Understanding the Selective Extraction of the Uranyl Ion from Seawater with Amidoxime-Functionalized Materials: Uranyl Complexes of Pyrimidine-2-amidoxime †
Abstract
1. Introduction
2. Results and Discussion
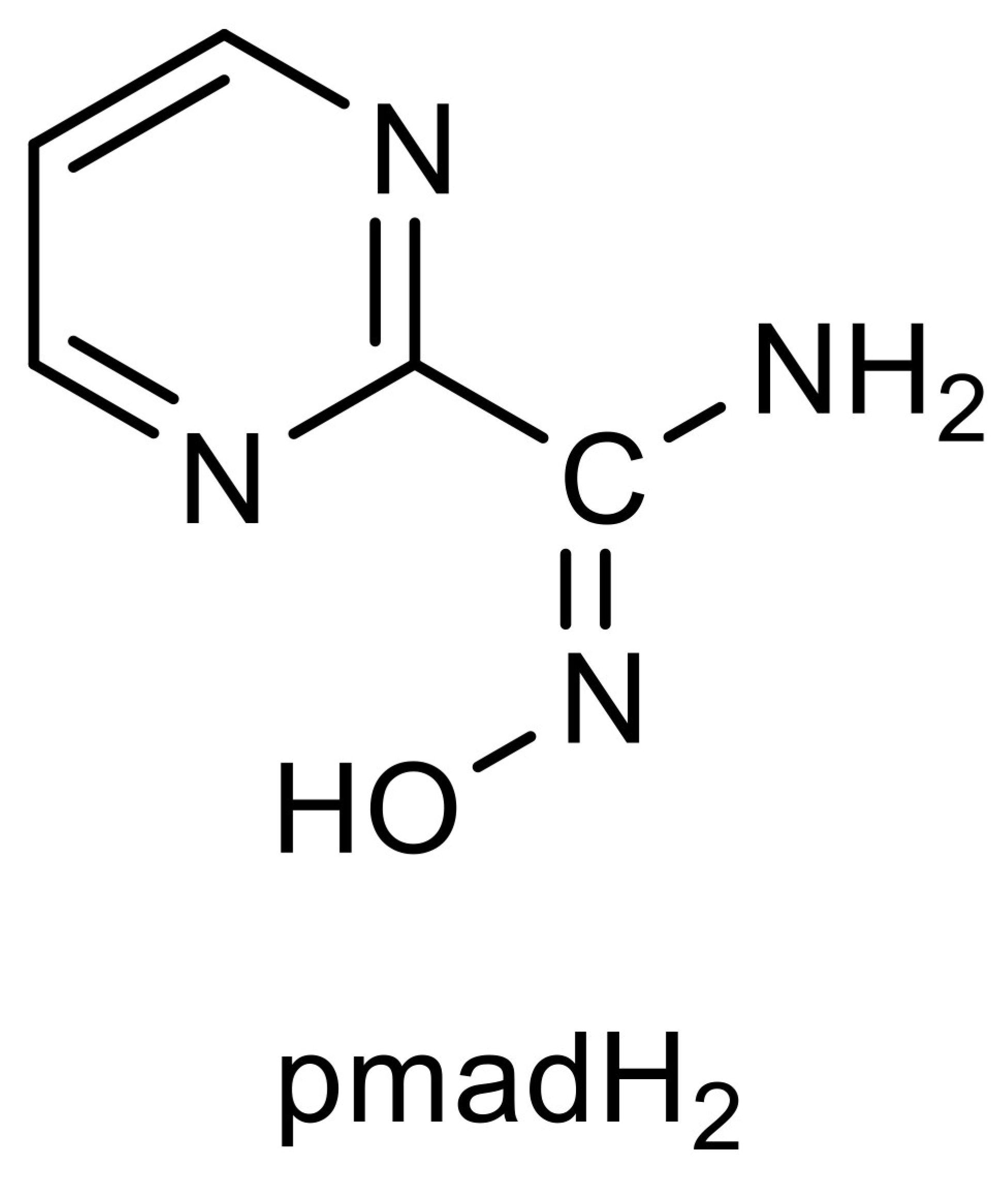
2.1. Synthesis of the Complex
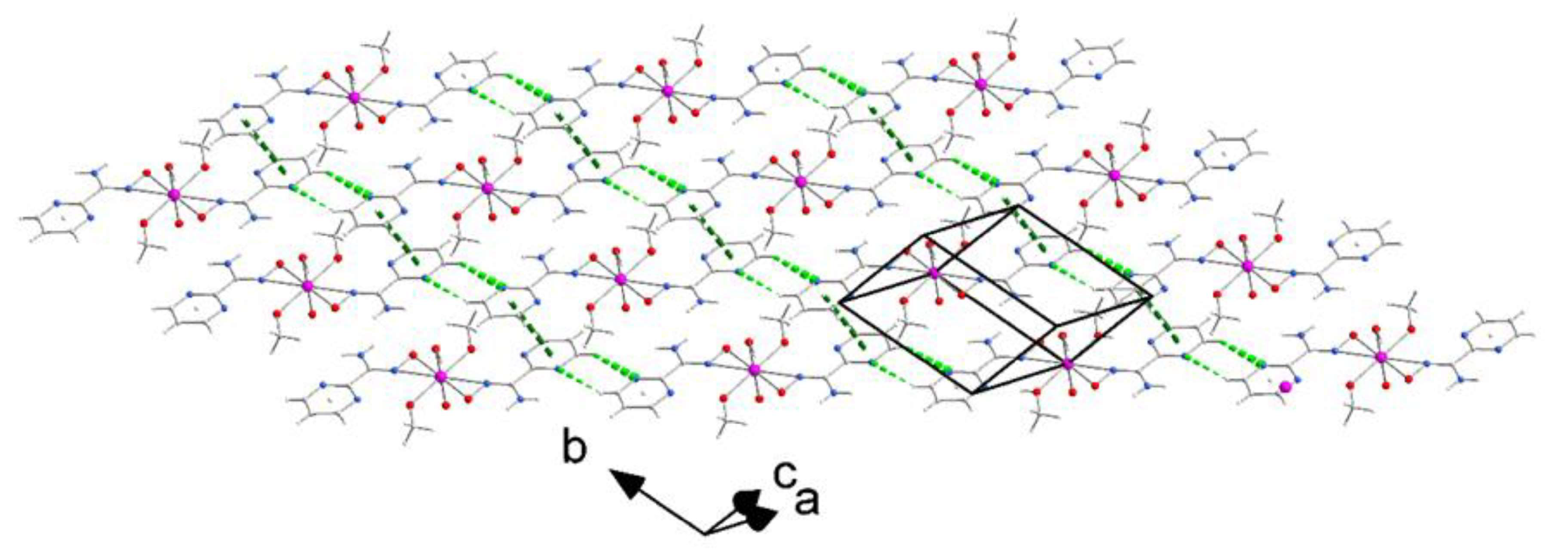
2.2. Structure Determination of the Complex
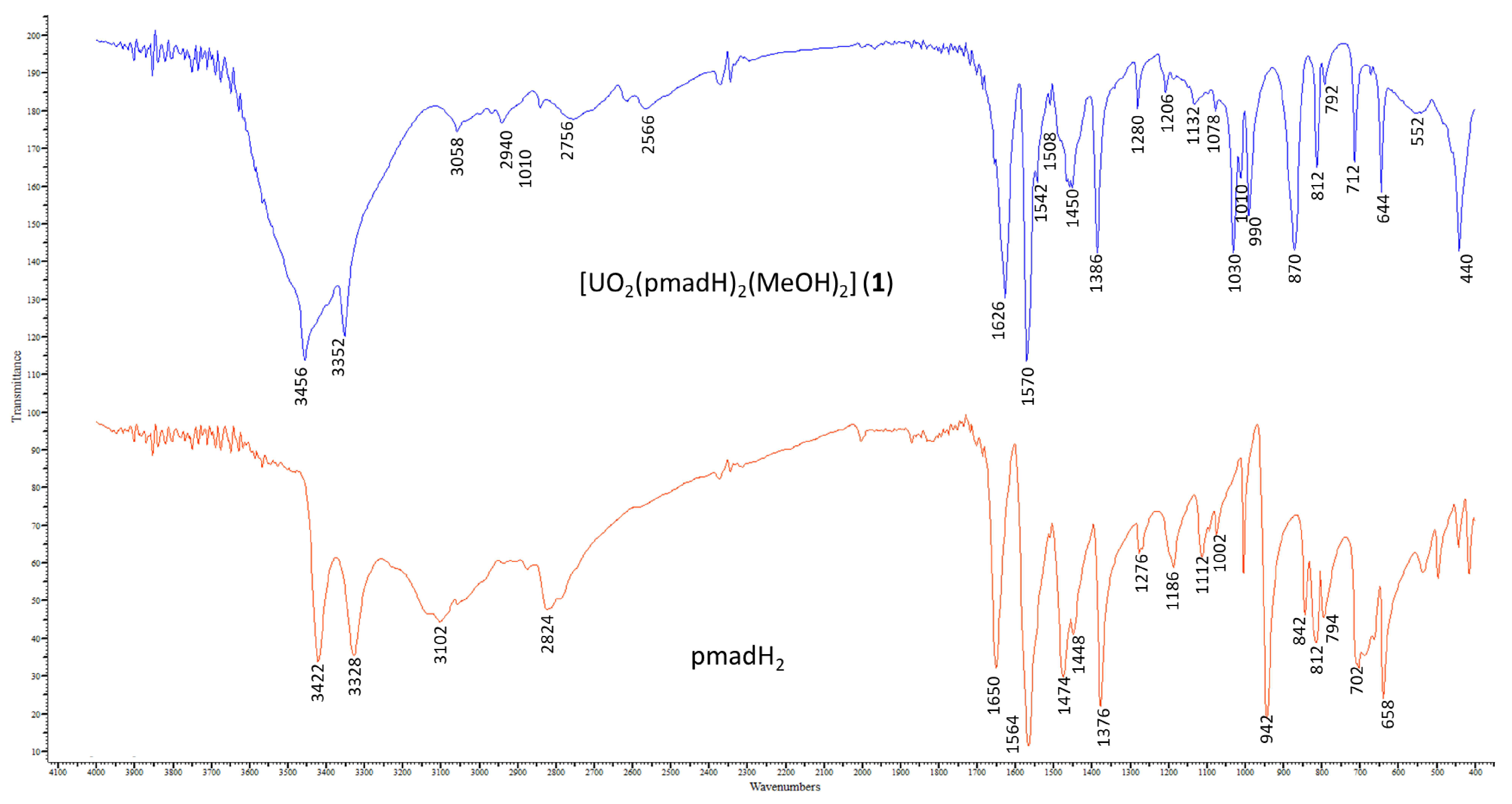
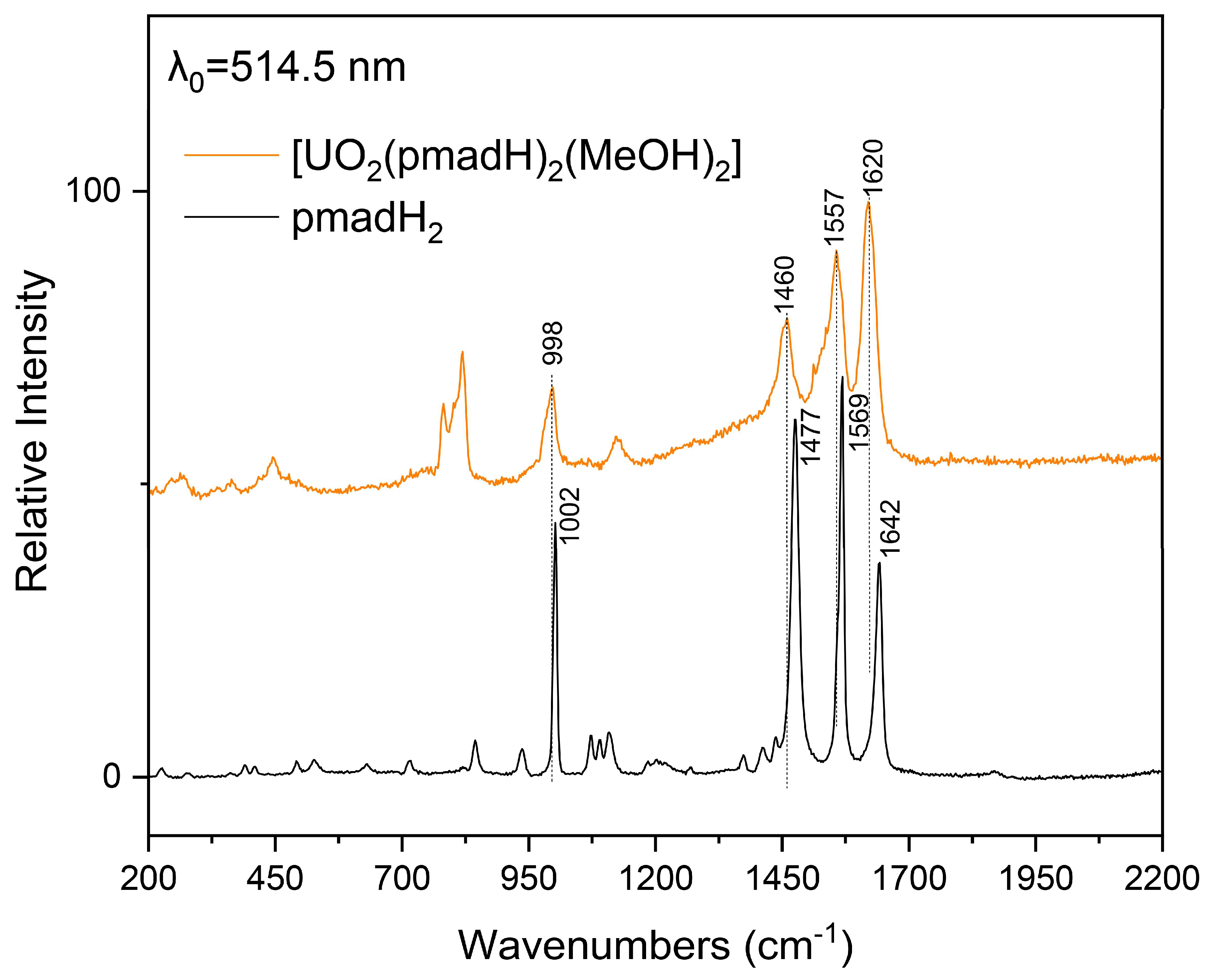
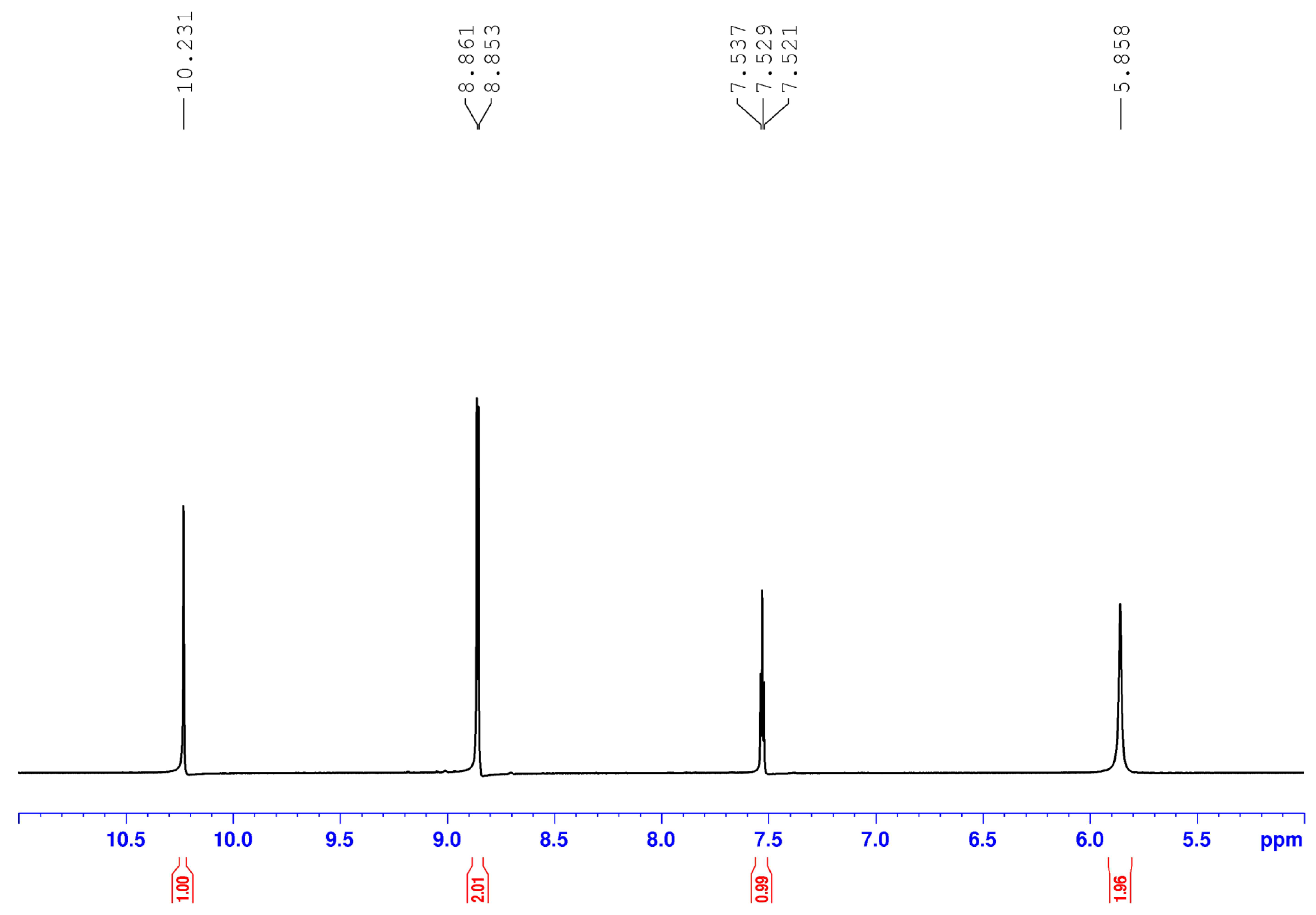
2.3. Spectroscopic Characterization
3. Materials and Methods
3.1. Materials and Instrumentation
3.2. Preparation of the Complex
3.3. Single-Crystal X-ray Crystallography
4. Concluding Comments and Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References and Note
- Rudkevich, D.M.; Verboom, W.; Brzozka, Z.; Palys, M.J.; Stauthamer, W.P.R.V.; van Hummel, G.J.; Franken, S.M.; Harkema, S.; Engbersen, J.F.J.; Reinhoudt, D.N. Functionalized UO2 Salenes: Neutral Receptors for Anions. J. Am. Chem. Soc. 1994, 116, 4341–4351. [Google Scholar] [CrossRef]
- Ai, J.; Chen, F.-Y.; Gao, C.-Y.; Tian, H.-R.; Pan, Q.-J.; Sun, Z.-M. Porous Anionic Uranyl–Organic Networks for Highly Efficient Cs+ Adsorption and Investigation of the Mechanism. Inorg. Chem. 2018, 57, 4419–4426. [Google Scholar] [CrossRef] [PubMed]
- Cowie, B.E.; Purkis, J.M.; Austin, J.; Love, J.B.; Arnold, P.L. Thermal and Photochemical Reduction and Functionalization Chemistry of the Uranyl Dication, [UVI O2]2+. Chem. Rev. 2019, 119, 10595–10637. [Google Scholar] [CrossRef] [PubMed]
- Fortier, S.; Hayton, T.W. Oxo Ligand Functionalization in the Uranyl Ion (UO22+). Coord. Chem. Rev. 2010, 254, 197–214. [Google Scholar] [CrossRef]
- Van Axel Castelli, V.; Dalla Cort, A.; Mandolini, L. Supramolecular Catalysis of 1,4-Thiol Addition by Salophen-Uranyl Complexes. J. Am. Chem. Soc. 1998, 120, 12688–12689. [Google Scholar] [CrossRef]
- Hawkins, C.A.; Bustillos, C.G.; Copping, R.; Scott, B.L.; May, I.; Nilsson, M. Challenging Conventional f-element Separation Chemistry-Reversing Uranyl(VI)/Lanthanide(III) Solvent Extraction Selectivity. Chem. Commun. 2014, 50, 8670–8673. [Google Scholar] [CrossRef] [PubMed]
- Behera, N.; Sethi, S. Unprecedented Catalytic Behavior of Uranyl(VI) Compounds in Chemical Reactions. Eur. J. Inorg. Chem. 2021, 2021, 95–111. [Google Scholar] [CrossRef]
- Abney, C.W.; Mayes, R.T.; Saito, T.; Dai, S. Materials for the Recovery of Uranium from Seawater. Chem. Rev. 2017, 117, 13935–14013. [Google Scholar] [CrossRef]
- Chen, D.; Li, Y.; Zhao, X.; Shi, M.; Shi, X.; Zhao, R.; Zhu, G. Self-Standing Porous Aromatic Framework Electrodes for Efficient Electrochemical Uranium Extraction. ACS Cent. Sci. 2023, 9, 2326–2332. [Google Scholar] [CrossRef]
- Wu, Y.; Xie, Y.; Liu, X.; Li, Y.; Wang, J.; Chen, Z.; Yang, H.; Hu, B.; Shen, C.; Tang, Z.; et al. Functional Nanomaterials for Selective Uranium Recovery from Seawater: Material Design, Extraction Properties and Mechanisms. Coord. Chem. Rev. 2023, 483, 215097. [Google Scholar] [CrossRef]
- Dungan, K.; Butler, G.; Livens, F.R.; Warren, L.M.M. Uranium from Seawater—Infinite Resource or Improbable Aspiration? Prog. Nucl. Energy 2017, 99, 81–85. [Google Scholar] [CrossRef]
- Tang, N.; Liang, J.; Niu, C.; Wang, H.; Luo, Y.; Xing, W.; Ye, S.; Liang, C.; Guo, H.; Guo, J.; et al. Amidoxime-Based Materials for Uranium Recovery and Removal. J. Mater. Chem. A 2020, 8, 7588–7625. [Google Scholar] [CrossRef]
- Barber, P.S.; Kelley, S.P.; Rogers, R.D. Highly Selective Extraction of the Uranyl Ion with Hydrophobic Amidoxime-Functionalized Ionic Liquids via η2 Coordination. RSC Adv. 2012, 2, 8526–8530. [Google Scholar] [CrossRef]
- Das, S.; Oyola, Y.; Mayes, R.T.; Janke, C.J.; Kuo, L.-J.; Gill, G.; Wood, J.R.; Dai, S. Extracting Uranium from Seawater: Promising AF Series Adsorbents. Ind. Eng. Chem. Res. 2016, 55, 4110–4117. [Google Scholar] [CrossRef]
- Ladshaw, A.P.; Wiechert, A.I.; Das, S.; Yiacoumi, S.; Tsouris, C. Amidoxime Polymers for Uranium Adsorption: Influence of Comonomers and Temperature. Materials 2017, 10, 1268. [Google Scholar] [CrossRef] [PubMed]
- Kravchuk, D.V.; Blanes Diaz, A.; Carolan, M.E.; Mpundu, E.A.; Cwiertny, D.M.; Forbes, T.Z. Uranyl Speciation on the Surface of Amidoximated Polyacrylonitrile Mats. Inorg. Chem. 2020, 59, 8134–8145. [Google Scholar] [CrossRef] [PubMed]
- Endrizzi, F.; Melchior, A.; Tolazzi, M.; Rao, L. Complexation of Uranium(VI) with Glutarimidoxioxime: Thermodynamic and Computational Studies. Dalton Trans. 2015, 44, 13835–13844. [Google Scholar] [CrossRef]
- Vukovic, S.; Watson, L.A.; Kang, S.O.; Custelcean, R.; Hay, B.P. How Amidoximate Binds the Uranyl Cation. Inorg. Chem. 2012, 51, 3855–3859. [Google Scholar] [CrossRef] [PubMed]
- Witte, E.G.; Schwochau, K.S.; Henkel, G.; Krebs, B. Uranyl Complexes of Acetamidoxime and Benzamidoxime. Preparation, Characterization, and Crystal Structure. Inorganica Chim. Acta 1984, 94, 323–331. [Google Scholar] [CrossRef]
- Tian, G.; Teat, S.J.; Zhang, Z.; Rao, L. Sequestering Uranium from Seawater: Binding Strength and Modes of Uranyl Complexes with Glutarimidedioxime. Dalton Trans. 2012, 41, 11579–11586. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, K.J.; Do-Thanh, C.L.; Penchoff, D.A.; Cramer, S.A.; Murdock, C.R.; Lu, Z.; Harrison, R.J.; Camden, J.P.; Jenkins, D.M. The Synthesis and Spectroscopic Characterization of an Aromatic Uranium Amidoxime Complex. Inorganica Chim. Acta 2014, 421, 374–379. [Google Scholar] [CrossRef]
- Kelley, S.P.; Barber, P.S.; Mullins, P.H.K.; Rogers, R.D. Structural Clues to UO22+/VO2+ Competition in Seawater Extraction Using Amidoxime-Based Extractants. Chem. Commun. 2014, 50, 12504–12507. [Google Scholar] [CrossRef]
- Sun, Q.; Aguila, B.; Perman, J.; Ivanov, A.S.; Bryantsev, V.S.; Earl, L.D.; Abney, C.W.; Wojtas, L.; Ma, S. Bio-Inspired Nano-Traps for Uranium Extraction from Seawater and Recovery from Nuclear Waste. Nat. Commun. 2018, 9, 1644. [Google Scholar] [CrossRef]
- Mishra, M.K.; Patil, Y.P.; Kelley, S.P.; Rogers, R.D. A Uranyl Metal Organic Framework Arising from the Coordination of a Partially Hydrolyzed Tetrauranyl Node with the Tautomerically Diverse 1,4-(Diamidoximyl)benzene Ligand. Cryst. Growth Des. 2019, 19, 5466–5470. [Google Scholar] [CrossRef]
- Decato, D.A.; Berryman, O.B. Structural and Computational Characterization of a Bridging Zwitterionic-Amidoxime Uranyl Complex. Org. Chem. Front. 2019, 6, 1038–1043. [Google Scholar] [CrossRef]
- Kennedy, Z.C.; Cardenas, A.J.P.; Corbey, J.F.; Warner, M.G. 2,6-Diiminopiperidin-1-ol: An Overlooked Motif Relevant to Uranyl and Transition Metal Binding on Poly(Amidoxime) Adsorbents. Chem. Commun. 2016, 52, 8802–8805. [Google Scholar] [CrossRef]
- For a recent, comprehensive and critical review about the chemistry, structures and technological implications of the to-date characterized uranyl-amidoxime complexes, see: Tsantis, S.T.; Iliopoulou, M.; Tzimopoulos, D.I.; Perlepes, S.P. Synthetic and Structural Chemistry of Uranyl-Amidoxime Complexes: Technological Implications. Chemistry, 2023; 5, 1419–1453.
- Zhang, L.; Qie, M.; Su, J.; Zhang, S.; Zhou, J.; Li, J.; Wang, Y.; Yang, S.; Wang, S.; Li, J.; et al. Tris-Amidoximate Uranyl Complexes via η2 Binding Mode Coordinated in Aqueous Solution Shown by X-ray Absorption Spectroscopy and Density Functional Theory Methods. J. Synchrotron Radiat. 2018, 25, 514–522. [Google Scholar] [CrossRef]
- Abney, C.W.; Mayes, R.T.; Piechowicz, M.; Lin, Z.; Bryantsev, V.S.; Veith, G.M.; Dai, S.; Lin, W. XAFS Investigation of Polyamidoxime-Bound Uranyl Contests the Paradigm from Small Molecule Studies. Energy Environ. Sci. 2016, 9, 448–453. [Google Scholar] [CrossRef]
- Tian, G.; Teat, S.J.; Rao, L. Thermodynamic Studies of U(VI) Complexation with Glutardiamidoxime for Sequestration of Uranium from Seawater. Dalton Trans. 2013, 42, 5690–5696. [Google Scholar] [CrossRef]
- Li, B.; Priest, C.; Jiang, D.E. Displacement of Carbonates in Ca2UO2(CO3)3 by Amidoxime-Based Ligands from Free-Energy Simulations. Dalton Trans. 2018, 47, 1604–1613. [Google Scholar] [CrossRef]
- Priest, C.; Li, B.; Jiang, D. Understanding the Binding of a Bifunctional Amidoximate–Carboxylate Ligand with Uranyl in Seawater. J. Phys. Chem. B 2018, 122, 12060–12066. [Google Scholar] [CrossRef]
- Wang, C.-Z.; Lan, J.-H.; Wu, Q.-Y.; Luo, Q.; Zhao, Y.-L.; Wang, X.-K.; Chai, Z.-F.; Shi, W.-Q. Theoretical Insights on the Interaction of Uranium with Amidoxime and Carboxyl Groups. Inorg. Chem. 2014, 53, 9466–9476. [Google Scholar] [CrossRef]
- Yin, X.-J.; Zhu, L.-G. One Ni(II) Polymer with Fluorescent Response Based on Mixed Ligands. ChemistrySelect 2018, 3, 10550–10552. [Google Scholar] [CrossRef]
- Yin, X.-J.; Zhu, L.-G. Structural Variation from Trinuclears to 1D Chains: Syntheses, Structures and Properties. Appl. Organomet. Chem. 2019, 33, e4796. [Google Scholar] [CrossRef]
- Palacios, M.A.; Mota, A.J.; Perea-Buceta, J.E.; White, F.J.; Brechin, E.K.; Colacio, E. Antiferromagnetic versus Ferromagnetic Exchange Interactions in Bis(μ-oximate)dinickel(II) Units for a Series of Closely Related Cube Shaped Carboxamideoximate-Bridged Ni4 Complexes. A Combined Experimental and Theoretical Magneto-Structural Study. Inorg. Chem. 2010, 49, 10156–10165. [Google Scholar] [CrossRef]
- Kalogridis, C.; Palacios, M.A.; Rodríguez-Diéguez, A.; Mota, A.J.; Choquesillo-Lazarte, D.; Brechin, E.K.; Colacio, E. Heterometallic Oximato-Bridged Linear Trinuclear NiII−MIII−NiII (MIII = Mn, Fe, Tb) Complexes Constructed with the fac-O3 [Ni(HL)3]− Metalloligand (H2L=pyrimidine-2-carboxamide oxime): A Theoretical and Experimental Magneto-Structural Study. Eur. J. Inorg. Chem. 2011, 2011, 5225–5232. [Google Scholar] [CrossRef]
- Yin, X.-J.; Zhu, L.-G. The Crystal Structure of Bis(acetato-κ1O)-Bis(N′-hydroxypyrimidine-2-carboximidamide-κ2N,N′) Manganese(II)—Methanol(1/2), C 14H18MnN8O6, 2(CH3OH)′. Z. Kristallogr. NCS 2019, 234, 925–926. [Google Scholar]
- Chen, K.; Wang, M.; Wang, D.; Zhou, X. A Bifunctional Fluorescence Sensing Coordination Polymer for Fe3+ and MnO4−. Inorg. Chim. Acta 2021, 525, 120467. [Google Scholar] [CrossRef]
- Saha, U.; Dolai, M.; Kumar, G.S.; Butcher, R.J.; Konar, S. New DNA-Interactive Manganese(II) Complex of Amidooxime: Crystal Structure, DFT Calculation, Biophysical and Molecular Docking Studies. J. Chem. Eng. Data 2020, 65, 5393–5404. [Google Scholar] [CrossRef]
- Dolai, M.; Saha, U.; Kumar, G.S.; Ali, M. Amidooxime-Based Mononuclear Mn(II) Complexes: Synthesis, Characterization, and Studies on DNA Binding and Nuclease Activity. ChemistrySelect 2018, 3, 6935–6941. [Google Scholar] [CrossRef]
- Gole, B.; Chakrabarty, R.; Mukherjee, S.; Song, Y.; Mukherjee, P.S. Use of 2-Pyrimidineamidoxime to Generate Polynuclear Homo-/Heterometallic Assemblies: Synthesis, Crystal Structures and Magnetic Study with Theoretical Investigations on the Exchange Mechanism. Dalton Trans. 2010, 39, 9766–9778. [Google Scholar] [CrossRef]
- Yin, X.J.; Zhu, L.G. Syntheses, Structures, and Properties of Two Coordination Polymers Based on 2-Pyrimidineamidoxime. Russ. J. Coord. Chem. 2018, 44, 448–453. [Google Scholar] [CrossRef]
- Yin, X.-J.; Lu, J.-Y.; Zhou, L.-G. Synthesis, Crystal Structure and Magnetic Studies of Two Mn(II) Complexes Based on 2-Pyrimidineamidoxime. Chin. J. Struct. Chem. 2020, 38, 1970–1976. [Google Scholar]
- Sahyoun, T.; Arrault, A.; Schneider, R. Amidoximes and Oximes: Synthesis, Structure, and Their Key Role as NO Donors. Molecules 2019, 24, 2470. [Google Scholar] [CrossRef]
- Aakeröy, C.B.; Sinha, A.S.; Epa, K.N.; Chopade, P.D.; Smith, M.M.; Desper, J. Structural Chemistry of Oximes. Cryst. Growth Des. 2013, 13, 2687–2695. [Google Scholar] [CrossRef]
- Tavakol, H.; Arshadi, S. Theoretical Investigation of Tautomerism in N-Hydroxy Amidines. J. Mol. Model. 2009, 15, 807–816. [Google Scholar] [CrossRef]
- Novikov, A.S.; Bolotin, D.S. Tautomerism of Amidoximes and Other Oxime Species. J. Phys. Org. Chem. 2018, 31, e3772. [Google Scholar] [CrossRef]
- Bolotin, D.S.; Bokach, N.A.; Kukushkin, V.Y. Coordination Chemistry and Metal-Involving Reactions of Amidoximes: Relevance to the Chemistry of Oximes and Oxime Ligands. Coord. Chem. Rev. 2016, 313, 62–93. [Google Scholar] [CrossRef]
- Coxall, R.A.; Harris, S.G.; Henderson, D.K.; Parsons, S.; Tasker, P.A.; Winpenny, R.E.P. Inter-Ligand Reactions: In Situ Formation of New Polydentate Ligands. J. Chem. Soc. Dalton Trans. 2000, 14, 2349–2356. [Google Scholar] [CrossRef]
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds, 4th ed.; Wiley: New York, NY, USA, 1986; pp. 109–113. [Google Scholar]
- Tsantis, S.T.; Bekiari, V.; Raptopoulou, C.P.; Tzimopoulos, D.I.; Psycharis, V.; Perlepes, S.P. Dioxidouranium(IV) Complexes with Schiff Bases Possessing an ONO Donor Set: Synthetic, Structural and Spectroscopic Studies. Polyhedron 2018, 152, 172–178. [Google Scholar] [CrossRef]
- Dollish, F.R.; Fateley, W.G.; Bentley, F.F. Characteristic Raman Frequencies of Organic Compounds; Wiley: New York, NY, USA, 1974; pp. 135–137. [Google Scholar]
- Holynska, M. Formation of Ni(II) Oxime-Bridged Basket-Like Complexes and Their Structural Aspects. Curr. Inorg. Chem. 2015, 5, 64–70. [Google Scholar] [CrossRef]
- Bullock, J.I. Raman and Infrared Spectroscopic Studies of the Uranyl Ion: The Symmetric Stretching Frequency, Force Constants, and Bond Lengths. J. Chem. Soc. A 1969, 781–784. [Google Scholar] [CrossRef]
- Silver, M.A.; Dorfner, W.L.; Cary, S.K.; Cross, J.N.; Lin, J.; Schelter, E.J.; Albrecht-Schmitt, T.E. Why Is Uranyl Formohydroxamate Red? Inorg. Chem. 2015, 54, 5280–5284. [Google Scholar] [CrossRef]
- Geary, W.J. The Use of Conductivity Measurements in Organic Solvents for the Characterisation of Coordination Compounds. Coord. Chem. Rev. 1971, 7, 81–122. [Google Scholar] [CrossRef]
- Fulmer, G.R.; Miller, A.J.M.; Sherden, N.H.; Gottlieb, H.E.; Nudelman, A.; Stoltz, B.M.; Bercaw, J.E.; Goldberg, K.I. NMR Chemical Shifts of Trace Impurities: Common Laboratory Solvents, Organics, and Gases in Deuterated Solvents Relevant to the Organometallic Chemist. Organometallics 2010, 29, 2176–2179. [Google Scholar] [CrossRef]
- Chen, Y.; Tang, T.; Chen, Y.; Xu, D. Novel 1,8-Naphthalimide Dye for Multichannel Sensing of H+ and Cu2+. Res. Chem. Intermed. 2018, 44, 2379–2393. [Google Scholar] [CrossRef]
- CrystalClear; Rigaku: The Woodlands, TX, USA; MSC Inc.: Mellville, NY, USA, 2005.
- Sheldrick, G.M. A Short History of SHELX. Acta Crystallogr. Sect. A 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. Sect. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Diamond—Crystal and Molecular Structure Visualization, Version 3.1; Crystal Impact: Bonn, Germany, 2018.


| Parameter | [UO2(pmadH)2(MeOH)2] (1) |
|---|---|
| Empirical formula | C12H18UN8O6 |
| Formula weight | 608.37 |
| Crystal system | triclinic |
| Space group | P |
| Color | orange |
| Crystal size, mm | 0.18 × 1.16 × 0.11 |
| Crystal habit | parallelepiped |
| a, Å | 7.4659(7) |
| b, Å | 7.9915(8) |
| c, Å | 8.7067(8) |
| α, ° | 108.854(3) |
| β, ° | 103.664(3) |
| γ, ° | 104.709(3) |
| Volume, Å3 | 445.82(7) |
| Z | 1 |
| Temperature, K | 180 |
| Radiation, Å | Mo Κα, 0.71073 |
| Calculated density, g∙cm−3 | 2.266 |
| Absorption coefficient, mm−1 | 9.15 |
| No. of measured, independent, and observed [Ι > 2σ(Ι)] reflections | 13,557, 1946, 1946 |
| Rint | 0.030 |
| Number of parameters | 152 |
| Final R indices [Ι > 2σ(Ι)] a | R1 = 0.0136, wR2 = 0.0332 |
| Goodness-of-fit on F2 | 1.12 |
| Largest differences peak and hole (e Å−3) | 1.07/−0.72 |
| Bond Lengths (Å) | Bond Angles (°) | ||
|---|---|---|---|
| U1-O3 | 1.803(2) | O3-U1-O3′ | 180.0(1) |
| U1-O1 | 2.324(2) | O3-U1-O1 | 90.3(1) |
| U1-N1 | 2.405(2) | O3-U1-O1′ | 89.7(1) |
| U1-O2 | 2.455(2) | O3-U1-N1 | 90.4(1) |
| O1-N1 | 1.379(3) | O3-U1-O2 | 90.8(1) |
| N1-C1 | 1.293(3) | O1-U1-N1 | 33.9(1) |
| N2-C1 | 1.356(3) | O1-U1-O2 | 106.8(1) |
| C1-C2 | 1.484(3) | O2-U1-N1 | 73.0(1) |
| N3-C2 | 1.334(3) | O1-N1-C1 | 115.9(2) |
| N3-C5 | 1.344(3) | N1-C1-N2 | 122.7(2) |
| N4-C2 | 1.341(3) | N1-C1-C2 | 118.4(2) |
| N4-C3 | 1.339(3) | N2-C1-C2 | 118.8(2) |
| D-H⋯A | d(D⋯A) | d(H⋯A) | <DHA | Symmetry Code of A |
|---|---|---|---|---|
| O2-H(O2)⋯N4 | 2.699(3) | 1.95(4) | 180(5) | |
| C3-H(C3)⋯O3 a | 3.446(3) | 2.59(4) | 158(3) | x + 1, y, z |
| C5-H(C5)⋯N3 a | 3.497(3) | 2.69(3) | 152(3) | −x + 3, −y − 1, −z + 1 |
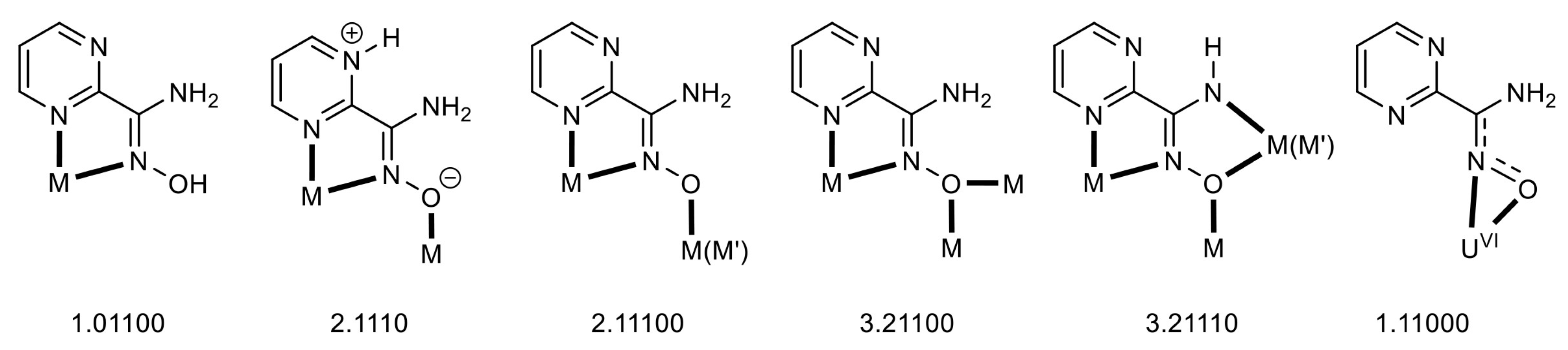
| Complex b,c | Coordination Mode(s) | Ref. |
|---|---|---|
| {[Cd(pmadH2)2(L1)]}n | 1.01100 | [43] |
| {[Cd(pmadH2)(L2)(H2O)]}n | 1.01100 | [43] |
| {[Cd2(L3)(pmadH2)2(H2O)2]}n | 1.01100 | [39] |
| [MnCl2(pmadH2)2] | 1.01100 | [41] |
| [MnCl(O2CMe)(pmadH2)2] | 1.01100 | [40] |
| [Mn(O2CMe)2(pmadH2)2] | 1.01100 | [38] |
| {[Co(pmadH2)2(L1)]}n | 1.01100 | [35] |
| {[Co(pmadH2)2(L4)]}n | 1.01100 | [35] |
| {[Co(pmadH2)(SO4)(H2O)2]}n | 1.01100 | [35] |
| {[Mn(pmadH2)2(L4)]}n | 1.01100 | [44] |
| {[Mn(pmadH2)(L2)(H2O)]}n | 1.01100 | [44] |
| [Co3(pmadH2)6](O2CMe)2(L4)2 | 2.1110 | [35] |
| [Co3(pmadH2)6](O2CMe)2(L1)2 | 2.1110 | [35] |
| {[Cu(pmadH2)(NO3)](NO3)}n | 1.01100 | [42] |
| {[Ni(pmadH2)2(L4)]}n | 1.01100 | [34] |
| [Ni2Mn(pmadH)6](ClO4) | 2.11100 | [37] |
| [Ni2Fe(pmadH)6](ClO4) | 2.11100 | [37] |
| [Ni2Tb(pmadH)6](NO3) | 2.11100 | [37] |
| [Ni4(pmadH)4(O2CPh)2(pyz)2(MeOH)2](ClO4) | 3.21100 | [36] |
| [Cu4(pmadH)2(pmad)2(NO3)](NO3) | 2.11100, 3.21110 | [42] |
| [Cu2Ni2(pmadH)2(pmad)2Cl2] | 2.11100, 3.21110 | [42] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsantis, S.T.; Lada, Z.G.; Skiadas, S.G.; Tzimopoulos, D.I.; Raptopoulou, C.P.; Psycharis, V.; Perlepes, S.P. Understanding the Selective Extraction of the Uranyl Ion from Seawater with Amidoxime-Functionalized Materials: Uranyl Complexes of Pyrimidine-2-amidoxime. Inorganics 2024, 12, 82. https://doi.org/10.3390/inorganics12030082
Tsantis ST, Lada ZG, Skiadas SG, Tzimopoulos DI, Raptopoulou CP, Psycharis V, Perlepes SP. Understanding the Selective Extraction of the Uranyl Ion from Seawater with Amidoxime-Functionalized Materials: Uranyl Complexes of Pyrimidine-2-amidoxime. Inorganics. 2024; 12(3):82. https://doi.org/10.3390/inorganics12030082
Chicago/Turabian StyleTsantis, Sokratis T., Zoi G. Lada, Sotiris G. Skiadas, Demetrios I. Tzimopoulos, Catherine P. Raptopoulou, Vassilis Psycharis, and Spyros P. Perlepes. 2024. "Understanding the Selective Extraction of the Uranyl Ion from Seawater with Amidoxime-Functionalized Materials: Uranyl Complexes of Pyrimidine-2-amidoxime" Inorganics 12, no. 3: 82. https://doi.org/10.3390/inorganics12030082
APA StyleTsantis, S. T., Lada, Z. G., Skiadas, S. G., Tzimopoulos, D. I., Raptopoulou, C. P., Psycharis, V., & Perlepes, S. P. (2024). Understanding the Selective Extraction of the Uranyl Ion from Seawater with Amidoxime-Functionalized Materials: Uranyl Complexes of Pyrimidine-2-amidoxime. Inorganics, 12(3), 82. https://doi.org/10.3390/inorganics12030082














