Tutorial on Development of 3D Vasculature Digital Phantoms for Evaluation of Photoacoustic Image Reconstruction Algorithms
Abstract
:1. Introduction
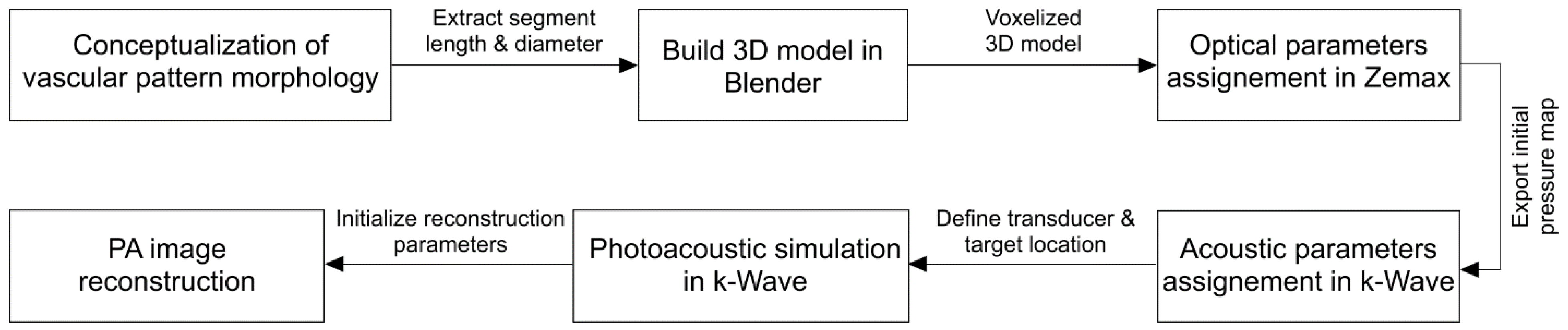
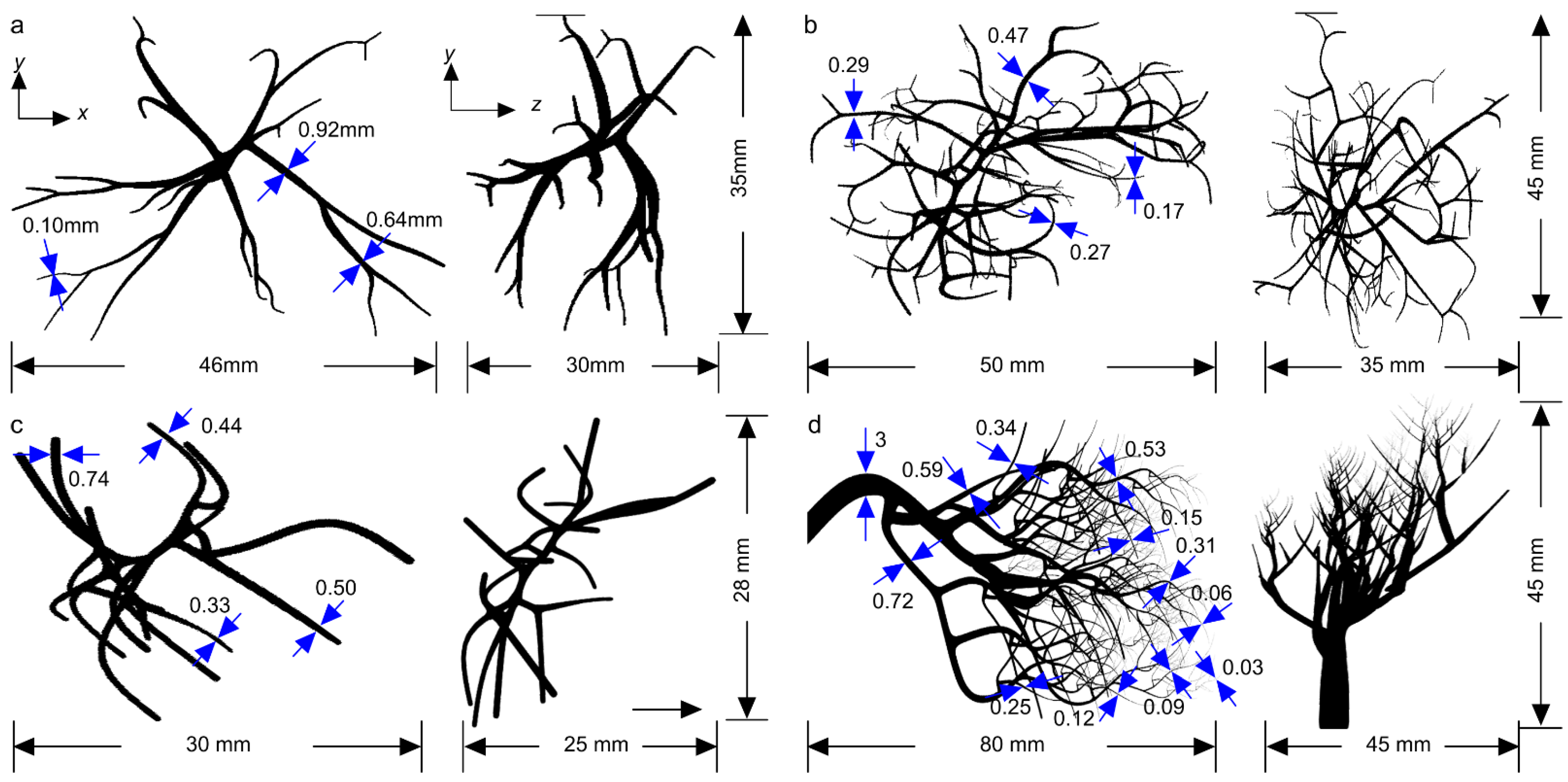
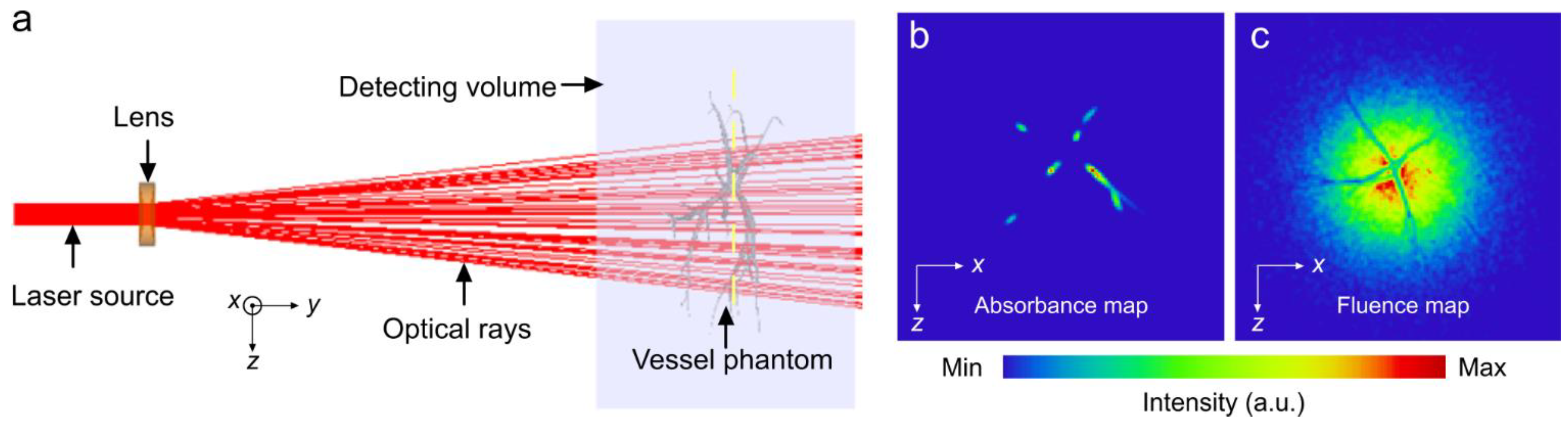
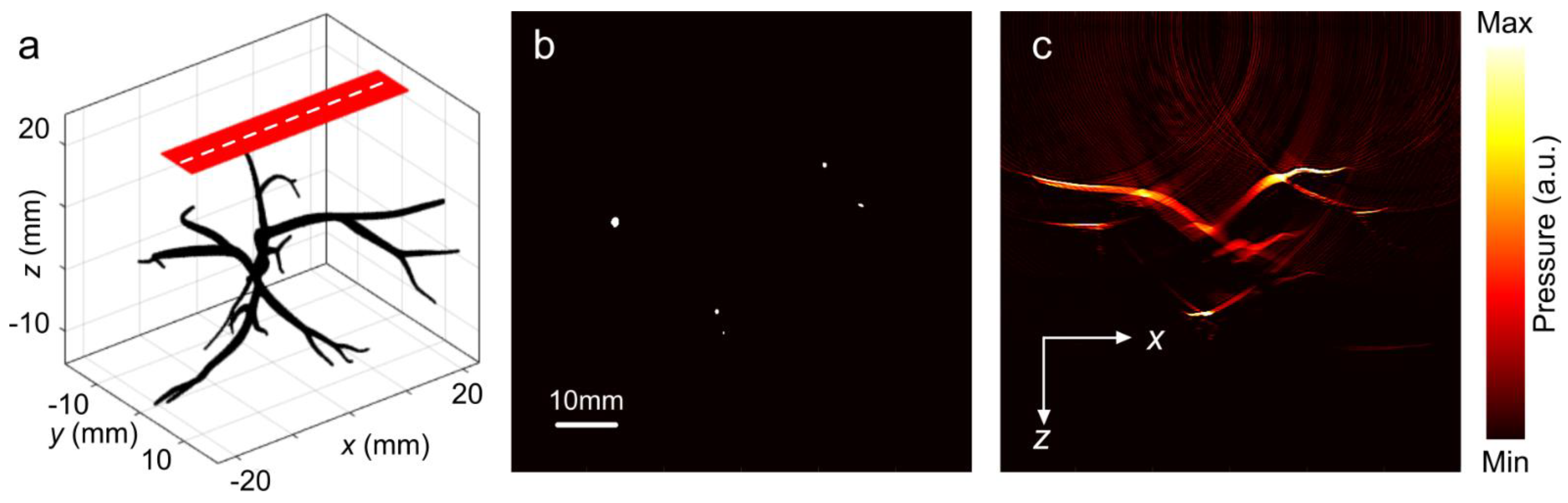
2. Methods
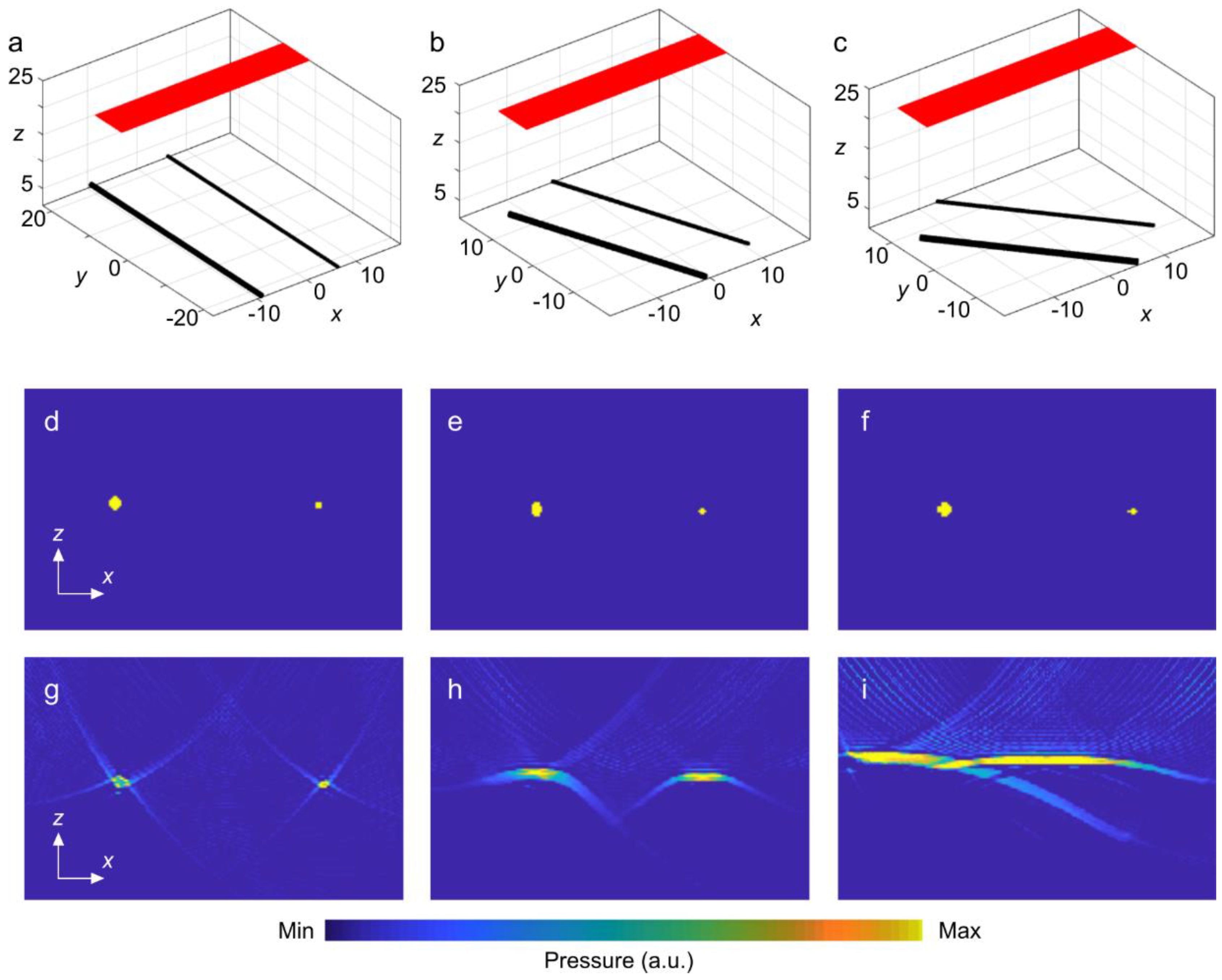
3. Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Fatima, A.; Kratkiewicz, K.; Manwar, R.; Zafar, M.; Zhang, R.; Huang, B.; Dadashzadeh, N.; Xia, J.; Avanaki, K.M. Review of cost reduction methods in photoacoustic computed tomography. Photoacoustics 2019, 15, 100137. [Google Scholar] [CrossRef] [PubMed]
- Manwar, A.; Kratkiewicz, K.; Nasiriavanaki, M.R. Photoacoustic Imaging: A Promising Alternative to Transcranial Ultrasound. Res. J. Photonics 2018, 2, 411–420. [Google Scholar]
- Zafar, M.; Kratkiewicz, K.; Manwar, R.; Avanaki, M. Low-cost fast photoacoustic computed tomography: Phantom study. In Proceedings of the Photons Plus Ultrasound: Imaging and Sensing 2019, San Francisco, CA, USA, 1–6 February 2019; p. 108785V. [Google Scholar]
- Mozaffarzadeh, M.; Mahloojifar, A.; Orooji, M.; Adabi, S.; Nasiriavanaki, M. Double-Stage Delay Multiply and Sum Beamforming Algorithm: Application to Linear-Array Photoacoustic Imaging. IEEE Trans. Biomed. Eng. 2017, 65, 31–42. [Google Scholar] [PubMed] [Green Version]
- Mozaffarzadeh, M.; Mahloojifar, A.; Orooji, M.; Kratkiewicz, K.; Adabi, S.; Nasiriavanaki, M. Linear-array photoacoustic imaging using minimum variance-based delay multiply and sum adaptive beamforming algorithm. J. Biomed. Opt. 2018, 23, 026002. [Google Scholar] [CrossRef]
- Omidi, P.; Zafar, M.; Mozaffarzadeh, M.; Hariri, A.; Haung, X.; Orooji, M.; Nasiriavanaki, M. A novel dictionary-based image reconstruction for photoacoustic computed tomography. Appl. Sci. 2018, 8, 1570. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi, L.; Behnam, H.; Tavakkoli, J.; Avanaki, M.R.N. Skull’s Photoacoustic Attenuation and Dispersion Modeling with Deterministic Ray-Tracing: Towards Real-Time Aberration Correction. Sensors 2019, 19, 345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Li, L.; Lin, L.; Shi, J.; Wang, L.V. Applications. In vivo superresolution photoacoustic computed tomography by localization of single dyed droplets. Light Sci. Appl. 2019, 8, 36. [Google Scholar] [CrossRef] [Green Version]
- Schoonover, R.W.; Anastasio, M.A. Image reconstruction in photoacoustic tomography involving layered acoustic media. JOSA A 2011, 28, 1114–1120. [Google Scholar] [CrossRef] [Green Version]
- Bar-Hillel, Y. An examination of information theory. Philos. Sci. 1955, 22, 86–105. [Google Scholar] [CrossRef]
- Xia, J.; Yao, J.; Wang, L.V. Photoacoustic tomography: Principles and advances. Electromagn. Waves Camb. Mass. 2014, 147, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.V. Tutorial on Photoacoustic Microscopy and Computed Tomography. IEEE J. Sel. Top. Quantum Electron. 2008, 14, 171–179. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.V.; Hu, S. Photoacoustic Tomography: In Vivo Imaging from Organelles to Organs. Science 2012, 335, 1458–1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, J.; Wang, L.V. Photoacoustic microscopy. Laser Photonics Rev. 2013, 7, 758–778. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yao, J.; Wang, L.V. Tutorial on photoacoustic tomography. J. Biomed. Opt. 2016, 21, 061007. [Google Scholar] [CrossRef] [Green Version]
- Manwar, R.; Hosseinzadeh, M.; Hariri, A.; Kratkiewicz, K.; Noei, S.; Avanaki, M.R.N. Photoacoustic Signal Enhancement: Towards Utilization of Low Energy Laser Diodes in Real-Time Photoacoustic Imaging. Sensors 2018, 18, 3498. [Google Scholar] [CrossRef] [Green Version]
- Zafar, M.; Kratkiewicz, K.; Manwar, R.; Avanaki, M. Development of Low-Cost Fast Photoacoustic Computed Tomography: System Characterization and Phantom Study. Appl. Sci. 2019, 9, 374. [Google Scholar] [CrossRef] [Green Version]
- Manwar, R.; McGuire, L.; Shoo, A.; Charbel, F.; Pillers, D.-A.; Avanaki, K. Cerebral blood oxygenation measurement in sheep brain in-vivo using transfontanelle photoacoustic spectroscopy. In Proceedings of the Photons Plus Ultrasound: Imaging and Sensing 2022, San Francisco, CA, USA, 3 March 2022; Volume 11960, pp. 248–255. [Google Scholar]
- Wang, L.V. Photoacoustic Imaging and Spectroscopy; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Wang, L.V.; Yao, J. A practical guide to photoacoustic tomography in the life sciences. Nat. Methods 2016, 13, 627. [Google Scholar] [CrossRef]
- Kratkiewicz, K.; Manwar, R.; Rajabi-Estarabadi, A.; Fakhoury, J.; Meiliute, J.; Daveluy, S.; Mehregan, D.; Avanaki, K.M. Photoacoustic/Ultrasound/Optical Coherence Tomography Evaluation of Melanoma Lesion and Healthy Skin in a Swine Model. Sensors 2019, 19, 2815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manwar, R.; Kratkiewicz, K.; Avanaki, K. Overview of Ultrasound Detection Technologies for Photoacoustic Imaging. Micromachines 2020, 11, 692. [Google Scholar] [CrossRef] [PubMed]
- MManwar, R.; Zafar, M.; Xu, Q. Signal and Image Processing in Biomedical Photoacoustic Imaging: A Review. Optics 2020, 2, 1–24. [Google Scholar] [CrossRef]
- Manwar, R.; Li, X.; Mahmoodkalayeh, S.; Asano, E.; Zhu, D.; Avanaki, K. Deep learning protocol for improved photoacoustic brain imaging. J. Biophotonics 2020, 13, e202000212. [Google Scholar] [CrossRef] [PubMed]
- Kratkiewicz, K.; Manwar, R.; Zafar, M.; Ranjbaran, S.M.; Mozaffarzadeh, M.; de Jong, N.; Ji, K.; Avanaki, K. Development of a Stationary 3D Photoacoustic Imaging System Using Sparse Single-Element Transducers: Phantom Study. Appl. Sci. 2019, 9, 4505. [Google Scholar] [CrossRef] [Green Version]
- Matchynski, J.I.; Manwar, R.; Kratkiewicz, K.J.; Madangopal, R.; Lennon, V.A.; Makki, K.M.; Reppen, A.L.; Woznicki, A.R.; Hope, B.T.; Perrine, S.A. Direct measurement of neuronal ensemble activity using photoacoustic imaging in the stimulated Fos-LacZ transgenic rat brain: A proof-of-principle study. Photoacoustics 2021, 24, 100297. [Google Scholar] [CrossRef] [PubMed]
- Mozaffarzadeh, M.; Mahloojifar, A.; Nasiriavanaki, M.; Orooji, M. Eigenspace-based minimum variance adaptive beamformer combined with delay multiply and sum: Experimental study. In Proceedings of the Photonics in Dermatology and Plastic Surgery 2018, San Francisco, CA, USA, 27–28 January 2018; p. 1046717. [Google Scholar]
- Zafar, M.; Manwar, R.; Avanaki, K. High-fidelity compression for high-throughput photoacoustic microscopy systems. J. Biophotonics 2022, 15, e202100350. [Google Scholar] [CrossRef] [PubMed]
- Manwar, R.; Kratkiewicz, K.; Avanaki, K. Investigation of the effect of the skull in transcranial photoacoustic imaging: A preliminary ex vivo study. Sensors 2020, 20, 4189. [Google Scholar] [CrossRef]
- Manwar, R.; Benavides Lara, J.; Prakash, R.; Ranjbaran, S.M.; Avanaki, K. Randomized Multi-Angle Illumination for Improved Linear Array Photoacoustic Computed Tomography in Brain. J. Biophotonics 2022, e202200016. [Google Scholar] [CrossRef] [PubMed]
- Hariri, A.; Omidi, P.; Nasiriavanaki, M. Resting-state functional connectivity measurement in the mouse brain using a low cost photoacoustic computed tomography. In Proceedings of the Laser Science, Rochester, NY, USA, 17–21 October 2016; p. JW4A.62. [Google Scholar]
- Mohammadi, L.; Behnam, H.; Tavakkoli, J.; Avanaki, K. Skull acoustic aberration correction in photoacoustic microscopy using a vector space similarity model: A proof-of-concept simulation study. Biomed. Opt. Express 2020, 11, 5542–5556. [Google Scholar] [CrossRef] [PubMed]
- Kratkiewicz, K.; Manwar, R.; Zhou, Y.; Mozaffarzadeh, M.; Avanaki, K. Technical considerations in the Verasonics research ultrasound platform for developing a photoacoustic imaging system. Biomed. Opt. Express 2021, 12, 1050–1084. [Google Scholar] [CrossRef]
- Hariri, A.; Fatima, A.; Mohammadian, N.; Bely, N.; Nasiriavanaki, M. Towards low cost photoacoustic microscopy system for evaluation of skin health. In Proceedings of the Imaging Spectrometry XXI, San Diego, CA, USA, 19 September 2016; p. 99760X. [Google Scholar]
- Hariri, A.; Bely, N.; Chen, C.; Nasiriavanaki, M. Towards ultrahigh resting-state functional connectivity in the mouse brain using photoacoustic microscopy. In Proceedings of the Photons Plus Ultrasound: Imaging and Sensing 2016, San Francisco, CA, USA, 14–16 February 2016; p. 97085A. [Google Scholar]
- Manwar, R.; Islam, T.; Ranjbaran, S.M.; Avanaki, K. Transfontanelle photoacoustic imaging: Ultrasound transducer selection analysis. Biomed. Opt. Express 2022, 13, 676–693. [Google Scholar] [CrossRef] [PubMed]
- Avanaki, K.; Gelovani, J.G. Ultrasound and Multispectral Photoacoustic Systems and Methods for Brain and Spinal Cord Imaging through Acoustic Windows. U.S. Patent 16/566,212, 12 March 2020. [Google Scholar]
- Mahmoodkalayeh, S.; Kratkiewicz, K.; Manwar, R.; Shahbazi, M.; Ansari, M.A.; Natarajan, G.; Asano, E.; Avanaki, K. Wavelength and pulse energy optimization for detecting hypoxia in photoacoustic imaging of the neonatal brain: A simulation study. Biomed. Opt. Express 2021, 12, 7458–7477. [Google Scholar] [CrossRef] [PubMed]
- Manwar, R.; Saint-Martin, L.; Avanaki, K. Couplants in Acoustic Biosensing Systems. Chemosensors 2022, 10, 181. [Google Scholar] [CrossRef]
- Garcia-Pardo, C.; Andreu, C.; Fornes-Leal, A.; Castelló-Palacios, S.; Perez-Simbor, S.; Barbi, M.; Vallés-Lluch, A.; Cardona, N. Ultrawideband technology for medical in-body sensor networks: An overview of the human body as a propagation medium, phantoms, and approaches for propagation analysis. IEEE Antennas Propag. Mag. 2018, 60, 19–33. [Google Scholar] [CrossRef]
- Lamouche, G.; Kennedy, B.F.; Kennedy, K.M.; Bisaillon, C.-E.; Curatolo, A.; Campbell, G.; Pazos, V.; Sampson, D.D. Review of tissue simulating phantoms with controllable optical, mechanical and structural properties for use in optical coherence tomography. Biomed. Opt. Express 2012, 3, 1381–1398. [Google Scholar] [CrossRef] [PubMed]
- Treeby, B.E.; Cox, B.T. k-Wave: MATLAB toolbox for the simulation and reconstruction of photoacoustic wave fields. J. Biomed. Opt. 2010, 15, 021314. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Zhou, W.; Matthews, T.P.; Appleton, C.M.; Anastasio, M.A. Generation of anatomically realistic numerical phantoms for photoacoustic and ultrasonic breast imaging. J. Biomed. Opt. 2017, 22, 041015. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ranjbaran, S.M.; Khan, A.; Manwar, R.; Avanaki, K. Tutorial on Development of 3D Vasculature Digital Phantoms for Evaluation of Photoacoustic Image Reconstruction Algorithms. Photonics 2022, 9, 538. https://doi.org/10.3390/photonics9080538
Ranjbaran SM, Khan A, Manwar R, Avanaki K. Tutorial on Development of 3D Vasculature Digital Phantoms for Evaluation of Photoacoustic Image Reconstruction Algorithms. Photonics. 2022; 9(8):538. https://doi.org/10.3390/photonics9080538
Chicago/Turabian StyleRanjbaran, Seyed Mohsen, Azam Khan, Rayyan Manwar, and Kamran Avanaki. 2022. "Tutorial on Development of 3D Vasculature Digital Phantoms for Evaluation of Photoacoustic Image Reconstruction Algorithms" Photonics 9, no. 8: 538. https://doi.org/10.3390/photonics9080538
APA StyleRanjbaran, S. M., Khan, A., Manwar, R., & Avanaki, K. (2022). Tutorial on Development of 3D Vasculature Digital Phantoms for Evaluation of Photoacoustic Image Reconstruction Algorithms. Photonics, 9(8), 538. https://doi.org/10.3390/photonics9080538




