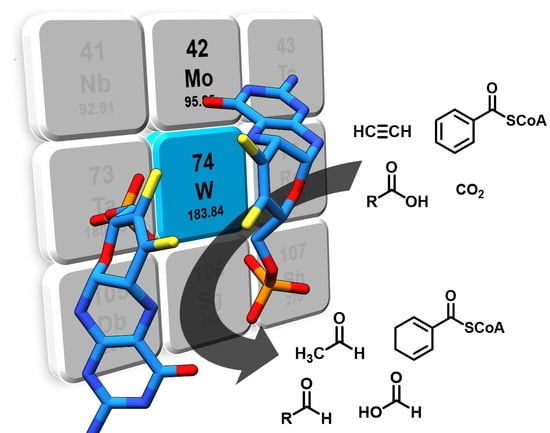
Tungstoenzymes: Occurrence, Catalytic Diversity and Cofactor Synthesis
Abstract
:1. Introduction
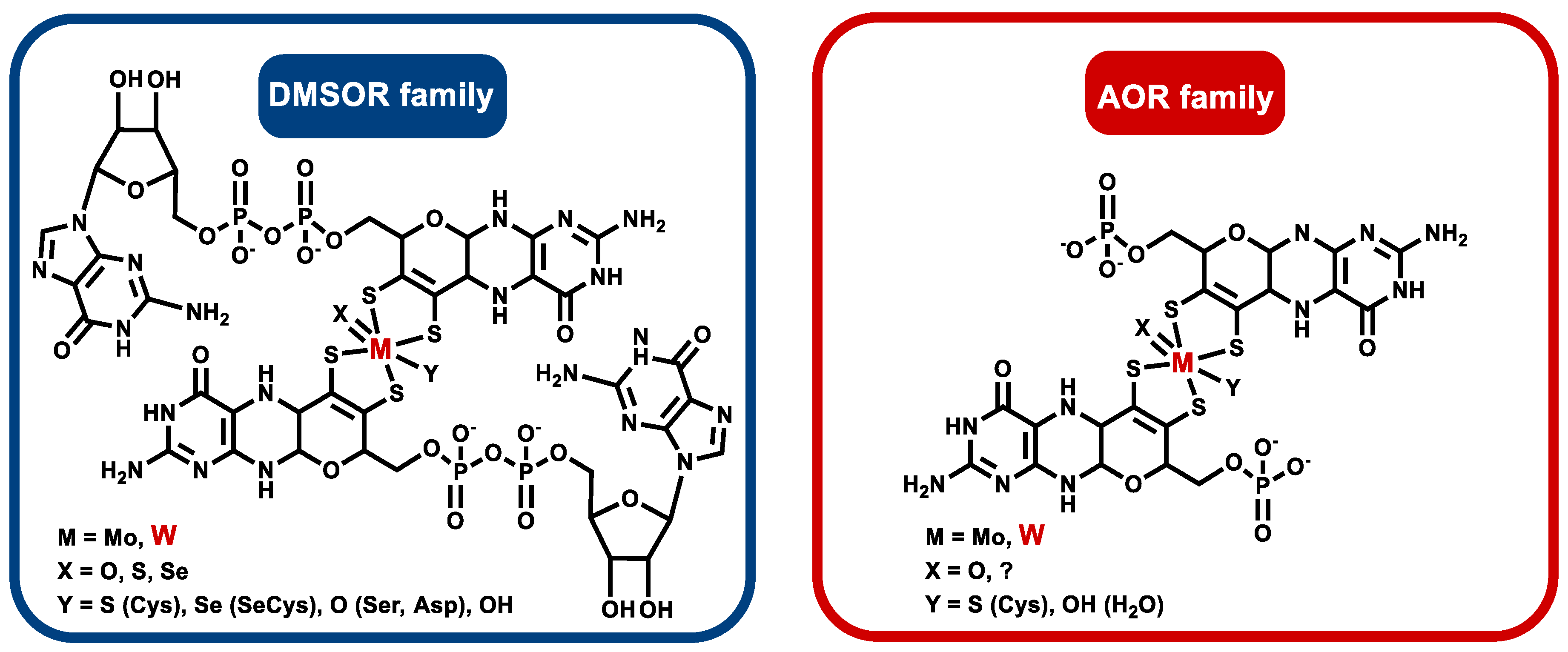
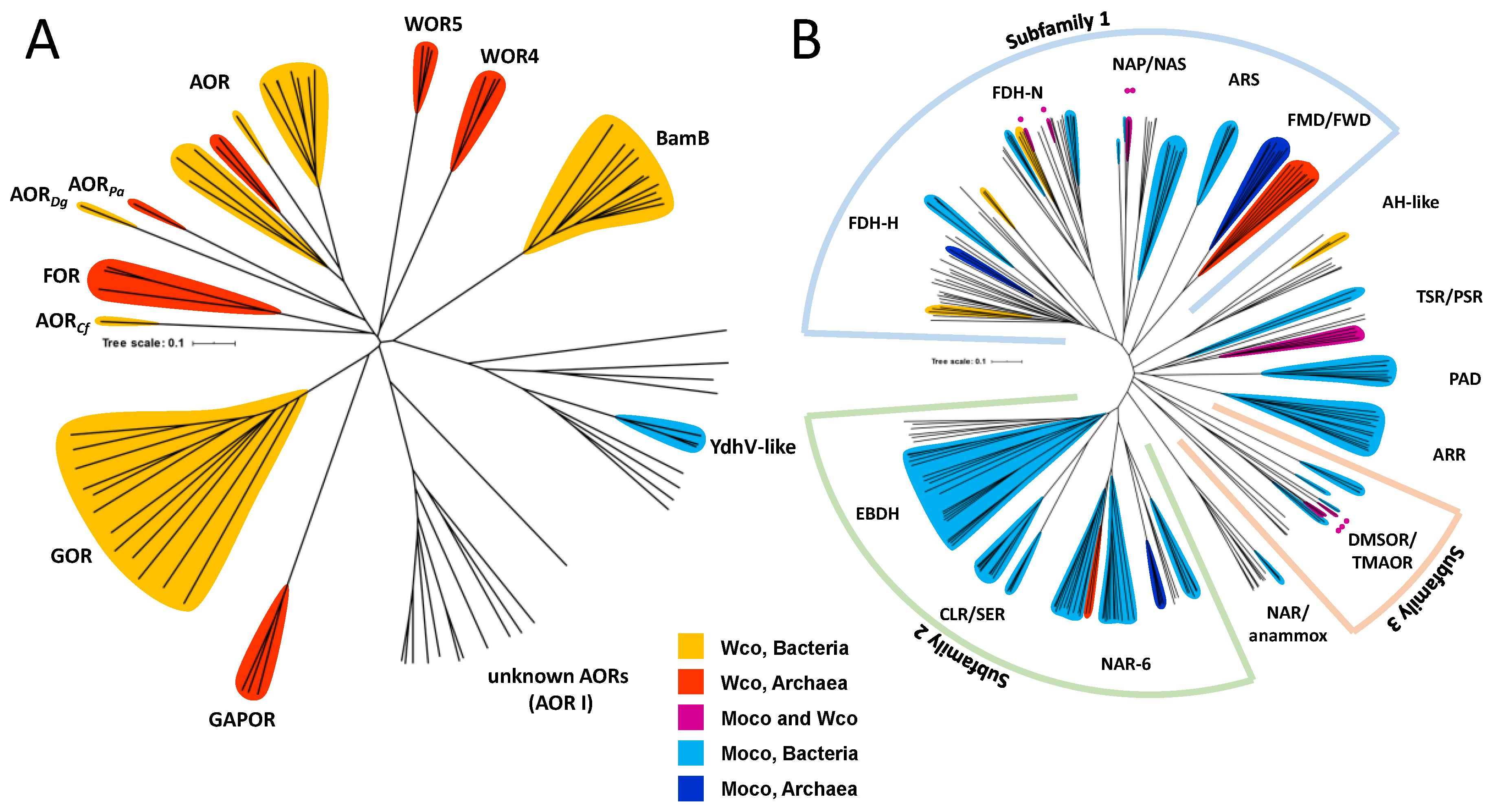
2. Affiliation of Tungsten Enzymes in the AOR and DMSOR Enzyme Families
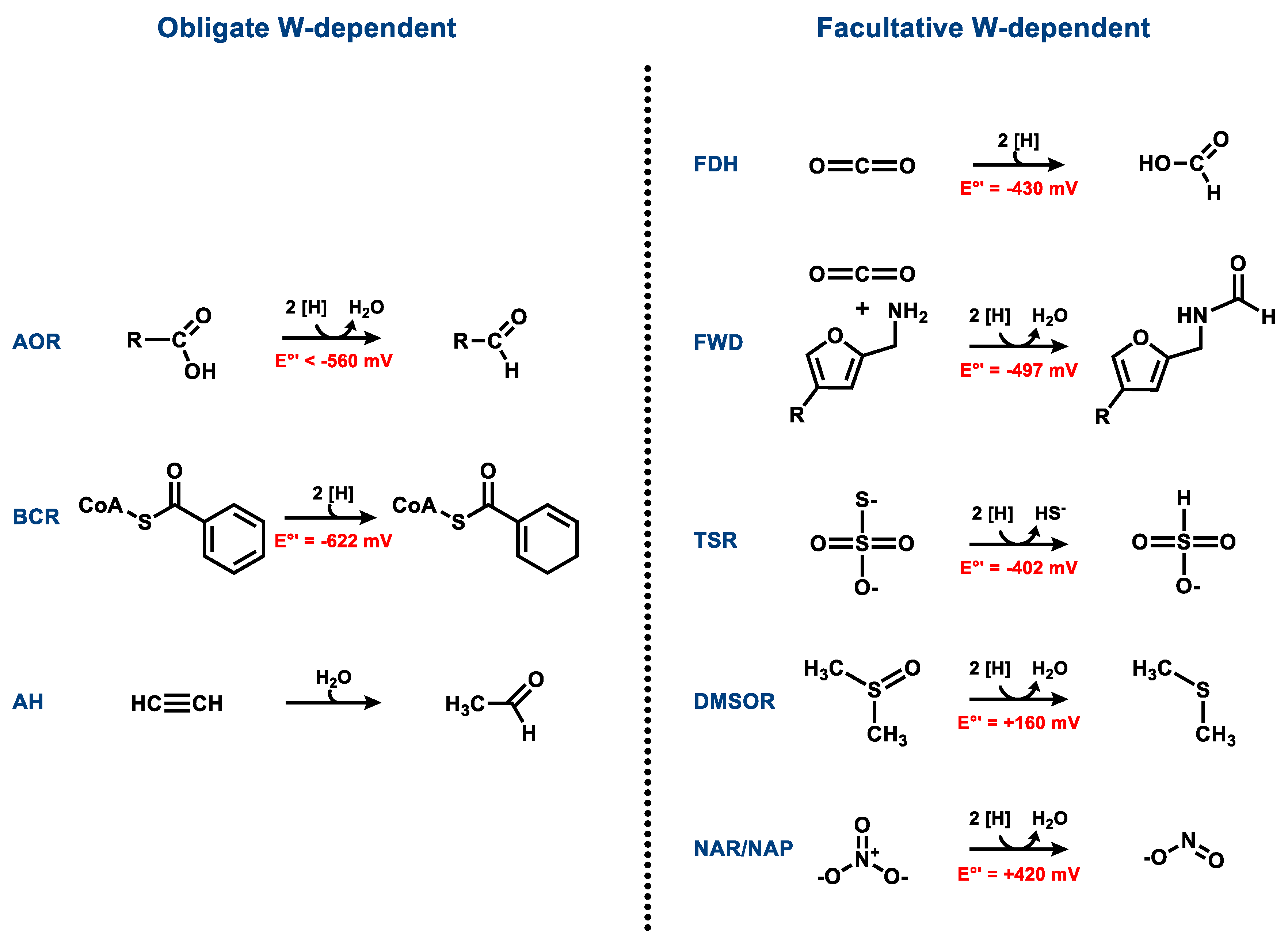
3. Obligately W-Containing Enzymes
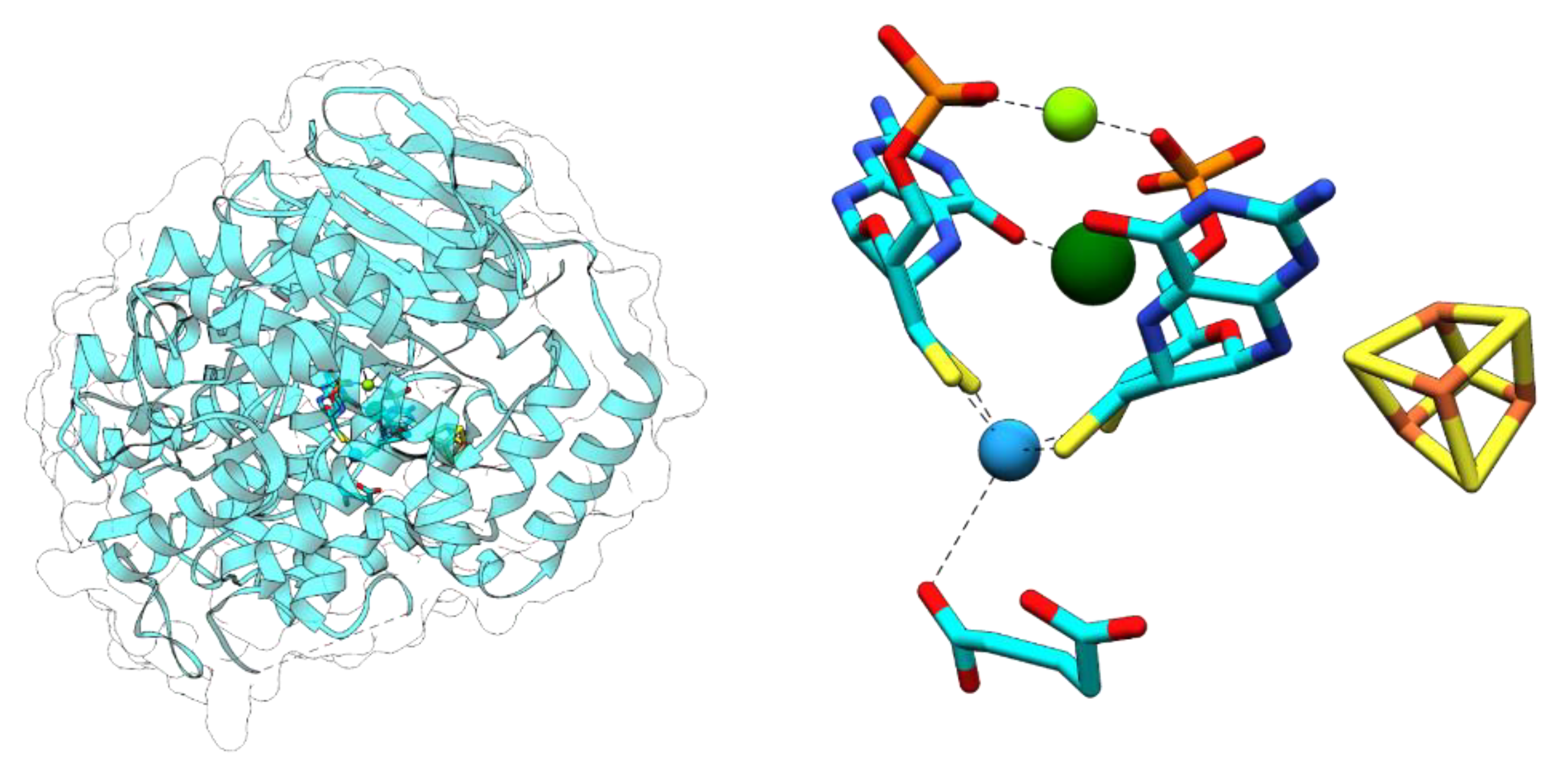
3.1. Aldehyde Oxidoreductases
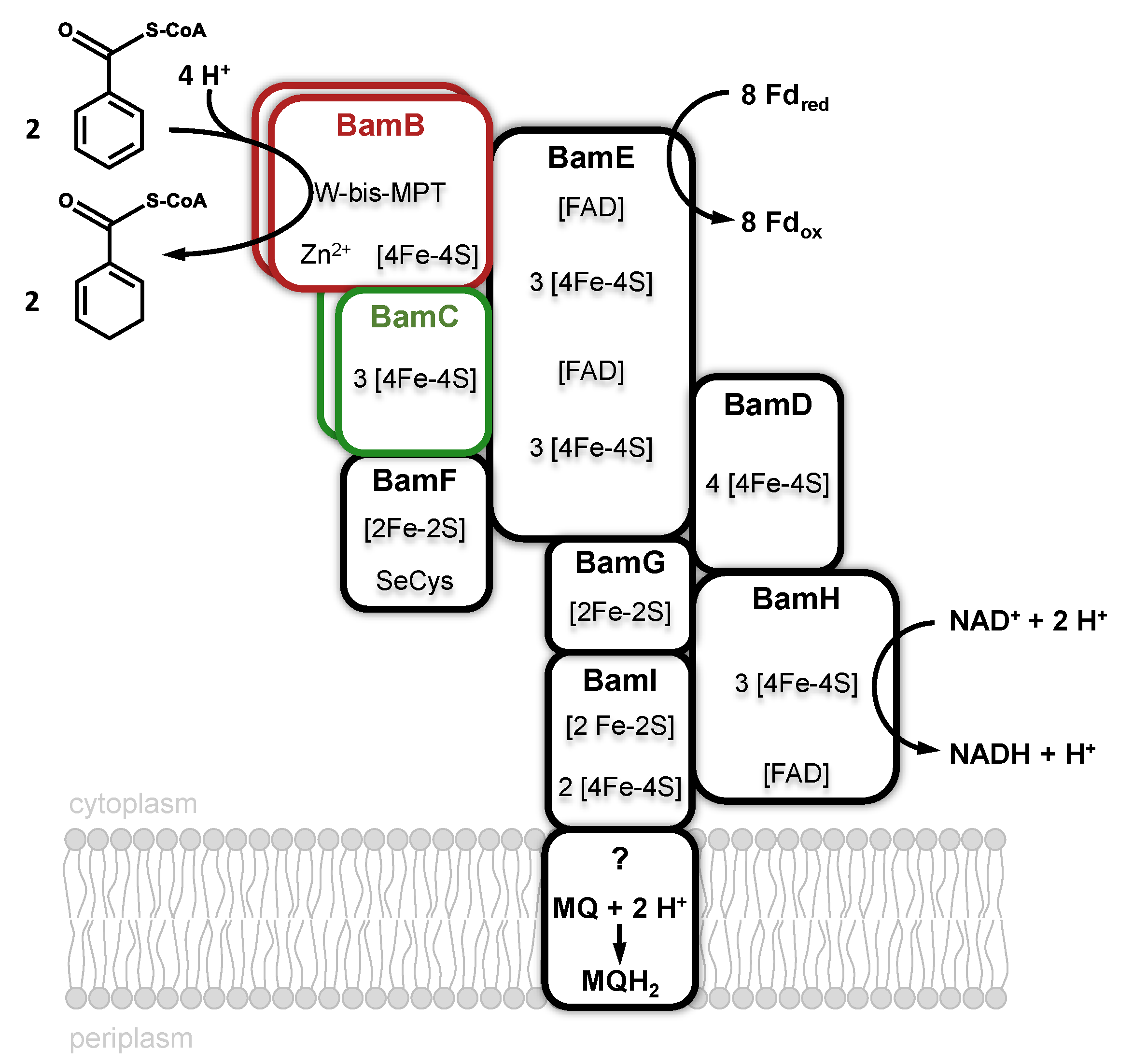
3.2. Class II Benzoyl-CoA Reductases
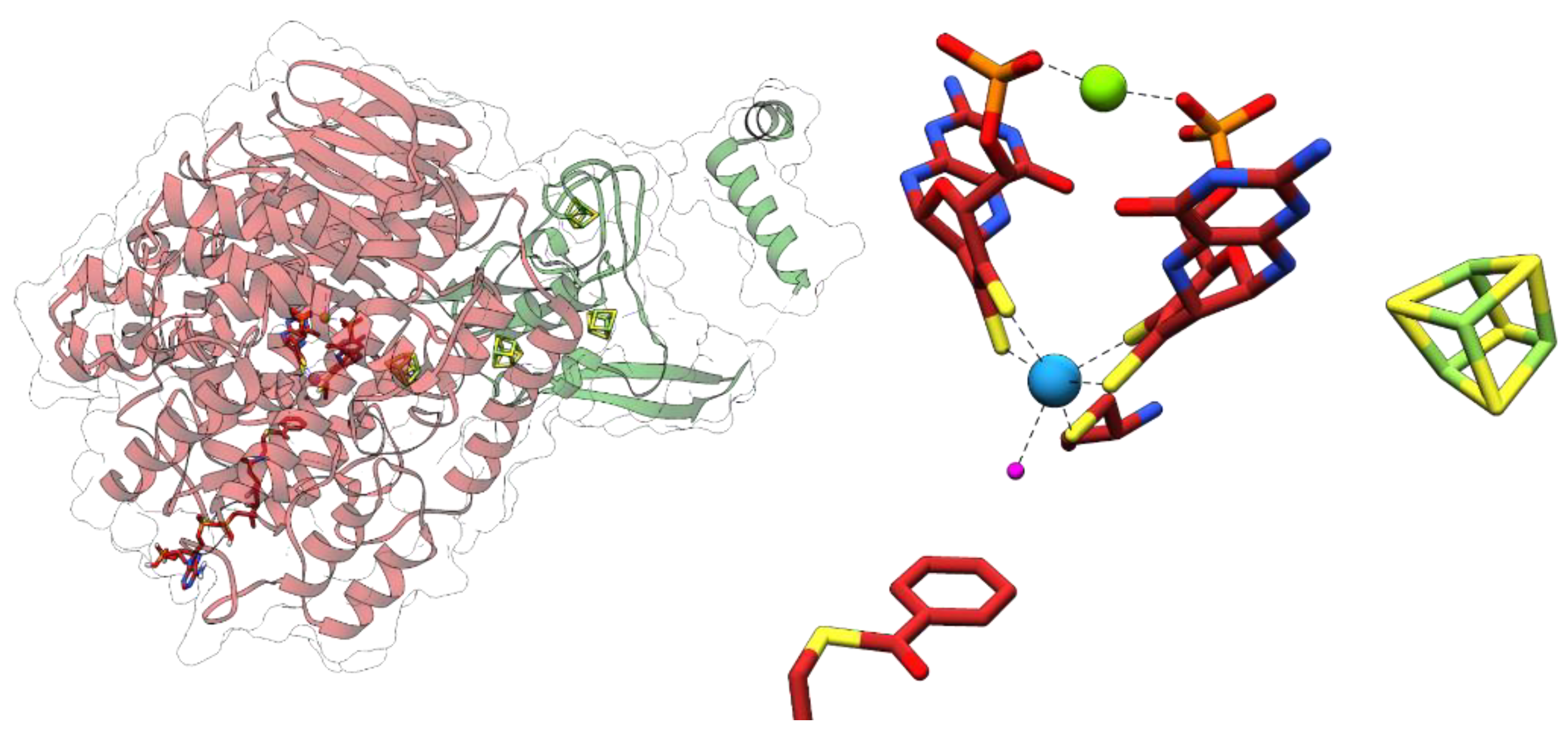
3.3. Acetylene Hydratases
4. Enzymes Containing Either Tungsten or Molybdenum
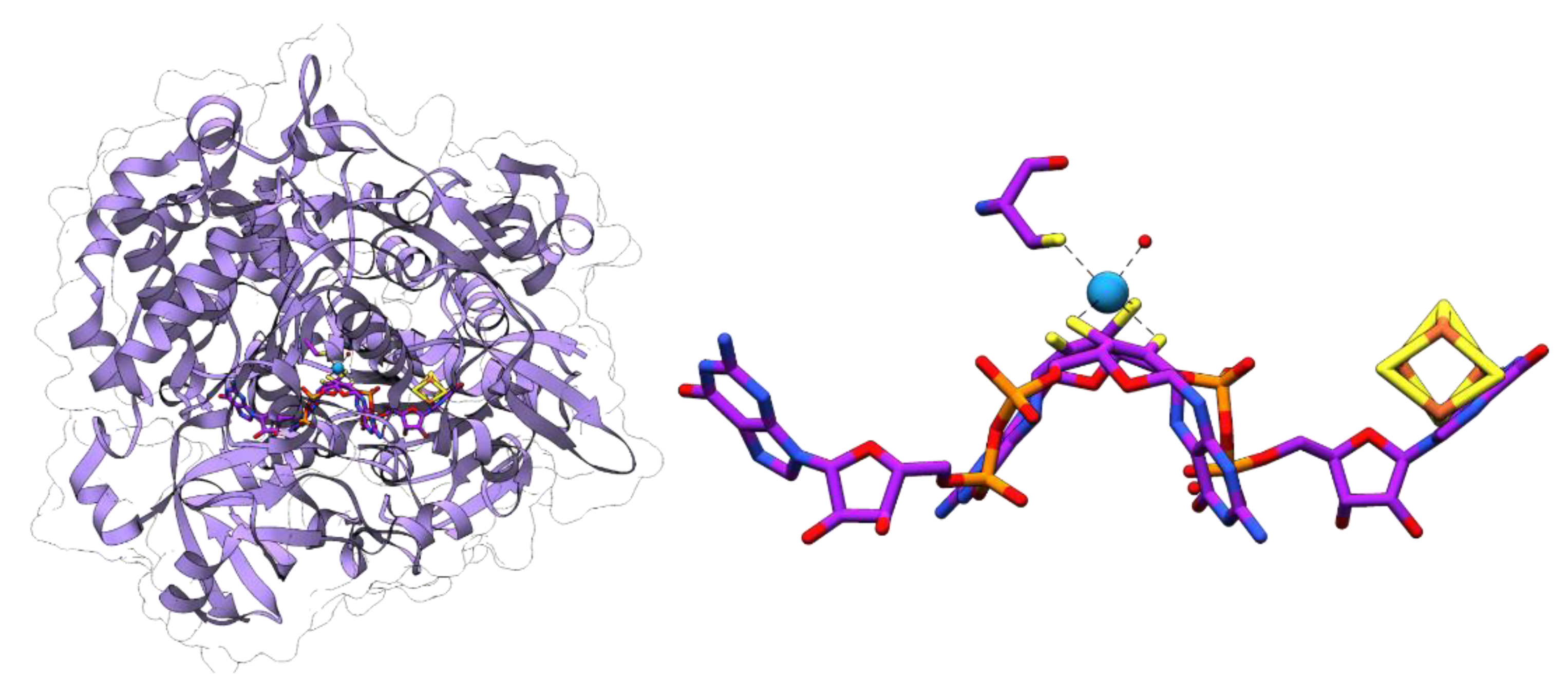
4.1. Formate Dehydrogenases
4.2. Formylmethanofuran Dehydrogenases
4.3. Respiratory Nitrate Reductases
4.4. Dimethyl Sulfoxide and Trimethylamine N-Oxide Reductases
4.5. Thiosulfate Reductases
5. Tungsten Uptake and Assembly of the Wco
5.1. Tungsten Uptake
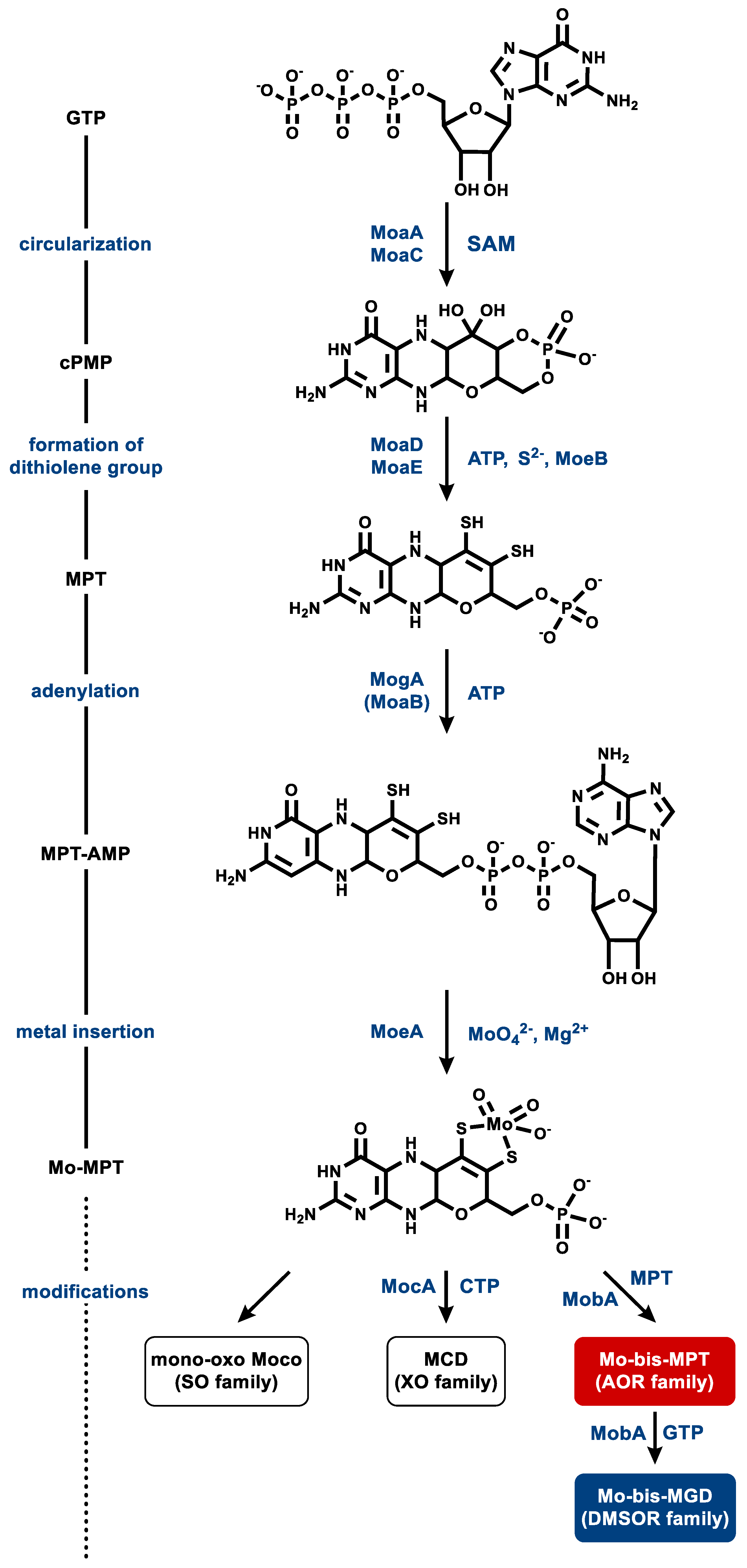
5.2. Metallopterin Cofactor Synthesis
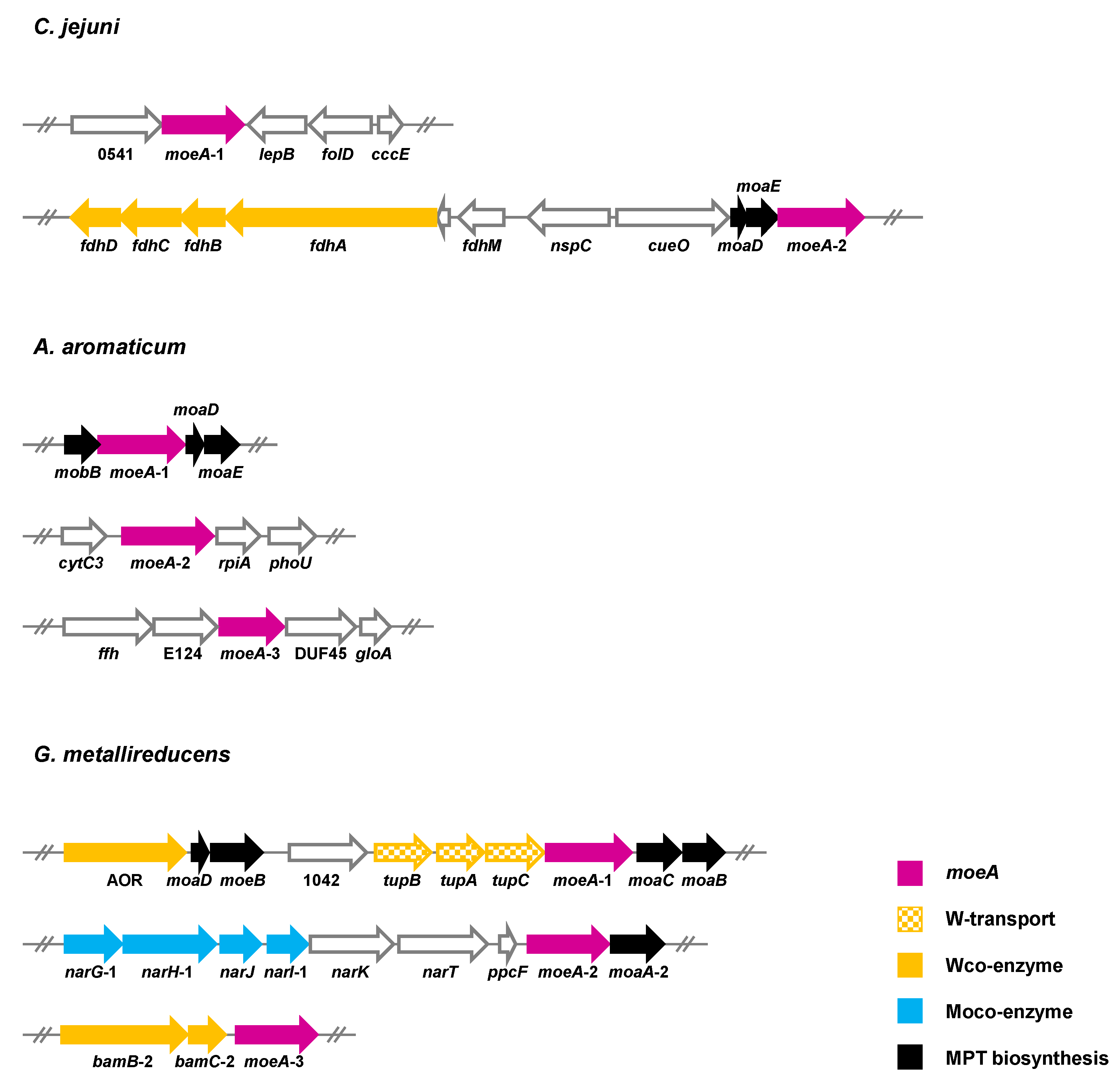
5.3. Strategies for the Selective Insertion of Molybdenum/Tungsten into Target Proteins
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hille, R. The molybdenum oxotransferases and related enzymes. Dalt. Trans. 2013, 42, 3029–3042. [Google Scholar] [CrossRef] [PubMed]
- Hille, R.; Hall, J.; Basu, P. The mononuclear molybdenum enzymes. Chem. Rev. 2014, 114, 3963–4038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leimkühler, S.; Iobbi-Nivol, C. Bacterial molybdoenzymes: Old enzymes for new purposes. FEMS Microbiol. Rev. 2015, 40, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Mendel, R.R.; Leimkühler, S. The biosynthesis of the molybdenum cofactors. J. Biol. Inorg. Chem. 2015, 20, 337–347. [Google Scholar] [CrossRef]
- Leimkühler, S. The biosynthesis of the molybdenum cofactors in Escherichia coli. Environ. Microbiol. 2020, 22, 2007–2026. [Google Scholar] [CrossRef] [Green Version]
- Hille, R.; Schulzke, C.; Kirk, M.L. Molybdenum and Tungsten Enzymes: Spectroscopic and Theoretical Investigations; Metallobiology; Royal Society of Chemistry: Cambridge, UK, 2016; ISBN 978-1-78262-878-1. [Google Scholar]
- Andreesen, J.R.; Makdessi, K. Tungsten, the surprisingly positively acting heavy metal element for prokaryotes. Ann. N. Y. Acad. Sci. 2008, 1125, 215–229. [Google Scholar] [CrossRef]
- Zhu, W.; Winter, M.G.; Byndloss, M.X.; Spiga, L.; Duerkop, B.A.; Hughes, E.R.; Büttner, L.; de Lima Romão, E.; Behrendt, C.L.; Lopez, C.A.; et al. Precision editing of the gut microbiota ameliorates colitis. Nature 2018, 553, 208–211. [Google Scholar] [CrossRef]
- Hille, R. Molybdenum and tungsten in biology. Trends Biochem. Sci. 2002, 27, 360–367. [Google Scholar] [CrossRef]
- Pushie, M.J.; Cotelesage, J.J.; George, G.N. Molybdenum and tungsten oxygen transferases-structural and functional diversity within a common active site motif. Metallomics 2014, 6, 15–24. [Google Scholar] [CrossRef]
- Romão, M.J. Molybdenum and tungsten enzymes: A crystallographic and mechanistic overview. Dalt. Trans. 2009, 4053–4068. [Google Scholar] [CrossRef]
- Bevers, L.E.; Hagedoorn, P.L.; Hagen, W.R. The bioinorganic chemistry of tungsten. Coord. Chem. Rev. 2009, 253, 269–290. [Google Scholar] [CrossRef]
- Neumann, M.; Mittelstädt, G.; Iobbi-Nivol, C.; Saggu, M.; Lendzian, F.; Hildebrandt, P.; Leimkühler, S. A periplasmic aldehyde oxidoreductase represents the first molybdopterin cytosine dinucleotide cofactor containing molybdo-flavoenzyme from Escherichia coli. FEBS J. 2009, 276, 2762–2774. [Google Scholar] [CrossRef] [PubMed]
- Romão, M.J.; Archer, M.; Moura, I.; Moura, J.J.; LeGall, J.; Engh, R.; Schneider, M.; Hof, P.; Huber, R. Crystal structure of the xanthine oxidase-related aldehyde oxido-reductase from D. Gigas. Science 1995, 270, 1170–1176. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, J.; Niks, D.; Hille, R. Kinetic and spectroscopic characterization of tungsten-substituted DMSO reductase from Rhodobacter sphaeroides. J. Biol. Inorg. Chem. 2018, 23, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Adams, M.W.W. Characterization of a fourth tungsten-containing enzyme from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 2002, 184, 6952–6956. [Google Scholar] [CrossRef] [Green Version]
- Mukund, S.; Adams, M.W. Molybdenum and vanadium do not replace tungsten in the catalytically active forms of the three tungstoenzymes in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 1996, 178, 163–167. [Google Scholar] [CrossRef] [Green Version]
- Sevcenco, A.-M.; Bevers, L.E.; Pinkse, M.W.H.; Krijger, G.C.; Wolterbeek, H.T.; Verhaert, P.D.E.M.; Hagen, W.R.; Hagedoorn, P.-L. Molybdenum incorporation in tungsten aldehyde oxidoreductase enzymes from Pyrococcus furiosus. J. Bacteriol. 2010, 192, 4143–4152. [Google Scholar] [CrossRef] [Green Version]
- Mukund, S.; Adams, M.W. Characterization of a novel tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon, Thermococcus litoralis. A role for tungsten in peptide catabolism. J. Biol. Chem. 1993, 268, 13592–13600. [Google Scholar]
- Chan, M.K.; Mukund, S.; Kletzin, A.; Adams, M.W.; Rees, D.C. Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase. Science 1995, 267, 1463–1469. [Google Scholar] [CrossRef]
- Arndt, F.; Schmitt, G.; Winiarska, A.; Saft, M.; Seubert, A.; Kahnt, J.; Heider, J. Characterization of an aldehyde oxidoreductase from the mesophilic bacterium Aromatoleum aromaticum Ebn1, a member of a new subfamily of tungsten-containing enzymes. Front. Microbiol. 2019, 10, 71. [Google Scholar] [CrossRef] [Green Version]
- Basen, M.; Schut, G.J.; Nguyen, D.M.; Lipscomb, G.L.; Benn, R.A.; Prybol, C.J.; Vaccaro, B.J.; Poole, F.L.; Kelly, R.M.; Adams, M.W.W. Single gene insertion drives bioalcohol production by a thermophilic archaeon. Proc. Natl. Acad. Sci. USA 2014, 111, 17618–17623. [Google Scholar] [CrossRef] [Green Version]
- Keller, M.W.; Lipscomb, G.L.; Nguyen, D.M.; Crowley, A.T.; Schut, G.J.; Scott, I.; Kelly, R.M.; Adams, M.W.W. Ethanol production by the hyperthermophilic archaeon Pyrococcus furiosus by expression of bacterial bifunctional alcohol dehydrogenases. Microb. Biotechnol. 2017, 10, 1535–1545. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Mukund, S.; Schut, G.J.; Dunn, D.M.; Weiss, R.; Adams, M.W. Purification and molecular characterization of the tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus: The third of a putative five-member tungstoenzyme family. J. Bacteriol. 1999, 181, 1171–1180. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Faham, S.; Roy, R.; Adams, M.W.; Rees, D.C. Formaldehyde ferredoxin oxidoreductase from Pyrococcus furiosus: The 1.85 Å resolution crystal structure and its mechanistic implications. J. Mol. Biol. 1999, 286, 899–914. [Google Scholar] [CrossRef] [PubMed]
- Mukund, S.; Adams, M.W.W. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase, a novel tungsten-containing enzyme with a potential glycolytic role in the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 1995, 270, 8389–8392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, I.M.; Rubinstein, G.M.; Poole, F.L.; Lipscomb, G.L.; Schut, G.J.; Williams-Rhaesa, A.M.; Stevenson, D.M.; Amador-Noguez, D.; Kelly, R.M.; Adams, M.W.W. The thermophilic biomass-degrading bacterium Caldicellulosiruptor bescii utilizes two enzymes to oxidize glyceraldehyde 3-phosphate during glycolysis. J. Biol. Chem. 2019, 294, 9995–10005. [Google Scholar] [CrossRef]
- Bevers, L.E.; Bol, E.; Hagedoorn, P.-L.; Hagen, W.R. WOR5, a novel tungsten-containing aldehyde oxidoreductase from Pyrococcus furiosus with a broad substrate specificity. J. Bacteriol. 2005, 187, 7056–7061. [Google Scholar] [CrossRef] [Green Version]
- Kletzin, A.; Adams, M.W. Tungsten in biological systems. FEMS Microbiol. Rev. 1996, 18, 5–63. [Google Scholar] [CrossRef]
- Hagedoorn, P.L.; Chen, T.; Schröder, I.; Piersma, S.R.; De Vries, S.; Hagen, W.R. Purification and characterization of the tungsten enzyme aldehyde:ferredoxin oxidoreductase from the hyperthermophilic denitrifier Pyrobaculum aerophilum. J. Biol. Inorg. Chem. 2005, 10, 259–269. [Google Scholar] [CrossRef]
- Reher, M.; Gebhard, S.; Schönheit, P. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase (GAPOR) and nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN), key enzymes of the respective modified Embden-Meyerhof pathways in the hyperthermophilic crenarchaeota Pyrobaculum aerophilum. FEMS Microbiol. Lett. 2007, 273, 196–205. [Google Scholar]
- Heider, J.; Boll, M.; Breese, K.; Breinig, S.; Ebenau-Jehle, C.; Feil, U.; Gad’on, N.; Laempe, D.; Leuthner, B.; Mohamed, M.E.S.; et al. Differential induction of enzymes involved in anaerobic metabolism of aromatic compounds in the denitrifying bacterium Thauera aromatica. Arch. Microbiol. 1998, 170, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, G.; Boll, M.; Heider, J. Microbial degradation of aromatic compounds-From one strategy to four. Nat. Rev. Microbiol. 2011, 9, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Löffler, C.; Kuntze, K.; Vazquez, J.R.; Rugor, A.; Kung, J.W.; Böttcher, A.; Boll, M. Occurrence, genes and expression of the W/Se-containing class II benzoyl-coenzyme A reductases in anaerobic bacteria. Environ. Microbiol. 2011, 13, 696–709. [Google Scholar] [CrossRef] [PubMed]
- Boll, M.; Löffler, C.; Morris, B.E.L.; Kung, J.W. Anaerobic degradation of homocyclic aromatic compounds via arylcarboxyl-coenzyme A esters: Organisms, strategies and key enzymes. Environ. Microbiol. 2014, 16, 612–627. [Google Scholar] [CrossRef]
- Kung, J.W.; Baumann, S.; von Bergen, M.; Müller, M.; Hagedoorn, P.-L.; Hagen, W.R.; Boll, M. Reversible biological birch reduction at an extremely low redox potential. J. Am. Chem. Soc. 2010, 132, 9850–9856. [Google Scholar] [CrossRef]
- Boll, M.; Fuchs, G. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. Eur. J. Biochem. 1995, 234, 921–933. [Google Scholar] [CrossRef]
- Wischgoll, S.; Heintz, D.; Peters, F.; Erxleben, A.; Sarnighausen, E.; Reski, R.; Van Dorsselaer, A.; Boll, M. Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol. Microbiol. 2005, 58, 1238–1252. [Google Scholar] [CrossRef]
- Huwiler, S.G.; Löffler, C.; Anselmann, S.E.L.; Stärk, H.-J.; von Bergen, M.; Flechsler, J.; Rachel, R.; Boll, M. One-megadalton metalloenzyme complex in Geobacter metallireducens involved in benzene ring reduction beyond the biological redox window. Proc. Natl. Acad. Sci. USA 2019, 116, 2259–2264. [Google Scholar] [CrossRef] [Green Version]
- Anselmann, S.E.L.; Löffler, C.; Stärk, H.; Jehmlich, N.; Bergen, M.; Brüls, T.; Boll, M. The class II benzoyl-coenzyme A reductase complex from the sulfate-reducing Desulfosarcina cetonica. Environ. Microbiol. 2019, 21, 4241–4252. [Google Scholar] [CrossRef] [Green Version]
- Kung, J.W.; Löffler, C.; Dorner, K.; Heintz, D.; Gallien, S.; Van Dorsselaer, A.; Friedrich, T.; Boll, M. Identification and characterization of the tungsten-containing class of benzoyl-coenzyme A reductases. Proc. Natl. Acad. Sci. USA 2009, 106, 17687–17692. [Google Scholar] [CrossRef] [Green Version]
- Buckel, W.; Thauer, R.K. Flavin-based electron bifurcation, a new mechanism of biological energy coupling. Chem. Rev. 2018, 118, 3862–3886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, F.; Rother, M.; Boll, M. Selenocysteine-containing proteins in anaerobic benzoate metabolism of Desulfococcus multivorans. J. Bacteriol. 2004, 186, 2156–2163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heintz, D.; Gallien, S.; Wischgoll, S.; Ullmann, A.K.; Schaeffer, C.; Kretzschmar, A.K.; Van Dorsselaer, A.; Boll, M. Differential membrane proteome analysis reveals novel proteins involved in the degradation of aromatic compounds in Geobacter metallireducens. Mol. Cel Proteom. 2009, 8, 2159–2169. [Google Scholar] [CrossRef] [Green Version]
- Weinert, T.; Huwiler, S.G.; Kung, J.W.; Weidenweber, S.; Hellwig, P.; Stärk, H.-J.; Biskup, T.; Weber, S.; Cotelesage, J.J.H.; George, G.N.; et al. Structural basis of enzymatic benzene ring reduction. Nat. Chem. Biol. 2015, 11, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Culka, M.; Huwiler, S.G.; Boll, M.; Ullmann, G.M. Breaking benzene aromaticity-computational insights into the mechanism of the tungsten-containing benzoyl-CoA reductase. J. Am. Chem. Soc. 2017, 139, 14488–14500. [Google Scholar] [CrossRef]
- Qian, H.X.; Liao, R.Z. QM/MM Study of tungsten-dependent benzoyl-coenzyme a reductase: Rationalization of regioselectivity and predication of W vs Mo selectivity. Inorg. Chem. 2018, 57, 10667–10678. [Google Scholar] [CrossRef]
- Birch, A.J. Reduction by dissolving metals. Nature 1946, 158. [Google Scholar] [CrossRef]
- Birch, A.J. The Birch reduction in organic synthesis. Pure Appl. Chem. 1996, 68, 553–556. [Google Scholar] [CrossRef]
- Chatterjee, A.; König, B. Birch-type photoreduction of arenes and heteroarenes by sensitized electron transfer. Angew. Chemie. Int. Ed. 2019, 58, 14289–14294. [Google Scholar] [CrossRef] [Green Version]
- Peters, B.K.; Rodriguez, K.X.; Reisberg, S.H.; Beil, S.B.; Hickey, D.P.; Kawamata, Y.; Collins, M.; Starr, J.; Chen, L.; Udyavara, S.; et al. Scalable and safe synthetic organic electroreduction inspired by Li-ion battery chemistry. Science 2019, 363, 838–845. [Google Scholar] [CrossRef]
- Schink, B. Fermentation of acetylene by an obligate anaerobe, Pelobacter acetylenicus sp. nov. Arch. Microbiol. 1985, 142, 295–301. [Google Scholar] [CrossRef] [Green Version]
- Meckenstock, R.U.; Krieger, R.; Ensign, S.; Kroneck, P.M.H.; Schink, B. Acetylene hydratase of Pelobacter acetylenicus. Molecular and spectroscopic properties of the tungsten iron-sulfur enzyme. Eur. J. Biochem. 1999, 264, 176–182. [Google Scholar] [CrossRef]
- Rosner, B.M.; Schink, B. Purification and characterization of acetylene hydratase of Pelobacter acetylenicus, a tungsten iron-sulfur protein. J. Bacteriol. 1995, 177, 5767–5772. [Google Scholar] [CrossRef] [Green Version]
- Seiffert, G.B.; Ullmann, G.M.; Messerschmidt, A.; Schink, B.; Kroneck, P.M.H.; Einsle, O. Structure of the non-redox-active tungsten/[4Fe:4S] enzyme acetylene hydratase. Proc. Natl. Acad. Sci. USA 2007, 104, 3073–3077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- tenBrink, F.; Schink, B.; Kroneck, P.M.H. Exploring the active site of the tungsten, iron-sulfur enzyme acetylene hydratase. J. Bacteriol. 2011, 193, 1229–1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boll, M.; Einsle, O.; Ermler, U.; Kroneck, P.M.H.P.M.H.; Ullmann, G.M.M. Structure and function of the unusual tungsten enzymes acetylene hydratase and class ii benzoyl-coenzyme a reductase. J. Mol. Microbiol. Biotechnol. 2016, 26, 119–137. [Google Scholar] [CrossRef]
- Hyman, M.R.; Daniel, A. Acetylene inhibition of metalloenzymes. Anal. Biochem. 1988, 173, 207–220. [Google Scholar] [CrossRef]
- Stewart, W.D.; Fitzgerald, G.P.; Burris, R.H. In situ studies on N2 fixation using the acetylene reduction technique. Proc. Natl. Acad. Sci. USA 1967, 58, 2071–2078. [Google Scholar] [CrossRef] [Green Version]
- Antony, S.; Bayse, C.A. Theoretical studies of models of the active site of the tungstoenzyme acetylene hydratase. Organometallics 2009, 28, 4938–4944. [Google Scholar] [CrossRef]
- Yadav, J.; Das, S.K.; Sarkar, S. A functional mimic of the new class of tungstoenzyme, acetylene hydratase. J. Am. Chem. Soc. 1997, 119, 4315–4316. [Google Scholar] [CrossRef]
- Schreyer, M.; Hintermann, L. Is the tungsten(IV) complex (NEt4)2[WO(mnt)2] a functional analogue of acetylene hydratase? Beilstein J. Org. Chem. 2017, 13, 2332–2339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidovič, C.; Peschel, L.M.; Buchsteiner, M.; Belaj, F.; Mösch-Zanetti, N.C. Structural mimics of acetylene hydratase: Tungsten complexes capable of intramolecular nucleophilic attack on acetylene. Chem. Eur. J. 2019, 25, 14267–14272. [Google Scholar] [CrossRef] [PubMed]
- Templeton, J.L.; Ward, B.C.; Chen, G.J.J.; Mcdonald, J.W.; Newton, W.E. Oxotungsten(IV)-acetylene complexes: Synthesis via intermetal oxygen atom transfer and nuclear magnetic resonance studies. Inorg. Chem. 1981, 20, 1248–1253. [Google Scholar] [CrossRef]
- Schobert, H. Production of acetylene and acetylene-based chemicals from coal. Chem. Rev. 2014, 114, 1743–1760. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, T.; Schwanhold, N.; Leimkühler, S. Assembly and catalysis of molybdenum or tungsten-containing formate dehydrogenases from bacteria. Biochim. Biophys. Acta Proteins Proteom. 2015, 1854, 1090–1100. [Google Scholar] [CrossRef]
- Maia, L.B.; Moura, J.J.G.; Moura, I. Molybdenum and tungsten-dependent formate dehydrogenases. J. Biol. Inorg. Chem. 2015, 20, 287–309. [Google Scholar] [CrossRef]
- Niks, D.; Hille, R. Molybdenum- and tungsten-containing formate dehydrogenases and formylmethanofuran dehydrogenases: Structure, mechanism, and cofactor insertion. Protein Sci. 2019, 28, 111–122. [Google Scholar] [CrossRef] [Green Version]
- Vorholt, J.A.; Thauer, R.K. Molybdenum and tungsten enzymes in C1 metabolism. Met. Ions Biol. Syst. 2002, 39, 571–619. [Google Scholar]
- Moura, J.J.G.; Brondino, C.D.; Trincão, J.; Romão, M.J. Mo and W bis-MGD enzymes: Nitrate reductases and formate dehydrogenases. J. Biol. Inorg. Chem. 2004, 9, 791–799. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, T.; Schrapers, P.; Utesch, T.; Nimtz, M.; Rippers, Y.; Dau, H.; Mroginski, M.A.; Haumann, M.; Leimkühler, S. The Molybdenum Active Site of Formate Dehydrogenase Is Capable of Catalyzing C-H Bond Cleavage and Oxygen Atom Transfer Reactions. Biochemistry 2016, 55, 2381–2389. [Google Scholar]
- Niks, D.; Duvvuru, J.; Escalona, M.; Hille, R. Spectroscopic and kinetic properties of the molybdenum-containing, NAD+-dependent formate dehydrogenase from Ralstonia eutropha. J. Biol. Chem. 2016, 291, 1162–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maia, L.B.; Fonseca, L.; Moura, I.; Moura, J.J.G. Reduction of carbon dioxide by a molybdenum-containing formate dehydrogenase: A kinetic and mechanistic study. J. Am. Chem. Soc. 2016, 138, 8834–8846. [Google Scholar] [CrossRef]
- Enoch, H.G.; Lester, R.L. Effects of molybdate, tungstate, and selenium compounds on formate dehydrogenase and other enzyme systems in Escherichia coli. J. Bacteriol. 1972, 110, 1032–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- May, H.D.; Patel, P.S.; Ferry, J.G. Effect of molybdenum and tungsten on synthesis and composition of formate dehydrogenase in Methanobacterium formicicum. J. Bacteriol. 1988, 170, 3384–3389. [Google Scholar] [CrossRef] [Green Version]
- Ljungdahl, L.G.; Andreesen, J.R. Formate Dehydrogenase, a selenium-tungsten enzyme from Clostridium thermoaceticum. Methods Enzymol. 1978, 53, 360–372. [Google Scholar]
- Strobl, G.; Feicht, R.; White, H.; Lottspeich, F.; Simon, H. The tungsten-containing aldehyde oxidoreductase from Clostridium thermoaceticum and its complex with a viologen-accepting NADPH oxidoreductase. Biol. Chem. Hoppe. Seyler 1992, 373, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Alissandratos, A.; Kim, H.K.; Matthews, H.; Hennessy, J.E.; Philbrook, A.; Easton, C.J. Clostridium carboxidivorans strain P7T recombinant formate dehydrogenase catalyzes reduction of CO2 to formate. Appl. Environ. Microbiol. 2013, 79, 741–744. [Google Scholar] [CrossRef] [Green Version]
- Graentzdoerffer, A.; Rauh, D.; Pich, A.; Andreesen, J.R. Molecular and biochemical characterization of two tungsten-and selenium-containing formate dehydrogenases from Eubacterium acidaminophilum that are associated with components of an iron-only hydrogenase. Arch. Microbiol. 2003, 179, 116–130. [Google Scholar] [CrossRef]
- Laukel, M.; Chistoserdova, L.; Lidstrom, M.E.; Vorholt, J.A. The tungsten-containing formate dehydrogenase from Methylobacterium extorquens AM1: Purification and properties. Eur. J. Biochem. 2003, 270, 325–333. [Google Scholar] [CrossRef] [Green Version]
- De Bok, F.A.M.; Hagedoorn, P.L.; Silva, P.J.; Hagen, W.R.; Schiltz, E.; Fritsche, K.; Stams, A.J.M. Two W-containing formate dehydrogenases (CO2-reductases) involved in syntrophic propionate oxidation by Syntrophobacter fumaroxidans. Eur. J. Biochem. 2003, 270, 2476–2485. [Google Scholar] [CrossRef]
- Almendra, M.J.; Brondino, C.D.; Gavel, O.; Pereira, A.S.; Tavares, P.; Bursakov, S.; Duarte, R.; Caldeira, J.; Moura, J.J.G.; Moura, I. Purification and characterization of a tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Biochemistry 1999, 38, 16366–16372. [Google Scholar] [CrossRef] [PubMed]
- Brondino, C.D.; Passeggi, M.C.G.; Caldeira, J.; Almendra, M.J.; Feio, M.J.; Moura, J.J.G.; Moura, I. Incorporation of either molybdenum or tungsten into formate dehydrogenase from Desulfovibrio alaskensis NCIMB 13491; EPR assignment of the proximal iron-sulfur cluster to the pterin cofactor in formate dehydrogenases from sulfate-reducing bacteria. J. Biol. Inorg. Chem. 2004, 9, 145–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Silva, S.M.; Pimentel, C.; Valente, F.M.A.; Rodrigues-Pousada, C.; Pereira, I.A.C. Tungsten and molybdenum regulation of formate dehydrogenase expression in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 2011, 193, 2909–2916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mota, C.S.; Valette, O.; González, P.J.; Brondino, C.D.; Moura, J.J.G.; Moura, I.; Dolla, A.; Rivas, M.G. Effects of molybdate and tungstate on expression levels and biochemical characteristics of formate dehydrogenases produced by Desulfovibrio alaskensis NCIMB 13491. J. Bacteriol. 2011, 193, 2917–2923. [Google Scholar] [CrossRef] [Green Version]
- Taylor, A.J.; Kelly, D.J. The function, biogenesis and regulation of the electron transport chains in Campylobacter jejuni: New insights into the bioenergetics of a major food-borne pathogen. Adv. Microb. Physiol. 2019, 74, 239–329. [Google Scholar]
- Reda, T.; Plugge, C.M.; Abram, N.J.; Hirst, J. Reversible interconversion of carbon dioxide and formate by an electroactive enzyme. Proc. Natl. Acad. Sci. USA 2008, 105, 10654–10658. [Google Scholar] [CrossRef] [Green Version]
- Schuchmann, K.; Müller, V. Direct and reversible hydrogenation of CO2 to formate by a bacterial carbon dioxide reductase. Science 2013, 342, 1382–1385. [Google Scholar] [CrossRef]
- Sokol, K.P.; Robinson, W.E.; Oliveira, A.R.; Zacarias, S.; Lee, C.Y.; Madden, C.; Bassegoda, A.; Hirst, J.; Pereira, I.A.C.; Reisner, E. Reversible and selective interconversion of hydrogen and carbon dioxide into formate by a semiartificial formate hydrogenlyase mimic. J. Am. Chem. Soc. 2019, 141, 17498–17502. [Google Scholar] [CrossRef] [Green Version]
- Sokol, K.P.; Robinson, W.E.; Oliveira, A.R.; Warnan, J.; Nowaczyk, M.M.; Ruff, A.; Pereira, I.A.C.; Reisner, E. Photoreduction of CO2 with a formate dehydrogenase driven by photosystem ii using a semi-artificial z-scheme architecture. J. Am. Chem. Soc. 2018, 140, 16418–16422. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.; Robinson, W.E.; Oliveira, A.R.; Heidary, N.; Kornienko, N.; Warnan, J.; Pereira, I.A.C.; Reisner, E. Interfacing formate dehydrogenase with metal oxides for the reversible electrocatalysis and solar-driven reduction of carbon dioxide. Angew. Chemie. Int. Ed. 2019, 58, 4601–4605. [Google Scholar] [CrossRef] [Green Version]
- Hochheimer, A.; Hedderich, R.; Thauer, R.K. The formylmethanofuran dehydrogenase isoenzymes in Methanobacterium wolfei and Methanobacterium thermoautotrophicum: Induction of the molybdenum isoenzyme by molybdate and constitutive synthesis of the tungsten isoenzyme. Arch. Microbiol. 1998, 170, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Bertram, P.A.; Schmitz, R.A.; Linder, D.; Thauer, R.K. Tungstate can substitute for molybdate in sustaining growth of Methanobacterium thermoautotrophicum—Identification and characterization of a tungsten isoenzyme of formylmethanofuran dehydrogenase. Arch. Microbiol. 1994, 161, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Matschiavelli, N.; Rother, M. Role of a putative tungsten-dependent formylmethanofuran dehydrogenase in Methanosarcina acetivorans. Arch. Microbiol. 2015, 197, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.; Ermler, U.; Shima, S. The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters. Science 2016, 354, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Gates, A.J.; Hughes, R.O.; Sharp, S.R.; Millington, P.D.; Nilavongse, A.; Cole, J.A.; Leach, E.R.; Jepson, B.; Richardson, D.J.; Butler, C.S. Properties of the periplasmic nitrate reductases from Paracoccus pantotrophus and Escherichia coli after growth in tungsten-supplemented media. FEMS Microbiol. Lett. 2003, 220, 261–269. [Google Scholar] [CrossRef] [Green Version]
- De Vries, S.; Momcilovic, M.; Strampraad, M.J.F.; Whitelegge, J.P.; Baghai, A.; Schröder, I. Adaptation to a high-tungsten environment: Pyrobaculum aerophilum contains an active tungsten nitrate reductase. Biochemistry 2010, 49, 9911–9921. [Google Scholar]
- Stewart, L.J.; Bailey, S.; Bennett, B.; Charnock, J.M.; Garner, C.D.; McAlpine, A.S. Dimethylsulfoxide reductase: An enzyme capable of catalysis with either molybdenum or tungsten at the active site. J. Mol. Biol. 2000, 299, 593–600. [Google Scholar] [CrossRef]
- Johnson, M.K.; Rees, D.C.; Adams, M.W.W. Tungstoenzymes. Chem. Rev. 1996, 96, 2817–2839. [Google Scholar] [CrossRef]
- Buc, J.; Santini, C.-L.; Giordani, R.; Czjzek, M.; Wu, L.-F.; Giordano, G. Enzymatic and physiological properties of the tungsten-substituted molybdenum TMAO reductase from Escherichia coli. Mol. Microbiol. 1999, 32, 159–168. [Google Scholar] [CrossRef]
- Haja, D.K.; Wu, C.H.; Poole, F.L.; Sugar, J.; Williams, S.G.; Jones, A.K.; Adams, M.W.W. Characterization of thiosulfate reductase from Pyrobaculum aerophilum heterologously produced in Pyrococcus furiosus. Extremophiles 2020, 24, 53–62. [Google Scholar] [CrossRef]
- Aguilar-Barajas, E.; Díaz-Pérez, C.; Ramírez-Díaz, M.I.; Riveros-Rosas, H.; Cervantes, C. Bacterial transport of sulfate, molybdate, and related oxyanions. BioMetals 2011, 24, 687–707. [Google Scholar] [CrossRef]
- Bevers, L.E.; Hagedoorn, P.L.; Krijger, G.C.; Hagen, W.R. Tungsten transport protein A (WtpA) in Pyrococcus furiosus: The first member of a new class of tungstate and molybdate transporters. J. Bacteriol. 2006, 188, 6498–6505. [Google Scholar] [CrossRef] [Green Version]
- Makdessi, K.; Fritsche, K.; Pich, A.; Andreesen, J.R. Identification and characterization of the cytoplasmic tungstate/molybdate-binding protein (Mop) from Eubacterium acidaminophilum. Arch. Microbiol. 2004, 181, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Makdessi, K.; Andreesen, J.R.; Pich, A. Tungstate uptake by a highly specific ABC transporter in Eubacterium acidaminophilum. J. Biol. Chem. 2001, 276, 24557–24564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smart, J.P.; Cliff, M.J.; Kelly, D.J. A role for tungsten in the biology of Campylobacter jejuni: Tungstate stimulates formate dehydrogenase activity and is transported via an ultra-high affinity ABC system distinct from the molybdate transporter. Mol. Microbiol. 2009, 74, 742–757. [Google Scholar] [CrossRef] [PubMed]
- Taveirne, M.E.; Sikes, M.L.; Olson, J.W. Molybdenum and tungsten in Campylobacter jejuni: Their physiological role and identification of separate transporters regulated by a single ModE-like protein. Mol. Microbiol. 2009, 74, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Otrelo-Cardoso, A.R.; Nair, R.R.; Correia, M.A.S.; Rivas, M.G.; Santos-Silva, T. TupA: A tungstate binding protein in the periplasm of Desulfovibrio alaskensis G20. Int. J. Mol. Sci. 2014, 15, 11783–11798. [Google Scholar] [CrossRef] [Green Version]
- Otrelo-Cardoso, A.R.; Nair, R.R.; Correia, M.A.S.; Cordeiro, R.S.C.; Panjkovich, A.; Svergun, D.I.; Santos-Silva, T.; Rivas, M.G. Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Gladyshev, V.N. Molybdoproteomes and evolution of molybdenum utilization. J. Mol. Biol. 2008, 379, 881–899. [Google Scholar] [CrossRef] [Green Version]
- Rajagopalan, K.V. Biosynthesis and processing of the molybdenum cofactors. Biochem. Soc. Trans. 1997, 25, 757–761. [Google Scholar] [CrossRef] [Green Version]
- Leimkühler, S. Shared function and moonlighting proteins in molybdenum cofactor biosynthesis. Biol. Chem. 2017, 398, 1009–1026. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, G.; Mendel, R.R.; Ribbe, M.W. Molybdenum cofactors, enzymes and pathways. Nature 2009, 460, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Bevers, L.E.; Hagedoorn, P.L.; Santamaria-Araujo, J.A.; Magalon, A.; Hagen, W.R.; Schwarz, G. Function of MoaB proteins in the biosynthesis of the molybdenum and tungsten cofactors. Biochemistry 2008, 47, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Wuebbens, M.M.; Rajagopalan, K.V. Structural characterization of a molybdopterin precursor. J. Biol. Chem. 1993, 268, 13493–13498. [Google Scholar] [PubMed]
- Hänzelmann, P.; Schindelin, H. Binding of 5′-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc. Natl. Acad. Sci. USA 2006, 103, 6829–6834. [Google Scholar] [CrossRef] [Green Version]
- Gutzke, G.; Fischer, B.; Mendel, R.R.; Schwarz, G. Thiocarboxylation of molybdopterin synthase provides evidence for the mechanism of dithiolene formation in metal-binding pterins. J. Biol. Chem. 2001, 276, 36268–36274. [Google Scholar] [CrossRef] [Green Version]
- Rudolph, M.J.; Wuebbens, M.M.; Rajagopalan, K.V.; Schindelin, H. Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation. Nat. Struct. Biol. 2001, 8, 42–46. [Google Scholar] [CrossRef]
- Wuebbens, M.M.; Rajagopalan, K.V. Mechanistic and mutational studies of Escherichia coli molybdopterin synthase clarify the final step of molybdopterin biosynthesis. J. Biol. Chem. 2003, 278, 14523–14532. [Google Scholar] [CrossRef] [Green Version]
- Pitterle, D.M.; Johnson, J.L.; Rajagopalan, K.V. In vitro synthesis of molybdopterin from precursor Z using purified converting factor. Role of protein-bound sulfur in formation of the dithiolene. J. Biol. Chem. 1993, 268, 13506–13509. [Google Scholar]
- Lake, M.W.; Wuebbens, M.M.; Rajagopalan, K.V.; Schindelin, H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature 2001, 414, 325–329. [Google Scholar] [CrossRef]
- Leimkühler, S.; Rajagopalan, K.V. A Sulfurtransferase is required in the transfer of cysteine sulfur in the in vitro synthesis of molybdopterin from precursor z in Escherichia coli. J. Biol. Chem. 2001, 276, 22024–22031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llamas, A.; Mendel, R.R.; Schwarz, G. Synthesis of adenylated molybdopterin: An essential step for molybdenum insertion. J. Biol. Chem. 2004, 279, 55241–55246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, M.S.; Johnson, J.L.; Rajagopalan, K. V Molybdenum cofactor biosynthesis in Escherichia coli mod and mog mutants. J. Bacteriol. 1996, 178, 4310–4312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, O.; Rajagopalan, K.V. Molybdopterin in carbon monoxide oxidase from carboxydotrophic bacteria. J. Bacteriol. 1984, 157, 643–648. [Google Scholar] [CrossRef] [Green Version]
- Reschke, S.; Duffus, B.R.; Schrapers, P.; Mebs, S.; Teutloff, C.; Dau, H.; Haumann, M.; Leimkühler, S. Identification of YdhV as the first molybdoenzyme binding a Bis-Mo-MPT cofactor in Escherichia coli. Biochemistry 2019, 58, 2228–2242. [Google Scholar]
- Johnson, J.L.; Bastian, N.R.; Rajagopalan, K. V Molybdopterin guanine dinucleotide: A modified form of molybdopterin identified in the molybdenum cofactor of dimethyl sulfoxide reductase from Rhodobacter sphaeroides forma specialis denitrificans. Proc. Natl. Acad. Sci. USA 1990, 87, 3190–3194. [Google Scholar] [CrossRef] [Green Version]
- Reschke, S.; Sigfridsson, K.G.V.; Kaufmann, P.; Leidel, N.; Horn, S.; Gast, K.; Schulzke, C.; Haumann, M.; Leimkühler, S. Identification of a bis-molybdopterin intermediate in molybdenum cofactor biosynthesis in Escherichia coli. J. Biol. Chem. 2013, 288, 29736–29745. [Google Scholar] [CrossRef] [Green Version]
- Johannes, J.; Bluschke, A.; Jehmlich, N.; Von Bergen, M.; Boll, M. Purification and characterization of active-site components of the putative p-cresol methylhydroxylase membrane complex from Geobacter metallireducens. J. Bacteriol. 2008, 190, 6493–6500. [Google Scholar] [CrossRef] [Green Version]
- Peters, F.; Heintz, D.; Johannes, J.; Van Dorsselaer, A.; Boll, M. Genes, enzymes, and regulation of para-cresol metabolism in Geobacter metallireducens. J. Bacteriol. 2007, 189, 4729–4738. [Google Scholar] [CrossRef] [Green Version]
- Neumann, M.; Stöcklein, W.; Leimkühler, S. Transfer of the molybdenum cofactor synthesized by Rhodobacter capsulatus MoeA to XdhC and MobA. J. Biol. Chem. 2007, 282, 28493–28500. [Google Scholar] [CrossRef] [Green Version]
- Thomé, R.; Gust, A.; Toci, R.; Mendel, R.; Bittner, F.; Magalon, A.; Walburger, A. A sulfurtransferase is essential for activity of formate dehydrogenases in Escherichia coli. J. Biol. Chem. 2012, 287, 4671–4678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnoux, P.; Ruppelt, C.; Oudouhou, F.; Lavergne, J.; Siponen, M.I.; Toci, R.; Mendel, R.R.; Bittner, F.; Pignol, D.; Magalon, A.; et al. Sulphur shuttling across a chaperone during molybdenum cofactor maturation. Nat. Commun. 2015, 6, 6148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwanhold, N.; Iobbi-Nivol, C.; Lehmann, A.; Leimkühler, S. Same but different: Comparison of two system-specific molecular chaperones for the maturation of formate dehydrogenases. PLoS ONE 2018, 13. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seelmann, C.S.; Willistein, M.; Heider, J.; Boll, M. Tungstoenzymes: Occurrence, Catalytic Diversity and Cofactor Synthesis. Inorganics 2020, 8, 44. https://doi.org/10.3390/inorganics8080044
Seelmann CS, Willistein M, Heider J, Boll M. Tungstoenzymes: Occurrence, Catalytic Diversity and Cofactor Synthesis. Inorganics. 2020; 8(8):44. https://doi.org/10.3390/inorganics8080044
Chicago/Turabian StyleSeelmann, Carola S., Max Willistein, Johann Heider, and Matthias Boll. 2020. "Tungstoenzymes: Occurrence, Catalytic Diversity and Cofactor Synthesis" Inorganics 8, no. 8: 44. https://doi.org/10.3390/inorganics8080044