, D.H. A functional activating protein 1 (AP-1) site regulates matrix metalloproteinase 2 (MMP-2) transcription by cardiac cells through interactions with JunB-Fra1 and JunB-FosB heterodimers. Biochem. J. 2003, 369, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Letra, A.; Silva, R.M.; Motta, L.G.; Blanton, S.H.; Hecht, J.T.; Granjeirol, J.M.; Vieira, A.R. Association of MMP3 and TIMP2 promoter polymorphisms with nonsyndromic oral clefts. Birth Defects Res. A Clin. Mol. Teratol. 2012, 94, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Garland, M.A.; Sun, B.; Zhang, S.; Reynolds, K.; McMahon, M.; Rajakumar, R.; Islam, M.S.; Liu, Y.; Chen, Y.; et al. Cellular and developmental basis of orofacial clefts. Birth Defects Res. 2020, 112, 1558–1587. [Google Scholar] [CrossRef]
- Hammond, N.L.; Dixon, M.J. Revisiting the embryogenesis of lip and palate development. Oral Dis. 2022, 28, 1306–1326. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yokota, T.; Iwata, J.; Chai, Y. Tgf-beta-mediated FasL-Fas-Caspase pathway is crucial during palatogenesis. J. Dent. Res. 2011, 90, 981–987. [Google Scholar] [CrossRef]
- Zhang, H.; Sweezey, N.B.; Kaplan, F. LGL1 modulates proliferation, apoptosis, and migration of human fetal lung fibroblasts. Am. J. Physiol. Lung Cell Mol. Physiol. 2015, 308, L391–L402. [Google Scholar] [CrossRef]
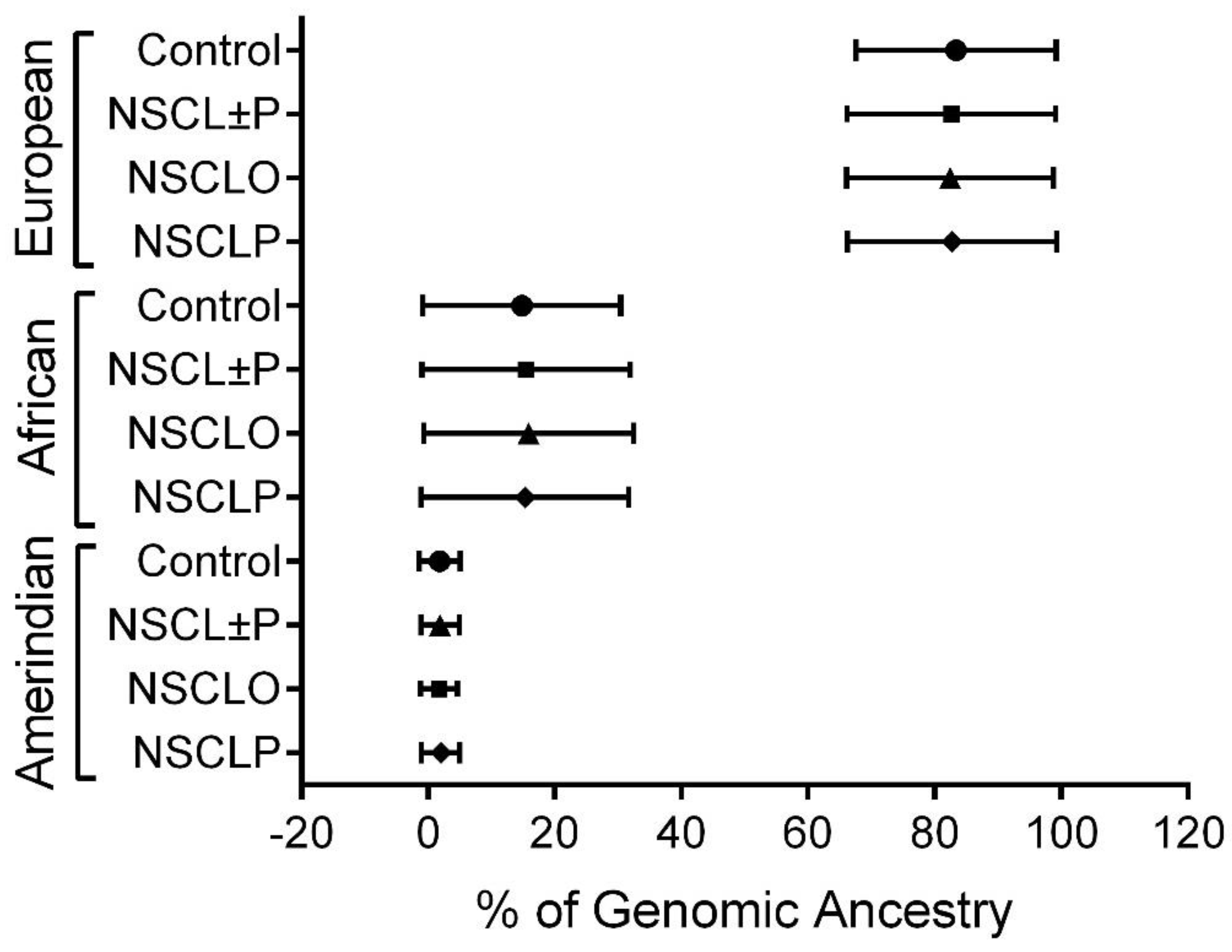
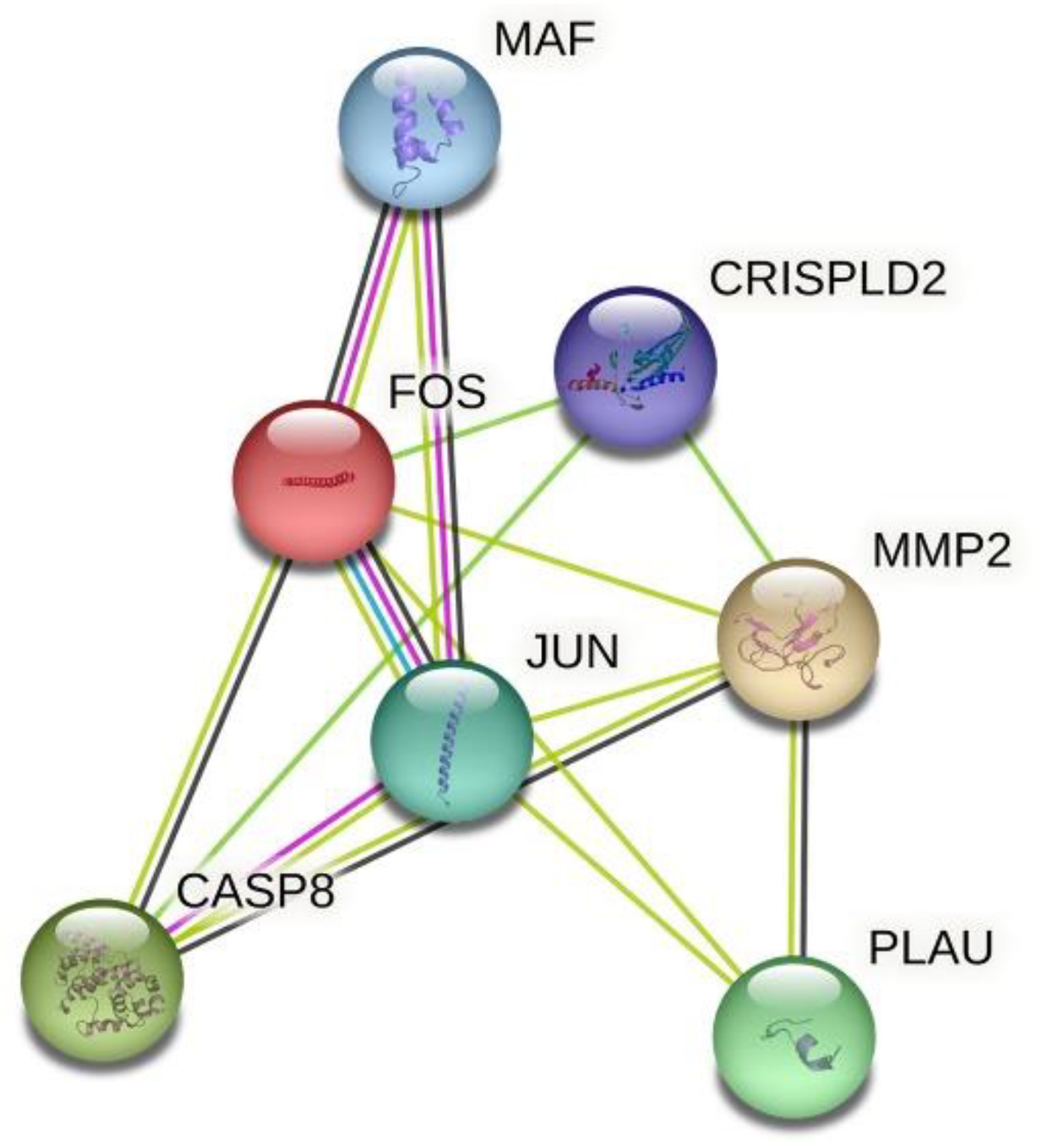
| Gene | SNP | Position | Allele * | MAF | HWE (p Value) | Call Rate |
|---|---|---|---|---|---|---|
| FOS (chromosome 14) | rs1046117 | 75279987 | T/C | 0.196 | 0.06 | 96.7% |
| CASP8 (chromosome 2) | rs3769825 | 201246657 | A/G | 0.460 | 0.46 | 98.3% |
| MMP2 (chromosome 16) | rs243836 | 55500324 | G/A | 0.464 | 0.74 | 98.8% |
| CRISPLD2 (chromosome 16) | rs1546124 | 84838445 | A/G | 0.322 | 0.57 | 99.6% |
| rs8061351 | 84849496 | T/C | 0.408 | 0.34 | 99.7% | |
| rs2326398 | 84869111 | A/G | 0.377 | 0.54 | 99.7% | |
| rs4783099 | 84907723 | C/T | 0.365 | 0.07 | 99.7% |
| Control | NSCL±P | OR (95% CI)/ p Value | NSCLO | OR (95% CI)/ p Value | NSCLP | OR (95% CI)/ p Value | |
|---|---|---|---|---|---|---|---|
| Allele | |||||||
| T | 80.4% | 78.0% | Reference | 76.5% | Reference | 79.5% | Reference |
| C | 19.6% | 22.0% | 1.19 (1.00–1.41)/0.04 | 23.5% | 1.28 (1.10–1.64)/0.004 | 20.5% | 1.15 (0.95–1.39)/0.13 |
| Genotype | |||||||
| TT | 65.9% | 60.6% | Reference | 56.4% | Reference | 62.4% | Reference |
| TC | 29.0% | 33.6% | 1.25 (1.01–1.55)/0.04 | 39.5% | 1.59 (1.16–2.18)/0.003 | 31.2% | 1.13 (0.89–1.44)/0.30 |
| CC | 5.1% | 5.8% | 1.22 (0.79–1.90)/0.36 | 4.1% | 0.95 (0.45–1.99)/0.90 | 6.4% | 1.35 (0.84–2.16)/0.21 |
| Dominant (TT/TC + CC) | 65.9%/34.1% | 60.6%/39.4% | 1.25 (1.02–1.53)/0.03 | 56.4%/43.6% | 1.50 (1.10–2.02)/0.007 | 62.4%/37.6% | 1.16 (0.93–1.46)/0.18 |
| Recessive (TT + TC/CC) | 94.9%/5.1% | 94.2%/5.8% | 1.14 (0.74–1.75)/0.55 | 95.9%/4.1% | 0.80 (0.39–1.67)/0.54 | 93.6%/6.4% | 1.30 (0.82–2.06)/0.27 |
| Control | NSCL±P | OR (95% CI)/ p Value | NSCLO | OR (95% CI)/ p Value | NSCLP | OR (95% CI)/ p Value | |
|---|---|---|---|---|---|---|---|
| Allele | |||||||
| A | 54.0% | 54.8% | Reference | 54.9% | Reference | 54.8% | Reference |
| G | 46.0% | 45.2% | 0.89 (0.77–1.02)/0.09 | 45.1% | 0.83 (0.67–1.02)/0.08 | 45.2% | 0.91 (0.78–1.06)/0.24 |
| Genotype | |||||||
| AA | 28.4% | 31.5% | Reference | 34.7% | Reference | 30.2% | Reference |
| AG | 51.0% | 50.6% | 0.91 (0.73–1.14)/0.38 | 47.6% | 0.76 (0.54–1.05)/0.09 | 51.9% | 0.98 (0.77–1.26)/0.86 |
| GG | 20.6% | 17.9% | 0.80 (0.60–1.06)/0.12 | 17.7% | 0.71 (0.46–1.09)/0.11 | 17.9% | 0.84 (0.61–1.15)/0.27 |
| Dominant (AA/AG + GG) | 28.4%/71.6% | 31.5%/68.5% | 0.88 (0.71–1.09)/0.22 | 34.7%/65.3% | 0.74 (0.54–1.02)/0.06 | 30.2%/69.8% | 0.94 (0.74–1.19)/0.61 |
| Recessive (AA + AG/GG) | 79.4%/20.6% | 82.1%/17.9% | 0.85 (0.66–1.09)/0.19 | 82.3%/17.7% | 0.85 (0.58–1.24)/0.38 | 82.1%/17.9% | 0.85 (0.65–1.12)/0.23 |
| Control | NSCL±P | OR (95% CI)/ p Value | NSCLO | OR (95% CI)/ p Value | NSCLP | OR (95% CI)/ p Value | |
|---|---|---|---|---|---|---|---|
| Allele | |||||||
| G | 53.6% | 53.2% | Reference | 53.6% | Reference | 52.9% | Reference |
| A | 46.4% | 46.8% | 1.04 (0.91–1.19)/0.53 | 46.4% | 0.99 (0.80–1.22)/0.95 | 47.1% | 1.06 (0.91–1.23)/0.40 |
| Genotype | |||||||
| GG | 28.4% | 28.0% | Reference | 29.6% | Reference | 27.4% | Reference |
| GA | 50.3% | 48.9% | 0.96 (0.76–1.21)/0.76 | 48.3% | 0.90 (0.64–1.27)/0.55 | 49.1% | 0.98 (0.76–1.26)/0.91 |
| AA | 21.3% | 23.1% | 1.06 (0.81–1.40)/0.73 | 22.1% | 0.98 (0.65–1.48)/0.91 | 23.5% | 1.10 (0.81–1.48)/0.64 |
| Dominant (GG/GA + AA) | 28.4%/71.6% | 28.0%/72.0% | 0.99 (0.80–1.23)/0.93 | 29.6%/70.4% | 0.92 (0.67–1.27)/0.62 | 27.4%/72.6% | 1.01 (0.80–1.29)/0.91 |
| Recessive (GG + GA/AA) | 78.7%/21.3% | 76.9%/23.1% | 1.09 (0.86–1.38)/0.47 | 77.9%/22.1% | 1.04 (0.73–1.48)/0.82 | 76.5%/23.5% | 1.11 (0.86–1.43)/0.42 |
| SNP1 | SNP2 | NH a | betaH b | NL c | betaL d | p Value e | Perm. p Value f | |
|---|---|---|---|---|---|---|---|---|
| NSCL±P | ||||||||
| rs1046117 (FOS) | rs3769825 (CASP8) | 2 | 0.5426 | 2 | −0.2482 | 0.0005 | 0.01 | |
| rs8061351 (CRISPLD2) | rs3769825 (CASP8) | 2 | 0.5479 | 0 | NA | 0.001 | 0.02 | |
| rs8061351 (CRISPLD2) | rs243836 (MMP2) | 1 | 0.7808 | 1 | −0.4374 | 0.003 | 0.06 | |
| rs4783099 (CRISPLD2) | rs243836 (MMP2) | 1 | 1.0643 | 0 | NA | 0.01 | 0.12 | |
| rs1546124 (CRISPLD2) | rs1046117 (FOS) | 2 | 0.6675 | 0 | NA | 0.01 | 0.11 | |
| rs2326398 (CRISPLD2) | rs243836 (MMP2) | 1 | 0.4507 | 0 | NA | 0.01 | 0.18 | |
| NSCLO | ||||||||
| rs1046117 (FOS) | rs243836 (MMP2) | 1 | 0.6455 | 0 | NA | 0.003 | 0.03 | |
| rs1046117 (FOS) | rs3769825 (CASP8) | 1 | 0.6822 | 1 | −0.4221 | 0.004 | 0.05 | |
| rs2326398 (CRISPLD2) | rs3769825 (CASP8) | 1 | 1.1374 | 0 | NA | 0.003 | 0.06 | |
| rs8061351 (CRISPLD2) | rs3769825 (CASP8) | 1 | 0.8242 | 0 | NA | 0.01 | 0.16 | |
| rs1546124 (CRISPLD2) | rs1046117 (FOS) | 1 | 0.9427 | 0 | NA | 0.03 | 0.21 | |
| rs8061351 (CRISPLD2) | rs243836 (MMP2) | 1 | 0.5800 | 1 | −0.3993 | 0.04 | 0.31 | |
| NSCLP | ||||||||
| rs8061351 (CRISPLD2) | rs3769825 (CASP8) | 2 | 0.6082 | 0 | NA | 0.0008 | 0.02 | |
| rs8061351 (CRISPLD2) | rs243836 (MMP2) | 2 | 0.6699 | 1 | −0.4583 | 0.002 | 0.04 | |
| rs1046117 (FOS) | rs3769825 (CASP8) | 1 | 0.9145 | 0 | NA | 0.007 | 0.10 | |
| rs2326398 (CRISPLD2) | rs243836 (MMP2) | 1 | 0.5372 | 0 | NA | 0.008 | 0.13 | |
| rs1546124 (CRISPLD2) | rs243836 (MMP2) | 1 | 0.5256 | 0 | NA | 0.01 | 0.16 | |
| rs4783099 (CRISPLD2) | rs243836 (MMP2) | 1 | 1.0860 | 1 | −0.3102 | 0.01 | 0.17 | |
| rs1546124 (CRISPLD2) | rs1046117 (FOS) | 1 | 0.9437 | 0 | NA | 0.03 | 0.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Machado, R.A.; de Oliveira, L.Q.R.; Rangel, A.L.C.A.; Reis, S.R.d.A.; Scariot, R.; Martelli, D.R.B.; Martelli-Júnior, H.; Coletta, R.D. Brazilian Multiethnic Association Study of Genetic Variant Interactions among FOS, CASP8, MMP2 and CRISPLD2 in the Risk of Nonsyndromic Cleft Lip with or without Cleft Palate. Dent. J. 2023, 11, 7. https://doi.org/10.3390/dj11010007
Machado RA, de Oliveira LQR, Rangel ALCA, Reis SRdA, Scariot R, Martelli DRB, Martelli-Júnior H, Coletta RD. Brazilian Multiethnic Association Study of Genetic Variant Interactions among FOS, CASP8, MMP2 and CRISPLD2 in the Risk of Nonsyndromic Cleft Lip with or without Cleft Palate. Dentistry Journal. 2023; 11(1):7. https://doi.org/10.3390/dj11010007
Chicago/Turabian StyleMachado, Renato Assis, Lilianny Querino Rocha de Oliveira, Ana Lúcia Carrinho Ayroza Rangel, Silvia Regina de Almeida Reis, Rafaela Scariot, Daniella Reis Barbosa Martelli, Hercílio Martelli-Júnior, and Ricardo D. Coletta. 2023. "Brazilian Multiethnic Association Study of Genetic Variant Interactions among FOS, CASP8, MMP2 and CRISPLD2 in the Risk of Nonsyndromic Cleft Lip with or without Cleft Palate" Dentistry Journal 11, no. 1: 7. https://doi.org/10.3390/dj11010007
APA StyleMachado, R. A., de Oliveira, L. Q. R., Rangel, A. L. C. A., Reis, S. R. d. A., Scariot, R., Martelli, D. R. B., Martelli-Júnior, H., & Coletta, R. D. (2023). Brazilian Multiethnic Association Study of Genetic Variant Interactions among FOS, CASP8, MMP2 and CRISPLD2 in the Risk of Nonsyndromic Cleft Lip with or without Cleft Palate. Dentistry Journal, 11(1), 7. https://doi.org/10.3390/dj11010007







