Interaction of Carbon Nanotubes, Capped Carbon Nanotubes, CNT2–5, C60, C70, HO-C60, [C60]2, and [C60]3 Fullerenes with Virulence Factors of Gram-Negative and Gram-Positive Bacteria: Potential Applications for 3D-Printed Scaffolds
Abstract
1. Introduction
2. Results
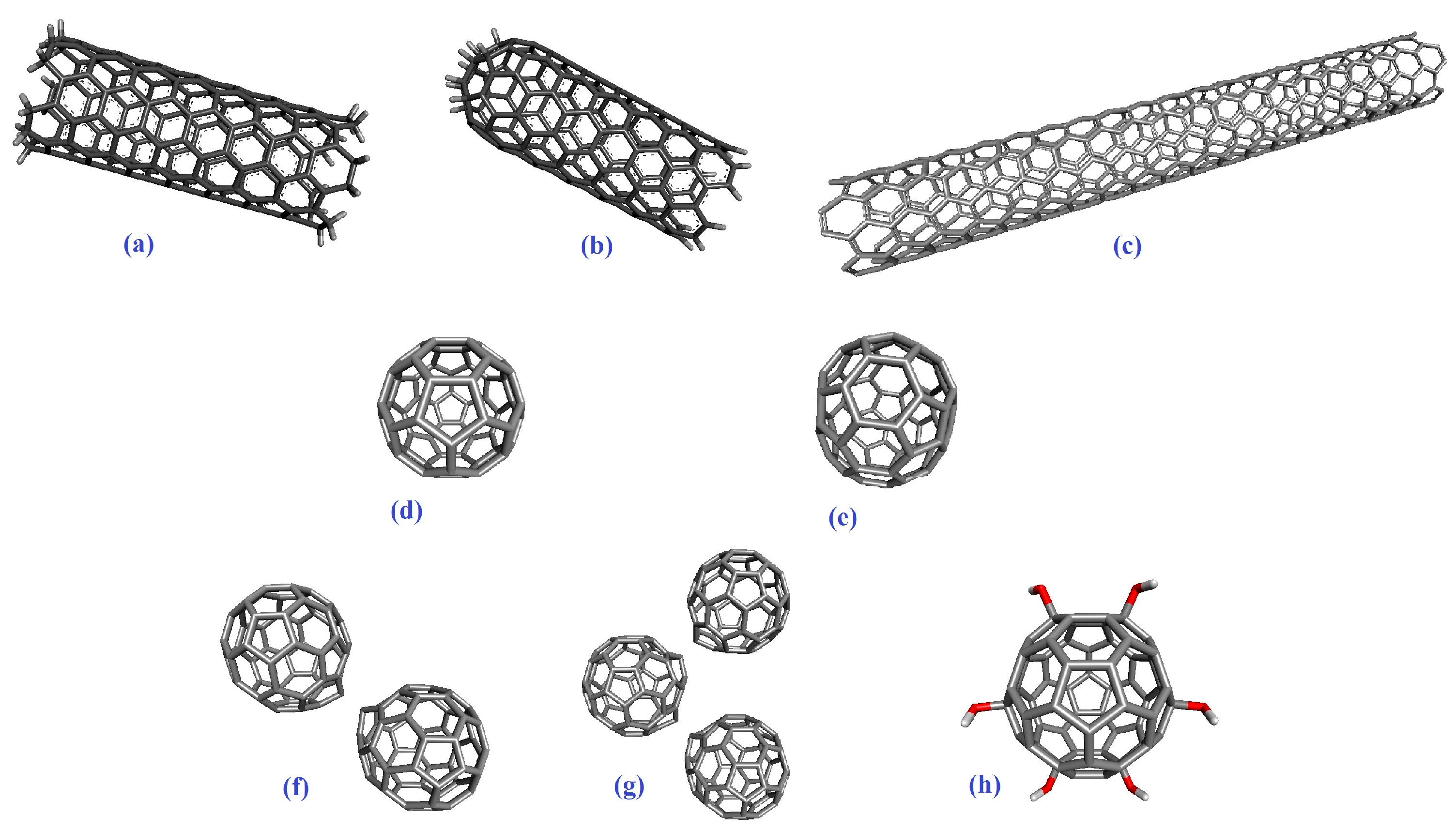
2.1. Molecular Docking
2.2. The Normal Mode Analysis
3. Discussion
4. Materials and Methods
4.1. Molecular Docking Preparation
4.2. The Normal Mode Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Amraei, S.; Eslami, G.; Taherpour, A.; Hashemi, A. Relationship between MOX genes and antibiotic resistance in Klebsiella pneumoniae strains in nosocomial infections. Micro Nano Bio Asp. 2022, 1, 12–17. [Google Scholar] [CrossRef]
- Amraei, S.; Eslami, G.; Taherpour, A.; Hashemi, A. The role of ACT and FOX genes in Klebsiella pneumoniae strains isolated from hospitalized patients. Micro Nano Bio Asp. 2022, 1, 18–25. [Google Scholar] [CrossRef]
- Amraei, S.; Ahmadi, S. Recent studies on antimicrobial and anticancer activities of saponins: A mini-review. Nano Micro Biosyst. 2022, 1, 22–26. [Google Scholar] [CrossRef]
- Alavi, M.; Jabari, E.; Jabbari, E. Functionalized carbon-based nanomaterials and quantum dots with antibacterial activity: A review. Expert Rev. Anti Infect. Ther. 2021, 19, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Tian, Y.; Ouyang, J.; Shen, Y.; Wang, X.; Luan, J. Carbon nanomaterials for drug delivery and tissue engineering. Front. Chem. 2022, 10, 990362. [Google Scholar] [CrossRef] [PubMed]
- Riha, S.M.; Maarof, M.; Fauzi, M.B. Synergistic Effect of Biomaterial and Stem Cell for Skin Tissue Engineering in Cutaneous Wound Healing: A Concise Review. Polymers 2021, 13, 1546. [Google Scholar] [CrossRef]
- Qiu, Y.L.; Chen, X.; Hou, Y.L.; Hou, Y.J.; Tian, S.B.; Chen, Y.H.; Yu, L.; Nie, M.H.; Liu, X.Q. Characterization of different biodegradable scaffolds in tissue engineering. Mol. Med. Rep. 2019, 19, 4043–4056. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, Z.; Husain, S.; Alotaibi, H.; Rehman, R.; Zafar, M.S.; Farooq, I.; Khan, A.S. Chapter 18-Novel Techniques of Scaffold Fabrication for Bioactive Glasses. In Biomedical, Therapeutic and Clinical Applications of Bioactive Glasses; Kaur, G., Ed.; Woodhead Publishing: Sawston, UK, 2019; pp. 497–519. [Google Scholar] [CrossRef]
- Suamte, L.; Tirkey, A.; Barman, J.; Jayasekhar Babu, P. Various manufacturing methods and ideal properties of scaffolds for tissue engineering applications. Smart Mater. Manuf. 2023, 1, 100011. [Google Scholar] [CrossRef]
- Kołbuk, D.; Heljak, M.; Choińska, E.; Urbanek, O. Novel 3D Hybrid Nanofiber Scaffolds for Bone Regeneration. Polymers 2020, 12, 544. [Google Scholar] [CrossRef]
- Chung, J.J.; Im, H.; Kim, S.H.; Park, J.W.; Jung, Y. Toward Biomimetic Scaffolds for Tissue Engineering: 3D Printing Techniques in Regenerative Medicine. Front. Bioeng. Biotechnol. 2020, 8, 586406. [Google Scholar] [CrossRef]
- Qu, M.; Wang, C.; Zhou, X.; Libanori, A.; Jiang, X.; Xu, W.; Zhu, S.; Chen, Q.; Sun, W.; Khademhosseini, A. Multi-Dimensional Printing for Bone Tissue Engineering. Adv. Healthc. Mater. 2021, 10, 2001986. [Google Scholar] [CrossRef]
- Shahrubudin, N.; Lee, T.C.; Ramlan, R. An Overview on 3D Printing Technology: Technological, Materials, and Applications. Procedia Manuf. 2019, 35, 1286–1296. [Google Scholar] [CrossRef]
- Bhagia, S.; Bornani, K.; Agrawal, R.; Satlewal, A.; Ďurkovič, J.; Lagaňa, R.; Bhagia, M.; Yoo, C.G.; Zhao, X.; Kunc, V.; et al. Critical review of FDM 3D printing of PLA biocomposites filled with biomass resources, characterization, biodegradability, upcycling and opportunities for biorefineries. Appl. Mater. Today 2021, 24, 101078. [Google Scholar] [CrossRef]
- Zhao, Y.-Q.; Yang, J.-H.; Ding, X.; Ding, X.; Duan, S.; Xu, F.-J. Polycaprolactone/polysaccharide functional composites for low-temperature fused deposition modelling. Bioact. Mater. 2020, 5, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Wasserfall, F.; Hendrich, N.; Ahlers, D.; Zhang, J. Topology-aware routing of 3D-printed circuits. Addit. Manuf. 2020, 36, 101523. [Google Scholar] [CrossRef]
- Cui, M.; Pan, H.; Su, Y.; Fang, D.; Qiao, S.; Ding, P.; Pan, W. Opportunities and challenges of three-dimensional printing technology in pharmaceutical formulation development. Acta Pharm. Sin. B 2021, 11, 2488–2504. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, T.; Qurban, R.O.; Bolarinwa, S.O.; Mirza, A.A.; Pasovic, M.; Memic, A. 3D Printing of Metal/Metal Oxide Incorporated Thermoplastic Nanocomposites with Antimicrobial Properties. Front. Bioeng. Biotechnol. 2020, 8, 568186. [Google Scholar] [CrossRef] [PubMed]
- Patila, M.; Chalmpes, N.; Dounousi, E.; Stamatis, H.; Gournis, D. Chapter Twelve—Use of functionalized carbon nanotubes for the development of robust nanobiocatalysts. In Methods Enzymology; Kumar, C.V., Ed.; Academic Press: Cambridge, MA, USA, 2020; Volume 630, pp. 263–301. [Google Scholar]
- Ardeshana, B.; Jani, U.; Patel, A. Influence of Bending Angle on Mechanical Performance of SWCNTs and DWCNTs Based on Molecular Mechanics: FE Approach. J. Vib. Eng. Technol. 2022, 11, 251–264. [Google Scholar] [CrossRef]
- Nadeem, M.F.; Azeem, M.; Farman, I. Comparative study of topological indices for capped and uncapped carbon nanotubes. Polycycl. Aromat. Compd. 2022, 42, 4666–4683. [Google Scholar] [CrossRef]
- Cui, H.; Yu, Y.; Li, X.; Sun, Z.; Ruan, J.; Wu, Z.; Qian, J.; Yin, J. Direct 3D printing of a tough hydrogel incorporated with carbon nanotubes for bone regeneration. J. Mater. Chem. B 2019, 7, 7207–7217. [Google Scholar] [CrossRef]
- Li, L.; Qin, S.; Peng, J.; Chen, A.; Nie, Y.; Liu, T.; Song, K. Engineering gelatin-based alginate/carbon nanotubes blend bioink for direct 3D printing of vessel constructs. Int. J. Biol. Macromol. 2020, 145, 262–271. [Google Scholar] [CrossRef]
- Young, Y.-F.; Lee, H.-J.; Shen, Y.-S.; Tseng, S.-H.; Lee, C.-Y.; Tai, N.-H.; Chang, H.-Y. Toxicity mechanism of carbon nanotubes on Escherichia coli. Mater. Chem. Phys. 2012, 134, 279–286. [Google Scholar] [CrossRef]
- Sui, M.; Zhang, L.; Sheng, L.; Huang, S.; She, L. Synthesis of ZnO coated multi-walled carbon nanotubes and their antibacterial activities. Sci. Total Environ. 2013, 452–453, 148–154. [Google Scholar] [CrossRef]
- Kang, S.; Pinault, M.; Pfefferle, L.D.; Elimelech, M. Single-Walled Carbon Nanotubes Exhibit Strong Antimicrobial Activity. Langmuir 2007, 23, 8670–8673. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Xia, X.; Xia, N.; Zhang, S.; Guo, X. Modification of Fatty Acids in Membranes of Bacteria: Implication for an Adaptive Mechanism to the Toxicity of Carbon Nanotubes. Environ. Sci. Technol. 2014, 48, 4086–4095. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Shi, G.; Wu, S.; Mo, J.; Shen, L.; Zhang, X.; Zhu, Y. Application of Fullerenes as Photosensitizers for Antimicrobial Photodynamic Inactivation: A Review. Front. Microbiol. 2022, 13, 957698. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, Y.; Shi, Y.; Song, H.; Yu, C. Antibiotic-Free Antibacterial Strategies Enabled by Nanomaterials: Progress and Perspectives. Adv. Mater. 2020, 32, 1904106. [Google Scholar] [CrossRef] [PubMed]
- Hada, V.; Chaturvedi, K.; Singhwane, A.; Siraj, N.; Gupta, A.; Sathish, N.; Chaurasia, J.P.; Srivastava, A.K.; Verma, S. Nanoantibiotic effect of carbon-based nanocomposites: Epicentric on graphene, carbon nanotubes and fullerene composites: A review. 3 Biotech 2023, 13, 147. [Google Scholar] [CrossRef] [PubMed]
- Krishna, R.H.; Chandraprabha, M.N.; Monika, P.; Br, T.; Chaudhary, V.; Manjunatha, C. Biomolecule conjugated inorganic nanoparticles for biomedical applications: A review. Biotechnol. Genet. Eng. Rev. 2022, 1–42. [Google Scholar] [CrossRef] [PubMed]
- Abdelsattar, A.S.; Dawoud, A.; Helal, M.A. Interaction of nanoparticles with biological macromolecules: A review of molecular docking studies. Nanotoxicology 2021, 15, 66–95. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, S.M.; Abd El-Baky, R.M.; Abourehab, M.A.S.; Fadl, G.F.M.; Gamil, N.G.F.M. Prevalence of Quorum Sensing and Virulence Factor Genes among Pseudomonas aeruginosa Isolated from Patients Suffering from Different Infections and Their Association with Antimicrobial Resistance. Infect. Drug Resist. 2023, 16, 2371–2385. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Gupta, R.K.; Harjai, K. Multiple virulence factors regulated by quorum sensing may help in establishment and colonisation of urinary tract by Pseudomonas aeruginosa during experimental urinary tract infection. Indian J. Med. Microbiol. 2013, 31, 29–33. [Google Scholar] [CrossRef]
- Seilie, E.S.; Bubeck Wardenburg, J. Staphylococcus aureus pore-forming toxins: The interface of pathogen and host complexity. Semin. Cell Dev. Biol. 2017, 72, 101–116. [Google Scholar] [CrossRef]
- Fishovitz, J.; Hermoso, J.A.; Chang, M.; Mobashery, S. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life 2014, 66, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, V.; Okon, B.P.; Satsangi, A.T.; Das, S.; Waturuocha, U.W.; Vashist, A.; Clark-Curtiss, J.E.; Saini, D.K. Mycobacterium tuberculosis PknK Substrate Profiling Reveals Essential Transcription Terminator Protein Rho and Two-Component Response Regulators PrrA and MtrA as Novel Targets for Phosphorylation. Microbiol. Spectr. 2022, 10, e01354-21. [Google Scholar] [CrossRef] [PubMed]
- Gijsbers, A.; Vinciauskaite, V.; Siroy, A.; Gao, Y.; Tria, G.; Mathew, A.; Sánchez-Puig, N.; López-Iglesias, C.; Peters, P.J.; Ravelli, R.B.G. Priming mycobacterial ESX-secreted protein B to form a channel-like structure. Curr. Res. Struct. Biol. 2021, 3, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, A.N. Emerging agents of gastroenteritis: Aeromonas, Plesiomonas, and the diarrheagenic pathotypes of Escherichia coli. Semin. Diagn. Pathol. 2019, 36, 187–192. [Google Scholar] [CrossRef]
- Johura, F.-T.; Parveen, R.; Islam, A.; Sadique, A.; Rahim, M.N.; Monira, S.; Khan, A.R.; Ahsan, S.; Ohnishi, M.; Watanabe, H.; et al. Occurrence of Hybrid Escherichia coli Strains Carrying Shiga Toxin and Heat-Stable Toxin in Livestock of Bangladesh. Front. Public Health 2017, 4, 287. [Google Scholar] [CrossRef][Green Version]
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef]
- Bhattacharjee, M.; Banerjee, M.; Mukherjee, A. In silico designing of a novel polyvalent multi-subunit peptide vaccine leveraging cross-immunity against human visceral and cutaneous leishmaniasis: An immunoinformatics-based approach. J. Mol. Model. 2023, 29, 99. [Google Scholar] [CrossRef]
- Bauer, J.A.; Pavlović, J.; Bauerová-Hlinková, V. Normal Mode Analysis as a Routine Part of a Structural Investigation. Molecules 2019, 24, 3293. [Google Scholar] [CrossRef]
- Al-Karmalawy, A.A.; Alnajjar, R.; Dahab, M.; Metwaly, A.; Eissa, I. Molecular docking and dynamics simulations reveal the potential of anti-HCV drugs to inhibit COVID-19 main protease. Pharm. Sci. 2021, 27, S109–S121. [Google Scholar] [CrossRef]
- Skariyachan, S.; Parveen, A.; Garka, S. Nanoparticle Fullerene (C60) demonstrated stable binding with antibacterial potential towards probable targets of drug resistant Salmonella typhi—A computational perspective and in vitro investigation. J. Biomol. Struct. Dyn. 2017, 35, 3449–3468. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, L.; Rasulev, B.; Turabekova, M.; Leszczynska, D.; Leszczynski, J. Receptor-and ligand-based study of fullerene analogues: Comprehensive computational approach including quantum-chemical, QSAR and molecular docking simulations. Org. Biomol. Chem. 2013, 11, 5798–5808. [Google Scholar] [CrossRef] [PubMed]
- Gopal, D.; Skariyachan, S.; Melappa, G. Chapter 8-Molecular interaction modeling of carbon nanotubes and fullerene toward prioritized targets of SARS-CoV-2 by computer-aided screening and docking studies. In Functionalized Carbon Nanomaterials for Theranostic Applications; Mallakpour, S., Hussain, C.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 157–179. [Google Scholar] [CrossRef]
- Teixeira-Santos, R.; Gomes, M.; Gomes, L.C.; Mergulhão, F.J. Antimicrobial and anti-adhesive properties of carbon nanotube-based surfaces for medical applications: A systematic review. iScience 2021, 24, 102001. [Google Scholar] [CrossRef] [PubMed]
- Azizi-Lalabadi, M.; Hashemi, H.; Feng, J.; Jafari, S.M. Carbon nanomaterials against pathogens; the antimicrobial activity of carbon nanotubes, graphene/graphene oxide, fullerenes, and their nanocomposites. Adv. Colloid Interface Sci. 2020, 284, 102250. [Google Scholar] [CrossRef] [PubMed]
- Zardini, H.Z.; Amiri, A.; Shanbedi, M.; Maghrebi, M.; Baniadam, M. Enhanced antibacterial activity of amino acids-functionalized multi walled carbon nanotubes by a simple method. Colloids Surf. B Biointerfaces 2012, 92, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; McDonald, T.J.; Kim, H.; Garg, V.K. Magnetic graphene–carbon nanotube iron nanocomposites as adsorbents and antibacterial agents for water purification. Adv. Colloid Interface Sci. 2015, 225, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Nie, C.; Yang, Y.; Cheng, C.; Ma, L.; Deng, J.; Wang, L.; Zhao, C. Bioinspired and biocompatible carbon nanotube-Ag nanohybrid coatings for robust antibacterial applications. Acta Biomater. 2017, 51, 479–494. [Google Scholar] [CrossRef]
- Hassani, M.; Tahghighi, A.; Rohani, M.; Hekmati, M.; Ahmadian, M.; Ahmadvand, H. Robust antibacterial activity of functionalized carbon nanotube-levofloxacine conjugate based on in vitro and in vivo studies. Sci. Rep. 2022, 12, 10064. [Google Scholar] [CrossRef]
- Alavi, M.; Hamblin, M.R. Interaction of copper oxide nanoparticles with bacterial nucleic acids: A mini-review. Micro Nano Bio Asp. 2023, 2, 20–25. [Google Scholar] [CrossRef]
- Aljelehawy, Q.; Maroufi, Y.; Javid, H.; Mohammadi, M.R.; Raji Mal Allah, O.; Taheri, S.V.; Mohammadzade, H. Anticancer, antineurodegenerative, antimicrobial, and antidiabetic activities of carvacrol: Recent advances and limitations for effective formulations. Nano Micro Biosyst. 2023, 2, 1–10. [Google Scholar] [CrossRef]
- Kostarelos, K.; Lacerda, L.; Pastorin, G.; Wu, W.; Wieckowski, S.; Luangsivilay, J.; Godefroy, S.; Pantarotto, D.; Briand, J.-P.; Muller, S.; et al. Cellular uptake of functionalized carbon nanotubes is independent of functional group and cell type. Nat. Nanotechnol. 2007, 2, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Wu, X.; Ouyang, P.; Shi, M.; Li, Q.; Maimaiti, T.; Lan, S.; Yang, S.-T.; Chang, X.-L. Surface modification mediates the interaction between fullerene and lysozyme: Protein structure and antibacterial activity. Environ. Sci. Nano 2021, 8, 76–85. [Google Scholar] [CrossRef]
- Prylutskyy, Y.I.; Petrenko, V.I.; Ivankov, O.I.; Kyzyma, O.A.; Bulavin, L.A.; Litsis, O.O.; Evstigneev, M.P.; Cherepanov, V.V.; Naumovets, A.G.; Ritter, U. On the Origin of C60 Fullerene Solubility in Aqueous Solution. Langmuir 2014, 30, 3967–3970. [Google Scholar] [CrossRef] [PubMed]
- Javed Ansari, M.; Soltani, A.; Ramezanitaghartapeh, M.; Singla, P.; Aghaei, M.; Khandan Fadafan, H.; Ardalan Khales, S.; Shariati, M.; Shirzad-Aski, H.; Balakheyli, H.; et al. Improved antibacterial activity of sulfasalazine loaded fullerene derivative: Computational and experimental studies. J. Mol. Liq. 2022, 348, 118083. [Google Scholar] [CrossRef]
- Lyon, D.Y.; Adams, L.K.; Falkner, J.C.; Alvarez, P.J. Antibacterial activity of fullerene water suspensions: Effects of preparation method and particle size. Environ. Sci. Technol. 2006, 40, 4360–4366. [Google Scholar] [CrossRef] [PubMed]
- Sabirov, D.S. Polarizability of C60 fullerene dimer and oligomers: The unexpected enhancement and its use for rational design of fullerene-based nanostructures with adjustable properties. RSC Adv. 2013, 3, 19430–19439. [Google Scholar] [CrossRef]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef]
- Zeng, J.; Platig, J.; Cheng, T.-Y.; Ahmed, S.; Skaf, Y.; Potluri, L.-P.; Schwartz, D.; Steen, H.; Moody, D.B.; Husson, R.N. Protein kinases PknA and PknB independently and coordinately regulate essential Mycobacterium tuberculosis physiologies and antimicrobial susceptibility. PLoS Pathog. 2020, 16, e1008452. [Google Scholar] [CrossRef]
- Zupetic, J.; Peñaloza, H.F.; Bain, W.; Hulver, M.; Mettus, R.; Jorth, P.; Doi, Y.; Bomberger, J.; Pilewski, J.; Nouraie, M.; et al. Elastase Activity from Pseudomonas aeruginosa Respiratory Isolates and ICU Mortality. Chest 2021, 160, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Chadha, J.; Harjai, K.; Chhibber, S. Revisiting the virulence hallmarks of Pseudomonas aeruginosa: A chronicle through the perspective of quorum sensing. Environ. Microbiol. 2022, 24, 2630–2656. [Google Scholar] [CrossRef] [PubMed]
- Bonifacius, A.; Goldmann, O.; Floess, S.; Holtfreter, S.; Robert, P.A.; Nordengrün, M.; Kruse, F.; Lochner, M.; Falk, C.S.; Schmitz, I.; et al. Staphylococcus aureus Alpha-Toxin Limits Type 1 While Fostering Type 3 Immune Responses. Front. Immunol. 2020, 11, 1579. [Google Scholar] [CrossRef]
- Ramstad, S.N.; Wasteson, Y.; Lindstedt, B.-A.; Taxt, A.M.; Bjørnholt, J.V.; Brandal, L.T.; Bohlin, J. Characterization of Shiga Toxin 2a Encoding Bacteriophages Isolated from High-Virulent O145:H25 Shiga Toxin-Producing Escherichia coli. Front. Microbiol. 2021, 12, 728116. [Google Scholar] [CrossRef]
- Angulo-Pineda, C.; Srirussamee, K.; Palma, P.; Fuenzalida, V.M.; Cartmell, S.H.; Palza, H. Electroactive 3D Printed Scaffolds Based on Percolated Composites of Polycaprolactone with Thermally Reduced Graphene Oxide for Antibacterial and Tissue Engineering Applications. Nanomaterials 2020, 10, 428. [Google Scholar] [CrossRef] [PubMed]
- Vidakis, N.; Petousis, M.; Velidakis, E.; Tzounis, L.; Mountakis, N.; Boura, O.; Grammatikos, S.A. Multi-functional polyamide 12 (PA12)/multiwall carbon nanotube 3D printed nanocomposites with enhanced mechanical and electrical properties. Adv. Compos. Mater 2022, 31, 630–654. [Google Scholar] [CrossRef]
- Gao, Z.; Varela, J.A.; Groc, L.; Lounis, B.; Cognet, L. Toward the suppression of cellular toxicity from single-walled carbon nanotubes. Biomater. Sci. 2016, 4, 230–244. [Google Scholar] [CrossRef]
- Costa, P.M.; Bourgognon, M.; Wang, J.T.W.; Al-Jamal, K.T. Functionalised carbon nanotubes: From intracellular uptake and cell-related toxicity to systemic brain delivery. J. Control. Release 2016, 241, 200–219. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Baek, K.; Shin, H.; Cho, M. Multiscale modeling of mechanical behaviors of Nano-SiC/epoxy nanocomposites with modified interphase model: Effect of nanoparticle clustering. Compos. Sci. Technol. 2021, 203, 108572. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Lopéz-Blanco, J.R.; Garzón, J.I.; Chacón, P. iMod: Multipurpose normal mode analysis in internal coordinates. Bioinformatics 2011, 27, 2843–2850. [Google Scholar] [CrossRef]
- Kovacs, J.A.; Chacón, P.; Abagyan, R. Predictions of protein flexibility: First-order measures. Proteins 2004, 56, 661–668. [Google Scholar] [CrossRef]
- Ghosh, P.; Bhakta, S.; Bhattacharya, M.; Sharma, A.R.; Sharma, G.; Lee, S.-S.; Chakraborty, C. A Novel Multi-Epitopic Peptide Vaccine Candidate against Helicobacter pylori: In-Silico Identification, Design, Cloning and Validation through Molecular Dynamics. Int. J. Pept. Res. Ther. 2021, 27, 1149–1166. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; Sanejouand, Y.-H. ElNémo: A normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004, 32, W610–W614. [Google Scholar] [CrossRef] [PubMed]
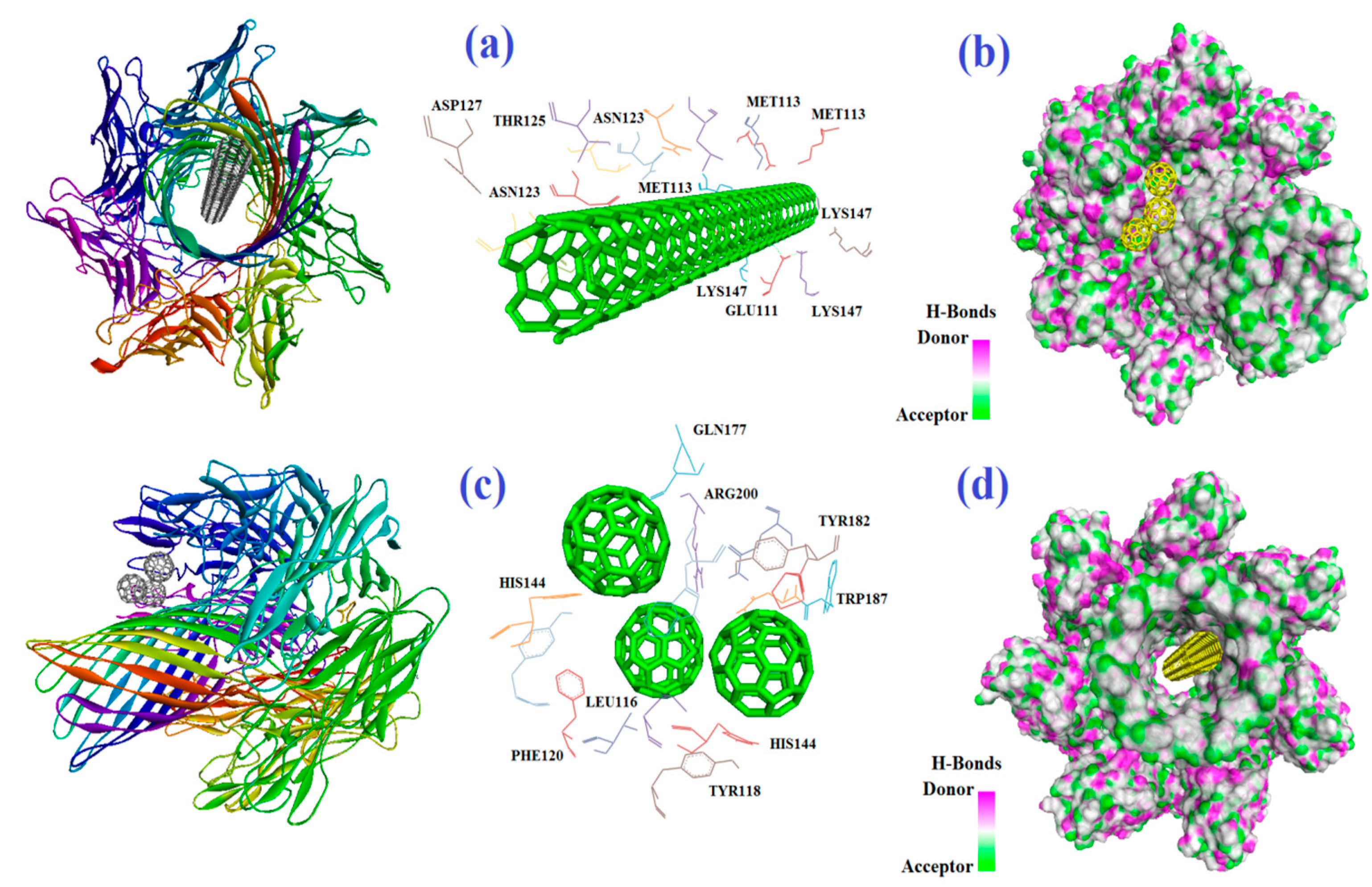
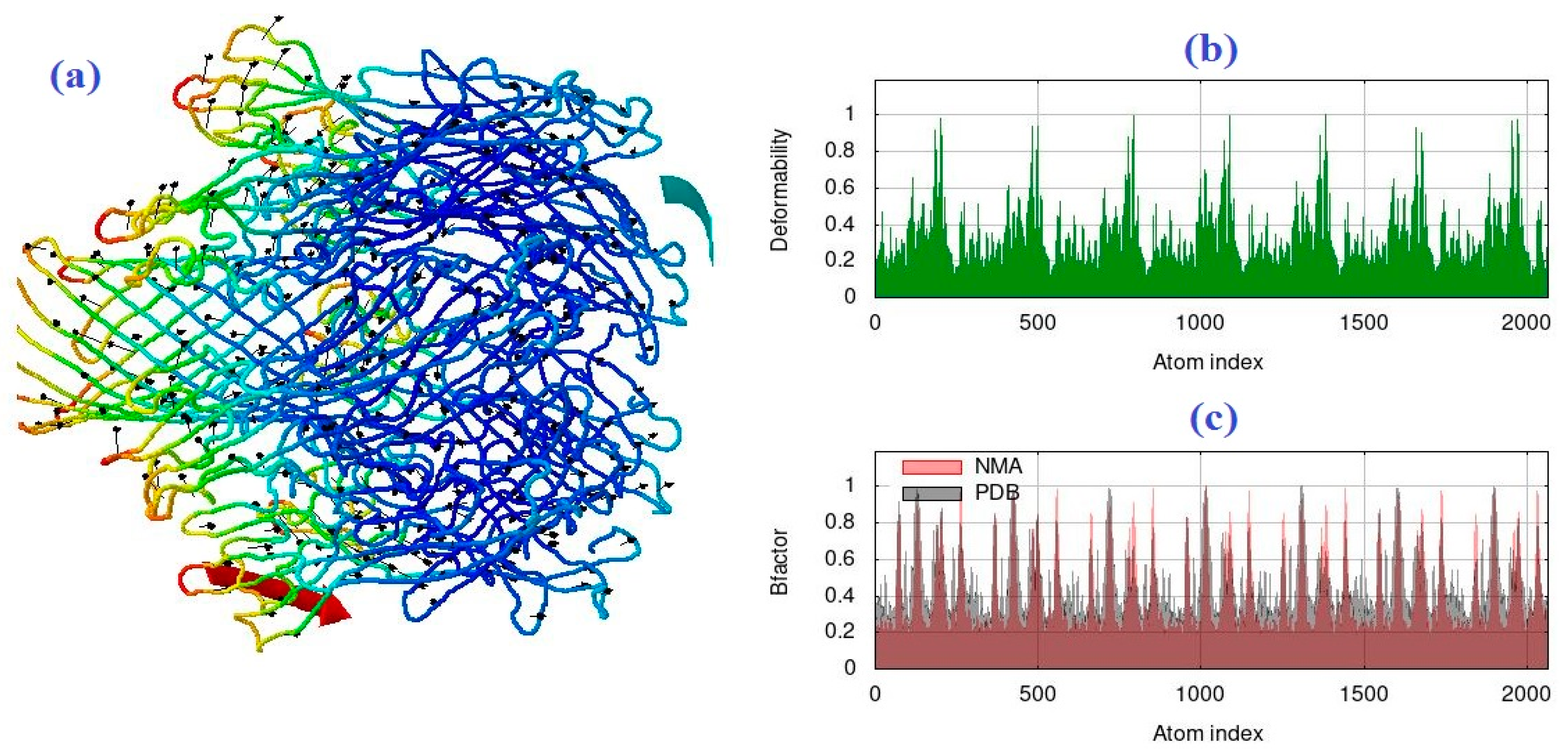
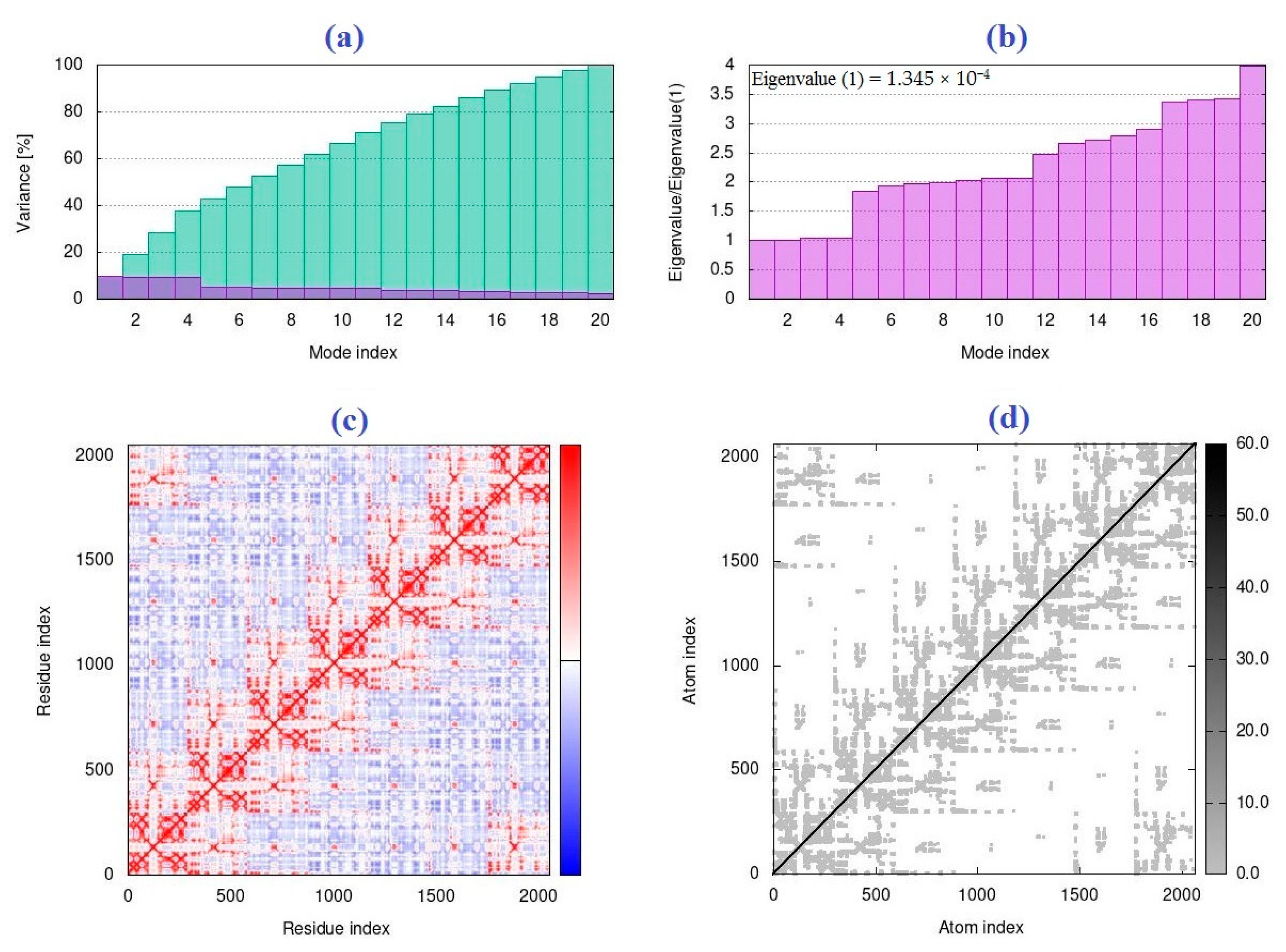

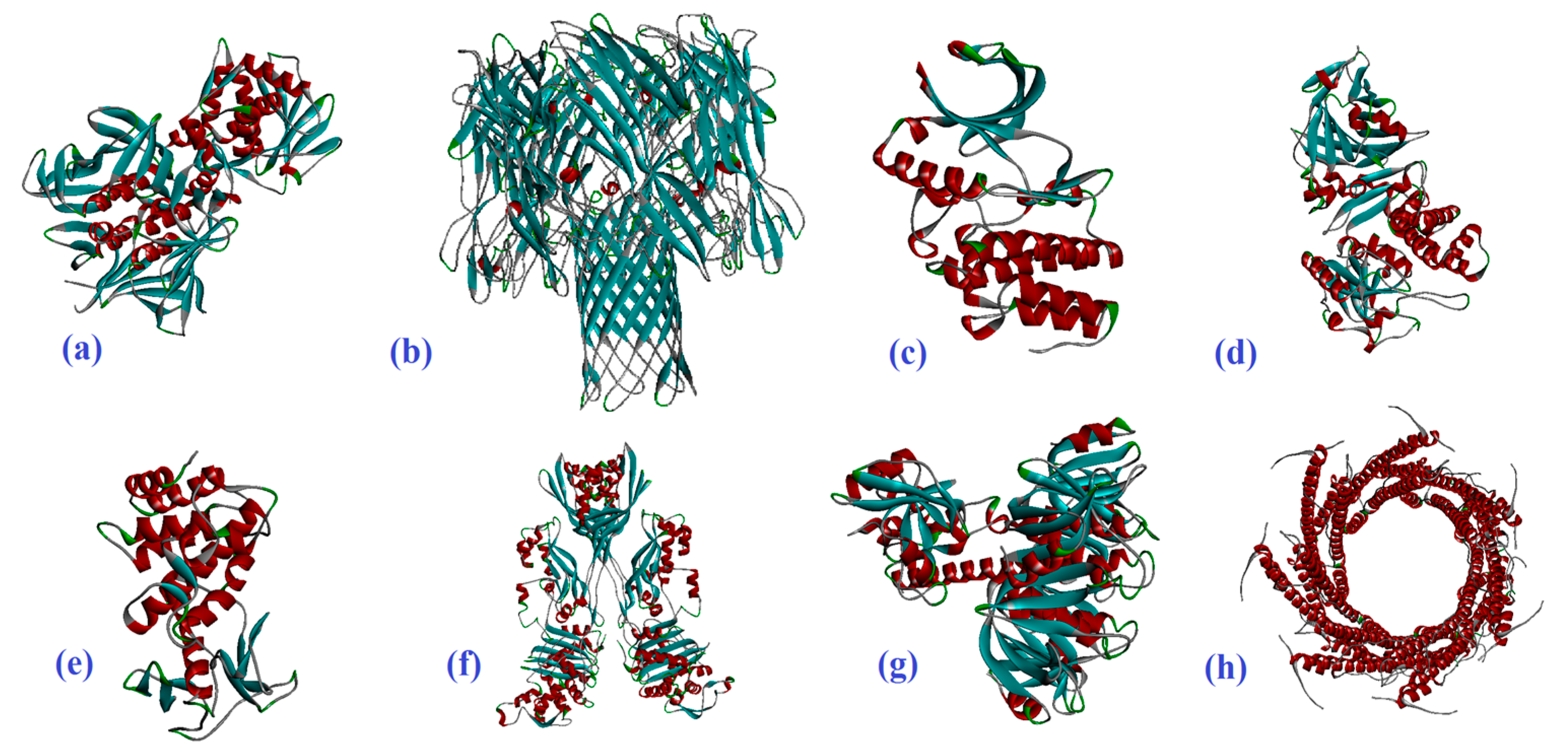
| R | CNT | IAA | Capped CNT | IAA | CNT2–5 | IAA |
|---|---|---|---|---|---|---|
| 7D6Q | −14.4 | ASP94, ASP111, SER113, PRO258, ASP70, LYS5, GLY6, LYS7, GLU9, ASP24 | −16.1 | ASP94, ASP111, SER113, PRO258, GLN261, ASN69, ASP70, LYS5, LYS7, GLU9, ASP24 | −19.8 | GLU177, GLN180, VAL188, TYR189 THR190, ASN226 ASP70, LYS5, GLY6, LYS7, ASP24 |
| 1TII | 97.4 | GLU22, THR24, LYS25, SER42, SER74, GLY75, MET76, ARG77, GLY1, ALA98, ARG15, ARG16, GLY18, ALA28, TYR29, GLU30, ARG31, LEU119, ARG141, ASP142 | 79 | GLU22, THR24, LYS25, SER42, SER74, GLY75, MET76, ARG77, GLY1, ALA98, ARG15, ARG16, GLY18, ALA28, GLU30, ARG31, LEU119, ARG141, ASP142 | - | - |
| 7AHL | −19.6 | THR125, LYS131, LEU135, ASN121, ASN123, LEU135, ASN121, ASN123, LEU135, ASN121, THR125, GLY126, ASP127 | −18.8 | LEU116, TYR118, VAL140, TYR112, HIS144, TRP179, PRO181, TYR182, SER186, TRP187, ASN188, PRO189, GLN194, ASN178, TRP179 | −32.7 | ASN123, THR125, ASP127, LEU135, LYS147, ASN121, MET113, GLU111 |
| 1MWT | −20.1 | ASN146, LYS148, GLU170, GLN199, GLN200, GLN203, TRP205, PRO213, ASN236, THR238, PRO258, ILE259, ASN260, SER261, ASP274, ASP275, ILE309 | −19.6 | ASN146, LYS148, GLU170, GLN199, GLN200 GLN203, TRP205 PRO213, THR238, PRO258 ILE259, ASP274, ASP275 ARG298, ILE309 | −19.4 | ASP82, LYS84, GLN98, ASN111, TYR169, SER225, LYS229, HIS232, LYS331, LYS334, GLU658 |
| 4OW8 | −13.0 | ARG112, SER212, LYS214, PRO216, ALA218, LYS228, PRO235, PRO238, ASP240 | −12.6 | ARG112, SER212, LYS214, PRO216, ALA218, LYS228, PRO235, PRO238, ASP240 | −13.5 | LYS108, THR110 GLY111, ARG112 SER212, GLY213 LYS214, ASP240 |
| 7P13 | −16.6 | ALA186, ASP189 GLN190, ASN274 GLN190, GLN193 HIS197, PRO276 LYS277, PRO279 PRO280 | −16.3 | ALA186, ASP189 GLN190, GLN190 GLN193, HIS197 PRO276, LYS277 PRO279, PRO280 | −26.3 | GLU263, ASN266 LYS267, HIS197 TYR211, GLU271 ASN274, PRO276 ALA186, ASP189 GLN190, GLN193 |
| 1IKQ | −12.9 | ARG213, ASN215, ASP218, GLU221, ASP403, GLU431 | −13.2 | ARG213, ASN215, ASP218, GLU221, and GLU431 | −18.8 | HIS128, ASP238, LYS240, ARG352, ALA464, ILE465, LEU535, PRO536, ARG538 |
| 1EZM | −11.9 | ASN112, TYR114, TRP115, ASP116, ASP136, GLU148, TYR155, GLU172, GLU175, ASP183, LEU185 | −12.3 | ASN112, TRP115, ASP116, ASP136, GLU148, TYR155, GLU172, GLU175, ASP183, LEU185 | −12.8 | ALA1, ILE25, VAL26, ASN27, ASP28, ASP34, GLY35 |
| R | C60 | IAA | C70 | IAA |
|---|---|---|---|---|
| 7D6Q | −10.1 | LYS270, ASN272, ASN273, LYS7, GLY46, ASN69, ASP70 | −10.0 | GLN118, ARG119, LEU123, GLU124, LYS5, GLN66, ASP70, GLU9, LYS22 |
| 1TII | −4.9 | THR24, LYS25, SER42, GLY1, ALA98, ARG141 | −2.5 | GLU22, THR24, LYS25, SER42, GLY1, ALA98 ARG15, ARG141, ASP142 |
| 7AHL | −12.8 | ARG104, ASN105, SER106, ILE107, TYR102, PRO103, THR155, PHE224, SER225, ASP227 | −13.6 | ARG104, ASN105, SER106, ILE107, TYR102, PRO103, THR155, PHE224, SER225, ASP227 |
| 1MWT | −10.4 | TYR255, ASN260 PHE371, GLY37 MET375, ASN377 TYR380 | −11.4 | TYR255, ASN260 LYS280, PHE371 GLY374, MET375 ASN377, TYR380 |
| 4OW8 | −8.3 | LEU190, HIS192, ILE230, LYS255, ASN256 | −8.1 | LEU190, HIS192, ILE230, LYS255, ASN256 |
| 7P13 | −9.7 | THR262, GLU263 ASN266, TYR211 GLN214, TRP218 | −10.5 | LYS259, THR262 GLU263, ASN266 TYR211, GLN214 LEU215, TRP218 GLU263, LYS267 |
| 1IKQ | −7.5 | HIS107, ASP139, ARG276, ARG279 | −7.6 | ASN215, GLU221, ASP403, GLN428 |
| 1EZM | −7.6 | TRP115, ASP116, GLY117, TYR155 | −7.7 | TRP115, ASP116, GLY117, TYR155 |
| R | HO-C60 | IAA | [C60]2 | IAA | [C60]3 | IAA |
|---|---|---|---|---|---|---|
| 7D6Q | −9.7 | LYS270, ILE271, ASN272, ASN273, LYS7, LEU44, THR45, GLY46, ASP70, ASN69, ASP70 | −17.5 | GLU211, ARG213, GLU215, ARG266, VAL268, LYS270, ASN272, ASN273, LEU275, LYS7, GLY46, ASN69, ASP70 | −20.6 | GLN173, ARG176, GLU177, GLN180, VAL188, TYR189, THR190, THR192, PRO193, GLY194, SER224, ASN226, ALA230, THR234, LYS5, LYS7, ASP24 |
| 1TII | −5.5 | THR24, LYS25, SER42, THR43, GLY1, ALA98, ARG15, ARG141 | 15.9 | GLU22, THR24, SER42, SER74, GLY75, MET76, ARG77, ALA98, ARG15 ARG16, GLY18, ALA28 ARG31, LEU119 ARG141, ASP142 | 185.6 | GLU22, THR24, SER41, SER42, THR43, SER74, GLY75, MET76, ARG77, GLY1, ARG77, GLU97, ALA98, ARG15, ARG16, GLY18, LEU119, ARG139, ARG141, ASP142 |
| 7AHL | −12.9 | ARG104, ASN105, SER106, ILE107, TYR102, PRO103, THR155, PHE224, ASP227 | −19 | ARG104, ASN105, SER106, ILE107 TYR102, PRO103, SER106, ILE107, ASP108, THR109, VAL149, THR155, PHE224, ASP227 | −26.6 | TYR118, PHE120, LEU116, TYR118, VAL140, HIS144, HIS144, GLN177, TRP179, PRO181, TYR182, TRP187, ASN188, GLN194, ARG200 |
| 1MWT | −9.8 | TYR255, PRO258, ASN260, LYS280, PHE371, MET372, TYR373, GLY374, MET375, ASN377, TYR380 | −17 | ASN104, TYR105, ASN146, TRP205, LYS273, ASP274, ASP275, TYR297, THR308, ILE309 | −22.7 | ASN104, TYR105, GLU170, GLN203, TRP205, PRO213, ASN236, THR238, LYS273, ASP274, ASP275, TYR297, ILE309 |
| 4OW8 | −8 | TYR208, SER212, GLY213, LYS214, PRO235, PRO236, LEU237, PRO238 | −12.6 | SER212, LYS214, LYS228, GLU232, PRO235, PRO236, PRO238 | −14.7 | THR21, GLY22, GLY23, LYS45, PHE48, VAL166, GLY175, MET176, VAL177, MET178, GLY179, THR180 GLY221, ALA222 |
| 7P13 | −9.7 | THR262, GLU263, ASN266, TYR211, GLN214, LEU215, TRP218, GLU263, LYS267 | −15.3 | LYS259, THR262, GLU263, ASN266, LYS267, LYS207, TYR211, GLN214, TRP218 | −20.2 | THR262, ASN266, TYR211, GLN214, TRP218, LYS259, THR262, GLU263, ASN266, LYS267 GLN214, TRP218 |
| 1IKQ | −8.4 | ASN215, GLU221, ASP403, GLN424, GLN428, ARG432 | −12.6 | GLU108, LYS185, ARG186, THR219, TRP281, GLU282, ASP406 | −15.6 | VAL351, ARG352, GLN353, GLU378, ILE465, PRO534, LEU535, PRO536, ARG538 |
| 1EZM | −7.9 | TRP115, ASP116, GLY117, HIS144, TYR155, ASN163 | −13.6 | ASP48, TYR106, TYR114, TRP115, ASP116, GLY117 LEU121, TYR155 | −18.1 | ASP48, SER49, LYS103, TYR106, TYR114, TRP115, ASP116, THR118 LEU121, TYR155 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alavi, M.; Ashengroph, M.; Mozafari, M.R. Interaction of Carbon Nanotubes, Capped Carbon Nanotubes, CNT2–5, C60, C70, HO-C60, [C60]2, and [C60]3 Fullerenes with Virulence Factors of Gram-Negative and Gram-Positive Bacteria: Potential Applications for 3D-Printed Scaffolds. Inorganics 2024, 12, 77. https://doi.org/10.3390/inorganics12030077
Alavi M, Ashengroph M, Mozafari MR. Interaction of Carbon Nanotubes, Capped Carbon Nanotubes, CNT2–5, C60, C70, HO-C60, [C60]2, and [C60]3 Fullerenes with Virulence Factors of Gram-Negative and Gram-Positive Bacteria: Potential Applications for 3D-Printed Scaffolds. Inorganics. 2024; 12(3):77. https://doi.org/10.3390/inorganics12030077
Chicago/Turabian StyleAlavi, Mehran, Morahem Ashengroph, and M. R. Mozafari. 2024. "Interaction of Carbon Nanotubes, Capped Carbon Nanotubes, CNT2–5, C60, C70, HO-C60, [C60]2, and [C60]3 Fullerenes with Virulence Factors of Gram-Negative and Gram-Positive Bacteria: Potential Applications for 3D-Printed Scaffolds" Inorganics 12, no. 3: 77. https://doi.org/10.3390/inorganics12030077
APA StyleAlavi, M., Ashengroph, M., & Mozafari, M. R. (2024). Interaction of Carbon Nanotubes, Capped Carbon Nanotubes, CNT2–5, C60, C70, HO-C60, [C60]2, and [C60]3 Fullerenes with Virulence Factors of Gram-Negative and Gram-Positive Bacteria: Potential Applications for 3D-Printed Scaffolds. Inorganics, 12(3), 77. https://doi.org/10.3390/inorganics12030077







