Abstract
Machine learning (ML) has an impressive capacity to learn and analyze a large volume of data. This study aimed to train different algorithms to discriminate between healthy and pathologic corneal images by evaluating digitally processed spectral-domain optical coherence tomography (SD-OCT) corneal images. A set of 22 SD-OCT images belonging to a random set of corneal pathologies was compared to 71 healthy corneas (control group). A binary classification method was applied where three approaches of ML were explored. Once all images were analyzed, representative areas from every digital image were also extracted, processed and analyzed for a statistical feature comparison between healthy and pathologic corneas. The best performance was obtained from transfer learning—support vector machine (TL-SVM) (AUC = 0.94, SPE 88%, SEN 100%) and transfer learning—random forest (TL- RF) method (AUC = 0.92, SPE 84%, SEN 100%), followed by convolutional neural network (CNN) (AUC = 0.84, SPE 77%, SEN 91%) and random forest (AUC = 0.77, SPE 60%, SEN 95%). The highest diagnostic accuracy in classifying corneal images was achieved with the TL-SVM and the TL-RF models. In image classification, CNN was a strong predictor. This pilot experimental study developed a systematic mechanized system to discern pathologic from healthy corneas using a small sample.
1. Introduction
In recent years, there has been a significant advancement in computerized corneal digital imaging analysis to develop objective and reproducible machine learning (ML) algorithms for preclinical detection and measurement of various corneal pathologic changes. Standardized quantitative measurement of different corneal structural alterations, such as stromal thinning and edema, inflammatory infiltration, fibrosis and scarring, are crucial for early detection, objective documentation, grading, disease progression, and therapeutic monitoring.
The great diversity of unspecific corneal pathologic lesions, and their significant overlap between disorders, represent a major disadvantage for analysis with spectral-domain optical coherence tomography (SD-OCT) [1]. Unlike other corneal imaging technologies, including corneal topography-tomography and aberrometry that analyze numerical data, the SD-OCT has difficulty providing precise measurement values over varied and unspecific morphologic patterns that could guide clinicians to more objective diagnostic analysis [1,2]. The latter represents a significant challenge for AI developers.
Nonetheless, artificial intelligence (AI) has an immense capacity to learn and analyze a large volume of data and, at the same time, autocorrect and continue learning to improve the sensitivity and specificity as a diagnostic and disease progression tool in ophthalmology [3,4]. Recently, supervised ML has been applied to systematic identification and diagnosis of different ocular pathologies, including diabetic retinopathy [5,6], age-related macular degeneration [7,8,9,10], glaucoma [11,12,13], keratoconus [14,15,16,17], corneal edema [18] and Fuchs endothelial corneal dystrophy (FECD) [19], among others. Different deep learning and conventional ML analysis methods are used in ophthalmology; among the most commonly applied are random forest (RF) [20], support vector machine (SVM) [21,22], convolutional neural network (CNN) [23,24] and transfer learning (TL) [25,26,27]. RF solves classification and regression problems based on rules to binary split data by assembling many decision trees for classification. In this model, the prediction is made by majority voting [28]. In SVM, a given labeled training data is submitted to an algorithm that outputs an optimal hyperplane that separates the elements in different groups [29]. CNN employs algorithms that use a cascade of multilayered artificial neural networks for feature extraction and transformation of data, and TL is a machine learning method where a model developed (e.g., fine-tuned weights) for a task is reused as the starting point for a model on a second task [30,31]. These deep learning algorithms provide an extraordinary amount of information, which is crucial for data analysis, but also, such information can be overwhelming and may significantly affect decision making.
Considering the challenge resulting from SD-OCT pathologic corneal imaging analysis, the purpose of this experimental pilot study was to train different known ML algorithms in smaller corneal SD-OCT imaging samples to know which of them are more reliable for accurate discrimination between healthy and pathologic corneas. Previous studies have focused on numerically measurable specific corneal pathologies like keratoconus and stromal edema.
2. Materials and Methods
(A) Design.
A prospective, cross-sectional, pilot exploratory cohort study was designed. Two participant groups were formed. The experimental group consisted of patients with various corneal diseases, and a control group of healthy patients with no corneal pathology. All patients read and signed informed consent to participate voluntarily in the study, which was previously approved by the Ethics and Research Committees of our institution (protocol registration No. CONBIOETICA-14-CEI-0003-2019 and 18-CI-14-120058, respectively), and conducted according to the tenets of the Declaration of Helsinki.
(B) Experimental procedures.
Figure 1 shows the workflow of experiments performed in the study. All corneal SD-OCT analyses were performed by the same technician (SIS). SD-OCT is a conventional imaging diagnostic tool in ophthalmology clinics that helps study the microstructural changes of different eye pathologies, including the cornea [32]. SD-OCT provided non-contact in-vivo corneal cross-sectional, high-resolution images. The Avanti RTVue XR (Optovue®, Fremont, CA, USA) SD-OCT corneal module permitted a high-speed acquisition of image frames (1024 axial scans in 0.4 s) with little motion artifacts, reducing background noise. This device worked at a wavelength of 830 nm and a speed of 26,000 A-scans per second [33]. All the experiments were performed on a dataset comprising cross-line morphologic corneal images belonging to a random set of corneal pathologies. This set of images was compared to healthy corneas (control group). The problem was confronted using binary classification methods. The quest illustrated three approaches:
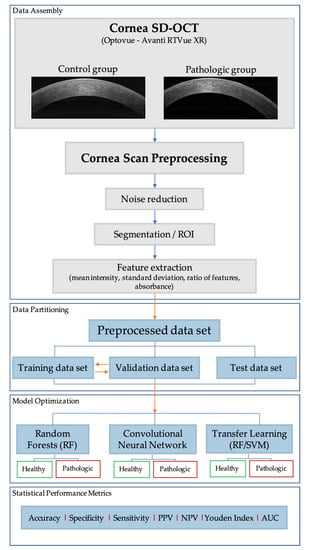
Figure 1.
Workflow diagram of the research methodology. Spectral-domain optical coherence tomography (SD-OCT), region of interest (ROI), random forest (RF), support vector machine (SVM), positive predictive value (PPV), NPV: negative predictive value (NPV) and area under the curve (AUC).
- Traditional machine learning, including RF and SVM.
- Deep learning using end-to-end CNN.
- TL using the pre-trained model (e.g., AlexNet) [34].
(C) Segmentation and Feature Extraction.
Cornea Scan Postprocessing: Digital SD-OCT images are usually contaminated with noise inherited from the sensor (Figure 2a). This problem is easily circumvented by applying a 2D median filter. Nevertheless, in order to extract statistical features from these images, one must do what is known in the imaging domain as image segmentation, which partitions the image into different segments (a.k.a., regions), which are, in our case, three segments (cornea region, background region and region of interest) as shown in Figure 2b–d, respectively. In the figure, the cornea region mask is obtained by applying contrast limited adaptive histogram equalization (CLAHE) followed by fixed thresholding (Figure 2b). The background region is simply the inverse of the mask (Figure 2c). The region of interest (ROI) was obtained using our fast image segmentation method based on Delaunay Triangulations [35,36], which was fully automatic and did not require specifying the number of clusters, as was the case with the k-means clustering [37] or the multilevel image thresholds using Otsu’s method [38] (Figure 2d).
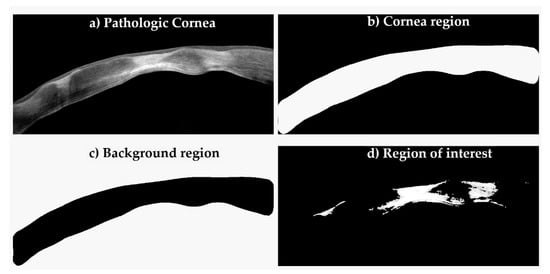
Figure 2.
Digital image segmentation process. In order to extract statistical features, image segmentation was performed into different portions.
Feature Extraction: In image processing and its intersection with ML, feature extraction plays a crucial role in pattern recognition [39]. The process starts by calculating a set of measured data from images intended to be informative and non-redundant, facilitating subsequent machine learning tasks. There is a plethora of features one can extract from images; however, in this study, we resorted to measuring a few simple statistical features, which are: (1) the mean intensity value, (2) the standard deviation of values, (3) the ratio of features (“/” denotes the ratio of two columns, e.g., H/J is the ratio of column H and column J) and (4) the absorbance (Table 1). Absorbance is a transform made to the image, calculated by using, where ln is the natural logarithm, A is the image and is the mean of the background region of A (Figure 2).

Table 1.
Sample of statistical features retrieved from all corneal SD-OCT digital images. Mean: mean intensity value; std: standard deviation value; “/”: ratio of two columns.
(D) Experimental set-up.
Random forest: The experimental set up to execute RF classification of data into healthy or pathologic images comprised two stages. The bag of decision trees was set to 50 growing trees (this number provides the right balance between the area under the curve (AUC), processing time and memory usage) in the training stage, which was used with a training set of 50 control and 15 pathologic images. The RF constructed decision trees based on the predictive characteristics of the features in Table 1. See also Figure 3 for a visualization of three out of the fifty trees. Figure 4 depicts how RF optimizes its performance across several growing trees (50 trees in our case) by performing out-of-bag (OBD) error calculation. The training phase converged into a model that we used, in the second stage, on a new validation test dataset comprising 21 controls and 7 pathologic images.
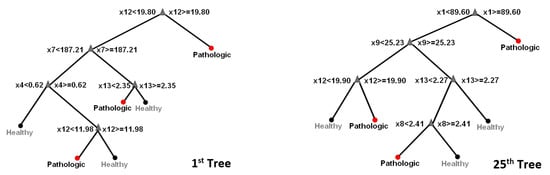
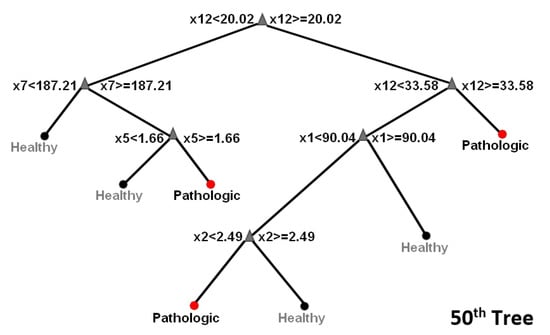
Figure 3.
Random forest tree visualization. Ensemble classifiers from the aggregation of multiple decision trees.
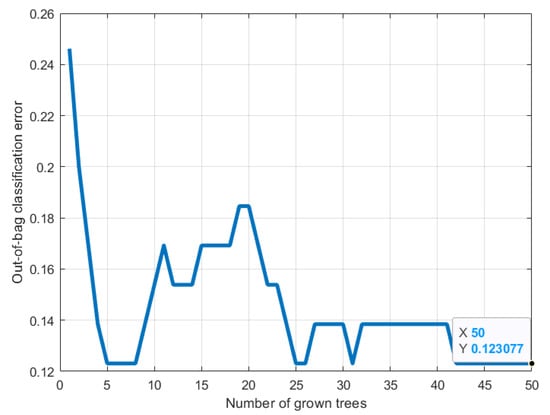
Figure 4.
Random forest classification error minimization across the growing trees during training. RF optimizes its performance across several growing trees (e.g., 50 trees) by performing out-of-bag (OBD) error calculation.
Convolutional Neural Network: CNN is a deep learning method and architecture that is well known in its capabilities for image classification [40,41]. Input image dimensions were fixed to [227 227 3] (to match that of the AlexNet pre-trained model input size requirement), and the fully connected layer was set to two classes (healthy/pathologic). The architecture embodied three convolution layers, each of which had a filter size of 5-by-5, the activation function was set to the rectified linear unit (ReLU) and the number of epochs was set to eight. Images were fed directly to the CNN classifier with the same training and testing proportions as in section A.
Transfer Learning: In TL, the statistical model we used for prediction had been pre-trained on an enormous data set of natural images (eg., millions of samples), and the weights were then used in local learning [41,42]. Features were augmented to yield 4096 features, which were then fed to classifiers with the same training and testing proportions as in section A. They used shallow learning classifiers such as the SVM and RF.
(E) Statistical metrics analysis.
We categorized the registries in cases with corneal pathology and healthy corneal OCT entries. We used traditional machine learning, including random forest (RF) and support vector machine (SVM), deep learning using the CNN and transfer learning (TL) using a pre-trained model (e.g., AlexNet). Then we applied the algorithm to another image matrix and finally measured the model precision, relative risk, sensitivity, specificity, negative predictive and positive predictive values of the algorithm. Receiver operating characteristic (ROC) curves were analyzed to determine the optimal cut-off values, sensitivity and specificity. The AUC was used as a measure of accuracy. Accuracy was measured on the test dataset (correctly predicted class/total testing class) × 100. All measurements were reported as the average of 10 runs on a random selection of samples to eliminate any training/test dataset selection bias. For all models, the data was divided according to the same ratio shown in Table 2.

Table 2.
Training and test data ratio.
All of the experiments, including the developed algorithms, were implemented using MATLAB ver. 9.5.0.944444 (R2018b) and IP Toolbox ver. 10.3 running on a 64-bit workstation with Windows 10 and 32.GB of RAM, 2.60 GHz.
3. Results
A total of 93 SD-OCT corneal images were registered in the study, 71 images formed part of the control group, and 22 pathologic images were included in the experimental group. The latter comprised 14 (63.6%) ectatic corneas and 8 (36.4%) corneas with infectious keratitis. The total analyzed corneas belonged to 55 (59.2%) women and 38 (40.8%) men. The mean age of patients in the experimental group was 38.68 ± 11.74 years, and in the control group, 45.56 ± 20.69 years.
We tested each model sequence’s accuracy to assign the entry as pathologic or healthy. The RF method (AUC = 0.77, SPE 60%, SEN 95%) had the lowest precision in the set (86.07%, ±5.44); however, the model only used 16 possible features extracted from the data, followed by CNN (AUC = 0.84, SPE 77%, SEN 91%). Figure 5 displays the importance of statistical features of images analyzed by RF. The measure represents the increase in prediction error for any given variable if that variable’s values are permuted across the out-of-bag observations. This measure is computed for every tree, then averaged over the entire ensemble, and divided by the standard deviation over the entire ensemble. It appears that features 12, 13 and 15 (i.e., corresponding to columns M, N, P in Table 1) are the most important in this classification model.
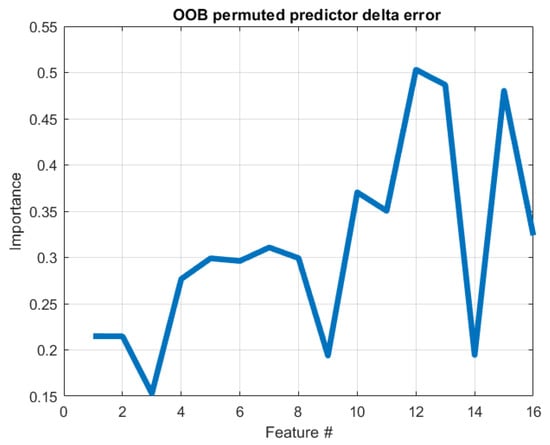
Figure 5.
Feature importance using the random forest method. The plot represents the increase in prediction error for any given variable if the values of that variable are permuted across the out-of-bag observations. Therefore, the higher the induced error is, the higher is the importance of that variable; out-of-bag (OOB).
Algorithms that used more (4096) had higher precision. The transfer learning-SVM model yielded the best results (TL-SVM. AUC = 0.94, SPE 88.12%, SEN 100%) followed by the transfer learning-RF model (TL-RF. AUC = 0.92, SPE 84%, SEN 100%). The visualization of this classification’s performance can be further examined in Figure 6 showing the best performing test’s confusion matrices from the 10 random tests for each algorithm. Table 3 summarizes the outcomes of the constructed models.
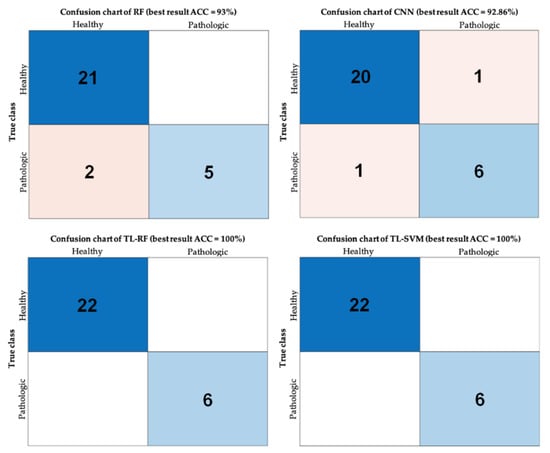
Figure 6.
Confusion charts. Performance visualization using confusion matrix charts for the examined classification problem using the four different approaches (best accuracy out of the ten tests); accuracy (ACC).

Table 3.
Performance of the classification methods using different algorithms. All metrics are the average over 10 runs (**) Area under the curve (-) convolutional neural network (CNN) extracts its own features automatically from images.
4. Discussion
AI facilitates the early detection of certain ophthalmic disorders, increasing their diagnostic accuracy. Additionally, it enables the objective evaluation of disease progression and detailed follow-up of therapeutic results, particularly in those disorders most related to images and numerical data from a distance, curvature, elevation, volume and thickness measurements of specific ocular structures, like the cornea, iris, lens and retina [30]. It is a powerful tool that permits us to increase the diagnostic sensitivity and specificity of ophthalmic pathology [43].
The corneal shape, volume, thickness, elevation and refractive power analysis has significantly evolved in recent years through the development of topography, tomography, pachymetry and aberrometry, which yield accurate corneal measurements. Because these devices generate multiple maps and images of the cornea, the amount of numerical data available from each acquisition may be overwhelming. Therefore, machine learning algorithms are being applied for early detection and accurate progression analysis of different corneal conditions, including keratoconus and endothelial health [16,43,44]. Currently, refractive surgery screening is the most fertile field for machine learning development in corneal disease. The reason for this is the increased risk of iatrogenic post-LASIK keratectasia due to unrecognized preoperative evaluation [16].
This study’s objective was to test and train different ML algorithms’ performance for the morphologic discrimination between pathologic and healthy corneas to optimize their role in early detection, differentiation and monitoring disease progression. There are few studies related to this challenging task, and a critical reason for this is the great morphologic variation and overlap of clinical manifestations among corneal disorders [18,19,45]. For example, when discriminating keratoconic corneas from healthy ones, metric parameters like the radius of curvature, elevation over a based-fit-sphere plane, volume and corneal thickness changes are considered for analysis [46]. The same is true for corneal stromal edema, where epithelial and stromal thickness and volume, measurable parameters are considered for analysis, but for discrimination and diagnostic differentiation of other non-measurable and unspecific clinical findings, like scarring, leukomas, inflammatory infiltrates, among other pathologic features of disorders like infectious keratitis, the application of ML algorithms becomes more complex and limited [18].
Artificial neural networks like CNN are strong predictors in image classification, with the advantage of dealing with noisy and missed clinical data, understanding complex data patterns in a way not possible for linear and non-linear calculations. However, this model requires massive clinical datasets (in the order of tens of thousands) for proper training [47], explaining why the CNN model performed poorly with the limited data set used for analysis in the present study [5]. Moreover, the availability of large training datasets is not always feasible, especially in corneal imaging analysis. There are specific difficulties, including the devices’ high costs, technical acquisition challenges and methodology differences that prevent building large datasets. Furthermore, when analyzing large datasets, there is a need for high computational power, limiting availability and increasing costs.
On the other hand, our results highlight the benefit of adopting the TL approach. A linear solution (two dimensional) is impossible in many ophthalmologic cases as discussed before; therefore, getting a solution in a higher-dimensional dataset is required. An advantage of the TL-RF method is that it can model non-linear class boundaries and may give variable importance, but at the same time, it may be slow, and it may be difficult to get insights into the decision rules. The TL-SVM solves the linearity problem with a relatively lower computational cost using the kernel trick, a function used to obtain non-linear variants of a selected algorithm [29]. In the present study, the highest diagnostic accuracy in classifying normal from pathologic corneal images was achieved with the TL-SVM and the TL-RF models in this order. Even though transfer learning models (e.g., AlexNet) are models that have been trained to extract reliable and unique features from millions of raw images, our study supports the hypothesis that their image descriptors can be extended to medical images.
Indeed, random forest learning models can achieve satisfactory outcomes with small datasets, but with the inconvenience of requiring a manual selection of specific visual features before classification. This condition can result in a set of suboptimal features that limits the algorithms’ application [43]. RF generates meta-models or ensembles of decision trees, and it is capable of fitting highly non-linear data given relatively small samples [48]. RF reached this performance with merely 16 features (values), a strong indication that if more statistical features (hypothetically driven by ophthalmology experts) are added, the model may, with overwhelming probability, outperform CNN in this small dataset. Additionally, unlike other deep learning models, RF models can pinpoint individual feature significance, which can help trace back diseases and trigger scientific discovery or physiological new findings.
Considering potential future research directions, we envision extending this work to classify different corneal disease sub-types; this will eventually aid clinicians in their diagnostic procedures. Furthermore, we aim to develop algorithms for risk prediction of corneal disease, which will help us identify individuals at higher risk of developing a corneal disease (e.g., personalized medicine), hence improving its earlier detection before the disease reaches a devastating stage. However, as mentioned before, the necessity of collecting a substantial amount of data to get more accurate predictions and also to be able to use RF algorithms that are more suitable for image analysis is imperative, but an arduous task [49,50].
Related work: In the last couple of years, several ophthalmology groups have been testing different ML algorithms to facilitate and improve the accuracy of clinical diagnosis of several corneal disorders (Table 4) [51]. Santos et al. reported CorneaNet, a CNN design for fast and robust segmentation of cornea SD-OCT images from healthy and keratoconic corneas with an accuracy of 99.5% and a segmentation time of less than 25 ms [46]. Kamiya et al. used deep learning in color-coded maps obtained by AS-OCT, achieving an accuracy of 0.991 in discriminating between keratoconic and healthy eyes and an accuracy of 0.874 in determining the keratoconus stage [52]. Shi et al. used Scheimpflug camera images and ultra-high-resolution optical coherence tomography (UHR-OCT) imaging data to improve subclinical keratoconus diagnostic capability using CNN. When only the UHR-OCT device was used for analysis, the diagnostic accuracy of subclinical keratoconus was 93%, the sensitivity reached 95.3% and the specificity 94.5%. These parameters were similar when both the Scheimpflug camera and the UHR-OCT were used in combination for analysis [53]. Treder et al. evaluated the performance of a deep learning-based algorithm in the detection of graft detachment in AS-OCT after Descemet membrane endothelial keratoplasty (DMEK) surgery with a sensitivity of 98%, a specificity of 94% and an accuracy of 96% [45]. Zéboulon et al. trained a CNN to classify each pixel in the corneal OCT images as normal or edematous and generate colored heat maps of the result, with an accuracy of 98.7%, a sensitivity of 96.4% and specificity of 100% [18]. Eleiwa et al. assessed the performance of a deep learning algorithm to detect Fuchs’ endothelial corneal dystrophy (FECD) and identify early-stage disease without clinically evident edema using OCT images. The final model achieved an AUC = 0.998 ± 0.001 with a specificity of 98% and sensitivity of 99% in discriminating healthy corneas from all FECD [19]. Yousefi et al. used AI to help the corneal surgeon identify higher-risk patients for future corneal or endothelial transplants using OCT-based corneal parameters [54]. After refined and made available through computerized ophthalmic diagnostic equipment, all these research developments will have a significantly impact in ophthalmic health care services around the world. Interestingly, none of these previous reports, particularly those involving corneal SD-OCT imaging analysis, used TL-SVM and TL-RF algorithms to analyze corneal pathology.

Table 4.
Comparative analysis of recent reports using different machine learning algorithms for the discrimination of corneal OCT images from different corneal pathologies.
5. Conclusions
Applying ML optimal algorithms to different corneal pathologies for early detection, accurate diagnosis and disease progression is challenging. There are economic limitations related to the high cost of equipment, technical acquisition challenges and differences in the methodologic analysis that make it difficult to build large and reliable datasets. Furthermore, without the availability of vast datasets to feed data-hungry ML algorithms, these algorithms would remain limited in their capability to yield reliable results.
In the present experimental pilot study with limited dataset samples, TL-SVM and TL-RF showed better sensitivity and specificity scores, and hence, they were more accurate to discriminate between healthy and pathologic corneas. On the contrary, the CNN algorithm showed less reliable results due to the limited training samples. The RF algorithm with handcrafted features had an acceptable performance, although, we trust that adding more hypothesis-driven features (16 features) would improve the performance furthermore. Additionally, RF is still a good option when one wants to explore specific imaging features and their physiological trace, for instance, we observed that features 12, 13 and 15 (i.e., corresponding to columns M, N, P in Table 1) are the most important in the RF classification model.
We believe that this revolutionary technology will mark the beginning of a new trend in image processing and corneal SD-OCT analysis, differing from current tendencies, where diverse anatomic characteristics like reflectivity, shadowing, thickness, among others, are subjectively analyzed. Nevertheless, this study may act as a proof of concept where external validations especially on large datasets are sought.
Author Contributions
Conceptualization, A.B.-A. and A.R.-G.; design of the work, A.B.-A., A.C. and A.R.-G.; analysis, A.C.; acquisition of data, A.B.-A.; interpretation of data, A.B.-A., A.C., J.C.J.-P. and A.R.-G.; drafting the work, A.B.-A., A.C., J.C.J.-P. and A.R.-G.; writing, A.B.-A., A.C., J.C.J.-P. and A.R.-G.; revising, A.B.-A., A.C., J.C.J.-P. and A.R.-G. All authors have read and agreed to the published version of the manuscript.
Funding
The Immuneye Foundation, Monterrey, Mexico.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by Centro de Retina Medica y Quirurgica, S.C. Ethics and Research Committees (protocol registration No. CONBIOETICA-14-CEI-0003-2019 and 18-CI-14-120058, respectively).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available because the study was approved under informed consent for the use of clinical records data for research purposes, protecting their privacy, prohibiting sharing information with third parties according to the Mexican General Law for the Protection of Personal Data in Possession of Obliged Parties, (published in the Federation Official Diary on January 26, 2017).
Acknowledgments
Susana Imperial Sauceda for corneal SD-OCT analyses.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yaqoob, Z.; Wu, J.; Yang, C. Spectral domain optical coherence tomography: A better oct imaging strategy. Biotechniques 2005, 39, S6–S13. [Google Scholar] [CrossRef]
- Lee, S.S.; Song, W.; Choi, E.S. Spectral domain optical coherence tomography imaging performance improvement based on field curvature aberration-corrected spectrometer. Appl. Sci. 2020, 10, 3657. [Google Scholar] [CrossRef]
- Riaz, H.; Park, J.; Choi, H.; Kim, H.; Kim, J. Deep and densely connected networks for classification of diabetic retinopathy. Diagnostics 2020, 10, 24. [Google Scholar] [CrossRef]
- Chaddad, A.; Kucharczyk, M.J.; Cheddad, A.; Clarke, S.E.; Hassan, L.; Ding, S.; Rathore, S.; Zhang, M.; Katib, Y.; Bahoric, B.; et al. Magnetic resonance imaging based radiomic models of prostate cancer: A narrative review. Cancers 2021, 13, 552. [Google Scholar] [CrossRef]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Cheung, C.Y.-L.; Lim, G.; Tan, G.S.W.; Quang, N.D.; Gan, A.; Hamzah, H.; Garcia-Franco, R.; San Yeo, I.Y.; Lee, S.Y.; et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 2017, 318, 2211–2223. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Baughman, D.M.; Lee, A.Y. Deep learning is effective for the classification of OCT images of normal versus age-related macular degeneration. Ophthalmol. Retin. 2017, 1, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Burlina, P.M.; Joshi, N.; Pekala, M.; Pacheco, K.D.; Freund, D.E.; Bressler, N.M. Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol. 2017, 135, 1170–1176. [Google Scholar] [CrossRef] [PubMed]
- Aslam, T.M.; Zaki, H.R.; Mahmood, S.; Ali, Z.C.; Ahmad, N.A.; Thorell, M.R.; Balaskas, K. Use of a neural net to model the impact of optical coherence tomography abnormalities on vision in age-related macular degeneration. Am. J. Ophthalmol. 2018, 185, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Grassmann, F.; Mengelkamp, J.; Brandl, C.; Harsch, S.; Zimmermann, M.E.; Linkohr, B.; Peters, A.; Heid, I.M.; Palm, C.; Weber, B.H.F. A deep learning algorithm for prediction of age-related eye disease study severity scale for age-related macular degeneration from color fundus photography. Ophthalmology 2018, 125, 1410–1420. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, U.; Fujita, H.; Bhandary, S.V.; Gudigar, A.; Tan, J.H.; Acharya, U.R. Deep convolution neural network for accurate diagnosis of glaucoma using digital fundus images. Inf. Sci. 2018, 441, 41–49. [Google Scholar] [CrossRef]
- Li, Z.; He, Y.; Keel, S.; Meng, W.; Chang, R.T.; He, M. Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs. Ophthalmology 2018, 125, 1199–1206. [Google Scholar] [CrossRef]
- Yousefi, S.; Kiwaki, T.; Zheng, Y.; Sugiura, H.; Asaoka, R.; Murata, H.; Lemij, H.; Yamanishi, K. Detection of longitudinal visual field progression in glaucoma using machine learning. Am. J. Ophthalmol. 2018, 193, 71–79. [Google Scholar] [CrossRef]
- Ambrósio, R.; Lopes, B.T.; Faria-Correia, F.; Salomão, M.Q.; Bühren, J.; Roberts, C.J.; Elsheikh, A.; Vinciguerra, R.; Vinciguerra, P. Integration of scheimpflug-based corneal tomography and biomechanical assessments for enhancing ectasia detection. J. Refract. Surg. 2017, 33, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, I.R.; Rodriguez, P.; Rozema, J.J.; Dhubhghaill, S.N.; Zakaria, N.; Tassignon, M.J.; Koppen, C. Evaluation of a machine-learning classifier for keratoconus detection based on Scheimpflug tomography. Cornea 2016, 35, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Lopes, B.T.; Ramos, I.C.; Salomão, M.Q.; Guerra, F.P.; Schallhorn, S.C.; Schallhorn, J.M.; Vinciguerra, R.; Vinciguerra, P.; Price, F.W.; Price, M.O.; et al. Enhanced tomographic assessment to detect corneal ectasia based on artificial intelligence. Am. J. Ophthalmol. 2018, 195, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Hwang, E.S.; Perez-Straziota, C.E.; Kim, S.W.; Santhiago, M.R.; Randleman, J.B. Distinguishing highly asymmetric keratoconus eyes using combined scheimpflug and spectral-domain OCT analysis. Ophthalmology 2018, 125, 1862–1871. [Google Scholar] [CrossRef] [PubMed]
- Zéboulon, P.; Ghazal, W.; Gatinel, D. Corneal edema visualization with optical coherence tomography using deep learning. Cornea 2020, in press. [Google Scholar] [CrossRef]
- Eleiwa, T.; Elsawy, A.; Özcan, E.; Abou Shousha, M. Automated diagnosis and staging of fuchs’ endothelial cell corneal dystrophy using deep learning. Eye Vis. 2020, 7, 44. [Google Scholar] [CrossRef]
- Meinshausen, N. Quantile regression forests. J. Mach. Learn. Res. 2006, 7, 983–999. [Google Scholar]
- Chang, C.C.; Lin, C.J. LIBSVM: A Library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Haffner, P.; Bottou, L.; Bengio, Y. Object Recognition with gradient-based learning. In Shape, Contour and Grouping in Computer Vision; Forsyth, D.A., Mundy, J.L., Gesú, V., Cipolla, R., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 1999; pp. 319–345. [Google Scholar]
- Freund, Y.; Schapire, R. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef]
- Ayodele, T.O. Types of machine learning algorithms. In New Advances in Machine Learning; Zhang, Y., Ed.; InTech: Rijeka, Croatia, 2010. [Google Scholar]
- Lu, W.; Tong, Y.; Yu, Y.; Xing, Y.; Chen, C.; Shen, Y. Applications of artificial intelligence in ophthalmology: General overview. J. Ophthalmol. 2018, 2018, 5278196. [Google Scholar] [CrossRef] [PubMed]
- Segal, M.R. Machine learning benchmarks and random forest regression. In UCSF: Center for Bioinformatics and Molecular Biostatistics; eScholarship Publishing, University of Califormia at San Franciso, 2004; Available online: https://escholarship.org/uc/item/35x3v9t4 (accessed on 9 February 2019).
- Schölkopf, B.; Smola, A.J. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond; MIT Press: Cambdrige, MA, USA, 2002. [Google Scholar]
- Ting, D.S.W.; Pasquale, L.R.; Peng, L.; Campbell, J.P.; Lee, A.Y.; Raman, R.; Tan, G.S.W.; Schmetterer, L.; Keane, P.A.; Wong, T.Y. Artificial intelligence and deep learning in ophthalmology. Br. J. Ophthalmol. 2019, 103, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Devi, M.A.; Ravi, S.; Vaishnavi, J.; Punitha, S. Classification of cervical cancer using artificial neural networks. Procedia. Comput. Sci. 2016, 89, 465–472. [Google Scholar] [CrossRef]
- Hahn, P.; Migacz, J.; O’Connell, R.; Maldonado, R.S.; Izatt, J.A.; Toth, C.A. The use of optical coherence tomography in intraoperative ophthalmic imaging. Ophthalmic Surg. Lasers Imaging 2011, 42, S85–S94. [Google Scholar] [CrossRef]
- Huang, D.; Duker, J.S.; Fujimoto, J.G.; Lumbroso, B.; Schuman, J.S.; Weinred, R.N. Imaging the Eye from Front to Back with RTVue Fourier-Domain Optical Coherence Tomography, 1st ed.; Huang, D., Duker, J.S., Fujimoto, J.G., Lumbroso, B., Eds.; Slack Incorporated: San Francisco, CA, USA, 2010. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the 2012 Annual Conference on Neural Information Processing Systems (NIPS), Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Cheddad, A.; Condell, J.; Curran, K.; Mc Kevitt, P.; Kevitt, P.M. On points geometry for fast digital image segmentation. In Proceedings of the 8th International Conference on Information Technology and Telecommunication; Galway Mayo Institute of Technology, Galway, Ireland, 24 October 2008; pp. 54–61. [Google Scholar]
- Cheddad, A.; Mohamad, D.; Manaf, A.A. Exploiting voronoi diagram properties in face segmentation and feature extraction. Pattern Recognit. 2008, 41, 3842–3859. [Google Scholar] [CrossRef]
- Lloyd, S.P. Least squares quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man. Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Nixon, M.S.; Aguado Gonzalez, A.S. Feature Extraction & Image Processing for Computer Vision, 4th ed.; Academic Press: London, UK, 2020. [Google Scholar]
- Lekamlage, C.D.; Afzal, F.; Westerberg, E.; Cheddad, A. Mini-DDSM: Mammography-based automatic age estimation. In Proceedings of the 3rd International Conference on Digital Medicine and Image Processing (DMIP 2020), ACM, Kyoto, Japan, 6–9 November 2020. [Google Scholar]
- Banerjee, I.; Ling, Y.; Chen, M.C.; Hasan, S.A.; Langlotz, C.P.; Moradzadeh, N.; Chapman, B.; Amrhein, T.; Mong, D.; Rubin, D.L.; et al. Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification. Artif. Intell. Med. 2019, 97, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Cheddad, A. On box-cox transformation for image normality and pattern classification. IEEE Access 2020, 8, 154975–154983. [Google Scholar] [CrossRef]
- Lopes, B.T.; Eliasy, A.; Ambrosio, R. Artificial intelligence in corneal diagnosis: Where are we? Curr. Ophthalmol. Rep. 2019, 7, 204–211. [Google Scholar] [CrossRef]
- Kolluru, C.; Benetz, B.; Joseph, N.; Lass, J.; Wilson, D.; Menegay, H. Machine learning for segmenting cells in corneal endothelium images. In Proceedings of the SPIE, San Diego, CA, USA, 13 March 2019; Volume 10950, p. 109504G. [Google Scholar] [CrossRef]
- Treder, M.; Lauermann, J.L.; Alnawaiseh, M.; Eter, N. Using deep learning in automated detection of graft detachment in descemet membrane endothelial keratoplasty: A pilot study. Cornea 2019, 38, 157–161. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, V.A.; Schmetterer, L.; Stegmann, H.; Pfister, M.; Messner, A.; Schmidinger, G.; Garhofer, G.; Werkmeister, R.M. CorneaNet: Fast segmentation of cornea OCT scans of healthy and keratoconic eyes using deep learning. Biomed. Opt. Express 2019, 10, 622. [Google Scholar] [CrossRef]
- Livingstone, D.J.; Manallack, D.T.; Tetko, I.V. Data modelling with neural networks: Advantages and limitations. J. Comput. Aided. Mol. Des. 1997, 11, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.K.; Cheddad, A.; Andersson, P. Random forest surrogate models to support design space exploration in aerospace use-case. In IFIP Advances in Information and Communication Technology, Proceedings of the International Conference Artificial Intelligence Applications and Innovations (AIAI’19), Hersonissos, Greece, 24–26 May 2019; MacIntyre, J., Maglogiannis, I., Iliadis, L., Pimenidis, E., Eds.; Springer: Crete, Greece, 2019; pp. 532–544. [Google Scholar]
- Cheddad, A.; Czene, K.; Eriksson, M.; Li, J.; Easton, D.; Hall, P.; Humphreys, K. Area and volumetric density estimation in processed full-field digital mammograms for risk assessment of breast cancer. PLoS ONE 2014, 9, e110690. [Google Scholar] [CrossRef]
- Cheddad, A.; Czene, K.; Shepherd, J.A.; Li, J.; Hall, P.; Humphreys, K. Enhancement of mammographic density measures in breast cancer risk prediction. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1314–1323. [Google Scholar] [CrossRef]
- Balyen, L.; Peto, T. Promising artificial intelligence-machine learning-deep learning algorithms in ophthalmology. Asia Pac. J. Ophthalmol. 2019, 8, 264–272. [Google Scholar] [CrossRef]
- Kamiya, K.; Ayatsuka, Y.; Kato, Y.; Fujimura, F.; Takahashi, M.; Shoji, N.; Mori, Y.; Miyata, K. Keratoconus detection using deep learning of colour-coded maps with anterior segment optical coherence tomography: A diagnostic accuracy study. BMJ Open 2019, 9, e031313. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Wang, M.; Zhu, T.; Zhang, Y.; Ye, Y.; Jiang, J.; Chen, S.; Lu, F.; Shen, M. Machine learning helps improve diagnostic ability of subclinical keratoconus using Scheimpflug and OCT imaging modalities. Eye Vis. 2020, 7, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, S.; Takahashi, H.; Hayashi, T.; Tampo, H.; Inoda, S.; Arai, Y.; Tabuchi, H.; Asbell, P. Predicting the likelihood of need for future keratoplasty intervention using artificial intelligence. Ocul. Surf. 2020, 18, 320–325. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).