Abstract
Nowadays, the ability to diagnose brain tumors intraoperatively and delineate their margins, as accurately as possible, is of primordial importance during surgery. However, the exact tumor boundaries and targets are difficult to find due to the similar visual appearances especially at the margins, leading in many cases to poor surgical outcomes and a high risk of recurrences. To solve this dogma, our group tried to determine different tissue optical signatures such as optical parameters and endogenous fluorescence. For that, we conducted a comparative study to differentiate between healthy and tumorous tissues under one- and two-photon excitations on optical properties to explore several optical parameters. In addition, the study of endogenous fluorescence can successfully help with the discrimination between tissue types using one- and two-photon excitations. This works suggests that the multimodal analysis of optical signatures of tissues could be a reliable technique that can be used to differentiate between healthy and tumorous tissues. This can help in the future with the implementation of such techniques in vivo during surgery to help the surgeon with the decision that needs to be taken as for tissue resection.
1. Introduction
The main challenge of any neurosurgical intervention today is based on the accurate identification of the tumor’s margins in order to achieve a complete and successful resection [1,2,3]. In fact, for some extra-axial lesions, the general appearance of the tumor under conventional imaging techniques such as computed tomography (CT) and magnetic resonance imaging (MRI) is sufficient to establish the tumor boundaries. However, these techniques are limited in terms of tumor grading and molecular characterization, making other types of lesions less easily distinguished due to their infiltrating characteristics. This inability to fully visualize invasive brain cancers results in subtotal surgical resection, increasing the chances of recurrences [4]. Similarly, unnecessary removal of brain tissue that does not contain cancerous cells can lead to major neurological deficits that affect the quality of life.
Biomedical optics in all of its fields is one of the research areas being developed to create applications and protocols to help with diagnosis and to treat such obstacles at the molecular level [5]. In fact, such technologies first started with the use of fluorescence imaging and tissue biomarkers for surgical guidance in brain tumor resection using 5-aminolevulinic acid-induced protoporphyrin IX (5-ALA-PpIX) fluorescence [6,7]. However, the use of exogenous dyes is limited by their infiltrating characteristics or even their potential side effects, thus requiring the recruitment of other tissue optical signatures [8,9].
On the other hand, autofluorescence spectroscopy has made significant progress in recent decades, allowing the exploration of tissues and organs for various biomedical applications [10,11,12]. Autofluorescence techniques, including spectroscopy and microscopy, have been utilized to visualize tissue structures and morphology, gain insights into cellular mechanisms and dynamics [13], and identify cancerous tissues [14,15,16,17]. Over the years, autofluorescence has been employed to characterize cancerous tissues in various organs such as the kidney [18], bladder [16], breast [19], brain [20,21,22], lungs, uterus, ovaries, and cervix [23,24]. Endogenous fluorescence, particularly the emission of fluorophores such as nicotinamide adenine dinucleotide (NADH) and flavins (FAD), has been closely monitored to distinguish between healthy, tumoral, and infiltrated tissues [16,25,26,27,28]. Consequently, the autofluorescence of these fluorophores and their related metabolic ratios have been extensively investigated to track tissue metabolism and to provide reliable information that helps to establish optical signatures specific to cancerous tissues [16,21,25].
In this context, our research team tried to study such signatures, either by analyzing human tissue endogenous fluorescence or by extracting specific optical parameters such as tissue absorption and scattering.
To achieve these objectives, our team previously tackled different optical properties’ determination using specific integrating-sphere and inverse Monte Carlo techniques on a large cohort of brain tissues [1]. The work mainly focused on establishing the absorption, scattering, and anisotropy coefficients using 405 nm as the excitation wavelength. The results showed that, by using such techniques, one is able to define several discriminatory indicators between control and tumor samples as well as distinguish between several tumor types.
To complete this multimodality, the present study aims to extend the work towards the near-infrared domain using the same optical techniques previously applied. The cohort accounted for in the current work includes a glioblastoma (GBM) group (a grade IV glioma and primary brain tumor) and a control group (epileptic cortex). As a first step, and using the same integrating-sphere technique, we quantitatively looked at the different optical properties both in the visible and in the near-infrared domains to study and analyze the effect of the wavelength change on the determination of different optical coefficients. These coefficients give an idea about the concentration of hemoglobin (either oxygenated or deoxygenated) and other molecules present in the tissue. Therefore, the extraction of these parameters from brain tissue samples can yield some reliable quantitative data to give a better idea about the penetration depth, the light path in the tissues, and the interactions between the different fluorophores, thus allowing the characterization of tissues and the accurate discrimination between healthy and tumorous tissues [1,2,29]. Second, using a multimodal microscope capable of alternating between confocal and two-photon microscopy, we were able to obtain a variety of contrasts at 405 and 800 nm excitation wavelengths, including non-linear fluorescence imaging, fluorescence spectra, and fluorescence lifetime and, thus, extract additional information related to the molecular interactions and physicochemical environment of the different tissue types. As such, a multimodal campaign that includes a study of different optical properties, endogenous fluorescence lifetime and spectral response, and excitation at different wavelengths, allows a better understanding of the tissue metabolic processes, structure, and microenvironment that all facilitate the transitioning between tumorous and healthy regions.
2. Materials and Methods
2.1. Samples
The samples used in the study were collected from the departments of Neurosurgery and Neuropathology at the GHU Paris Psychiatrie et Neuroscience Hospital, upon the approval of GHU Paris Psychiatrie et Neuroscience Hospital—University Paris Descartes Review Board (CPP Ile de France 3, S.C.3227). All methods were performed in accordance with the relevant guidelines and regulations of this protocol and written informed consent was obtained from all patients involved in the study. A total of 14 samples were used in the following experiments, belonging to different tissue types. Six glioblastoma samples were compared to eight healthy cortex samples mainly coming from an epileptic surgery. These samples were stored at −80 °C after being received from the hospital. A few hours before cutting, the tissues were stored at −20 °C, after which they were cut at −18 °C into 200 µm slices using a cryostat (CM 1950, Leica Microsystems). The tissues were then fixed with ethanol 100° and stored at 4 °C until experimentation.
2.2. Integrating Sphere Setup
A standard optical setup was deployed to measure the transmittance T(λ) and reflectance R(λ), from which one can determine the coefficients of absorption (μa) and scattering (μs), as well as the anisotropy (g) of all samples. To measure each, we used an integrating sphere (model: IS200-4 Thorlabs) that includes four identical ports, each one having a 12.7 mm diameter, and an additional port with a 3 mm diameter was used to collect light from the sphere to a spectrometer (HR2000-Ocean optics) through an optical fiber (QP600, Ocean insight). Light that is generated by the laser (Chameleon ultra II, coherent) first passes by a couple of pinholes that will optimize the power and the shape of the laser light going into the integrating sphere. As for the samples, these were placed at the front of the sphere, where we measured transmittance, and at the back of the sphere, where we measured reflectance. When it comes to measuring the collimated transmittance, this requires a setup independent of the use of the sphere. In fact, to measure anisotropy (or collimated transmittance), three apertures perfectly aligned in front of the laser were used. The setup was optimized prior to measurements with the use of an absorbent filter (OD = 0.85) that reduced the intensity coming from the laser.
The work was focused on the near-infrared domain so, for this reason, an 800 nm excitation wavelength was used. Briefly, and for each of the samples in hand, four to five regions of interest (ROI) were chosen in which transmittance, reflectance, and collimated transmittance were measured. The average of these measurements was taken into consideration when determining the optical properties of each sample.
After undergoing the necessary measurements of the transmittance, reflectance, and collimated transmittance, the inverse adding doubling (IAD) algorithm developed by Scott Prahl (http://omlc.ogi.edu/software/iad/ accessed on 5 April 2017) was used in order to extract the reduced scattering (μs’) and the absorption (μa) coefficients as well as the anisotropy factor g of the samples in hand. Further details about the functioning of this algorithm have been previously presented [1].
2.3. Multimodal Microscopic and Fluorescence Lifetime Imaging (FLIM) Setup
From the above setup, three optical signals on specific ROI were recorded: (1) one- and two-photon spectral analysis using, respectively, 405 nm and 800 nm as excitation wavelengths; (2) two-photon fluorescence lifetime imaging measurements; and (3) fluorescence + second harmonic generation (SHG) imaging under two-photon excitation.
More details about this multimodal microscope setup were published elsewhere [30]. The analysis of the different optical signals obtained were carried out using the Leica software (LAS X version 3.5, Leica microsystems). For the spectra at 405 and 800 nm excitation wavelengths, a specific position per sample was considered, and with the use of the spectral acquisition option in the Leica software, these spectra were generated.
In addition, Matlab (R2017b) and ImageJ programs were also used to generate fluorescence images. As for FLIM measurements, the same position as that of the spectra was considered and analyzed using an 800 nm excitation wavelength. On each fluorescence lifetime image, nine ROIs were selected and then analyzed with the Symphotime software (Version 2.3, PicoQuant, GmbH, Berlin, Germany). The total phasor histogram was extracted from fluorescence lifetime images of the different tissue types, which permits calculation of the long lifetime intensity fraction (LLIF) and is represented by the alpha coefficient. More details about the phasor histogram and the alpha calculation method are available in our previously published works [2,29].
Briefly, each fluorescence decay curve I(t) of every pixel of the FLIM images was converted into two coordinates in a Cartesian plot, following Equations (1) and (2) below:
The index “i” corresponds to a pixel in the FLIM image while S and G are the x and y coordinates of each phasor. ω corresponds to the laser repetition angular frequency and is related to the sampling period (Ts) and to the signal length (L) as ω = 2π/L.Ts.
After regrouping, in a global phasor histogram, all phasor counts of the same tissue type and local maxima of the global histogram were localized and a polynomial fitting line was plotted. The two intersection points between the phasor circle segment and this fitting line are related to the fluorescence lifetime values of the two components of FAD or NADH (free and protein-bound). Afterwards, each phasor count was projected on the fitting line in order to calculate the LLIF (alpha) histogram.
3. Results
3.1. Optical Properties
Figure 1 represents the absorption and scattering as well as the anisotropy coefficients of the control versus tumor brain tissue samples studied at different excitation wavelengths. The presence of previous data from the group on the 405 nm excitation made it possible to compare our results to what was previously done by our team [1]. It is clear that, regardless of the nature of the tissue, there is a large decrease in both the absorption and scattering when shifting between 405 nm and the near-infrared domain. However, the contrary is observed when it comes to anisotropy, whereby there is a remarkable increase in g values when going from the visible to the near IR domain. Moreover, when comparing the boxplots of both the absorption and scattering of each group of tissues, it is clearly visible that there is a large discrepancy in the range of values in µs, which is particularly noticed in the tumorous tissue types (Figure 1b). Furthermore, these measurements give a broad idea about the mean absorption and scattering coefficients in each tissue type. From Figure 1a, which represents the absorption coefficient of the control and tumor groups, one can estimate an average µa value of around 0.2 mm−1 for the control group and an average of around 0.4 mm−1 for the tumorous group both at 800 nm excitation. This shows a difference of around 0.2 mm−1 between control and tumor (p-value = 7.38 × 10−7 < 0.05). The same applies for the 405 nm excitation (p-value = 9.84 × 10−5 < 0.05). On the other hand, from Figure 1b, which represents the scattering coefficients of the corresponding tissue types, one can approximate an average µs value of around 17.2 mm−1 for control tissue and an average of around 20 mm−1 for the tumorous tissue both at 800 nm excitation. This suggests a difference of more than 3 mm−1 when it comes to scattering but this difference is not significant where the calculated p-value = 0.13 > 0.05. However, the difference between the two tissue types is more obvious and significantly discriminative under the 405 nm excitation wavelength in the case of µs (p-value = 6.93 × 10−9 < 0.05). When it comes to the anisotropy coefficient, one can differentiate between the different tissue types at both wavelengths. For instance, by looking at Figure 1c, one can estimate an average of 0.89 for control tissues against 0.91 for tumor tissues at 800 nm excitation with a significant difference (p-value = 5.59 × 10−3 < 0.05) and an average of 0.86 for control against 0.89 for tumor tissues at 405 nm with a significant difference too (p-value = 2.48 × 10−9 < 0.05).
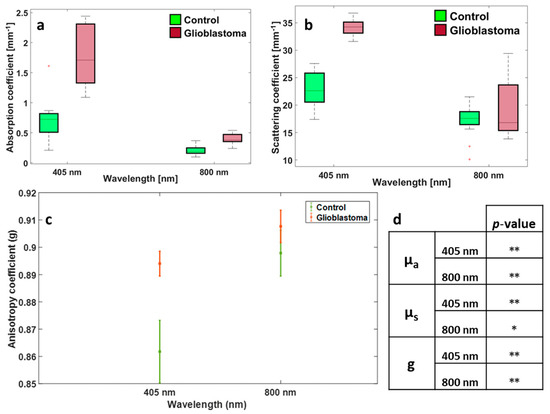
Figure 1.
Distribution of absorption (a), scattering (b), and anisotropy coefficients (c) for the control and tumorous tissues using 405 and 800 nm excitation wavelengths, (d) summary table of p-values obtained by realizing a student for control vs. GBM for each parameter and for each wavelength; **: indicates that p-value < 0.05; *: indicates that p-value > 0.05.
3.2. NIR Spectral Analysis
Figure 2a (control) and b (GBM) illustrate the fluorescence + SHG microscopic images acquired from the nonlinear microscope set-up. The control image shows a homogenous tissue structure with no presence of dense vasculature or collagen fiber structure, while the GBM image shows a denser tissue structure. In addition, it shows typical large vessels surrounded by a dense collagen network presented by the SHG signal (in green).
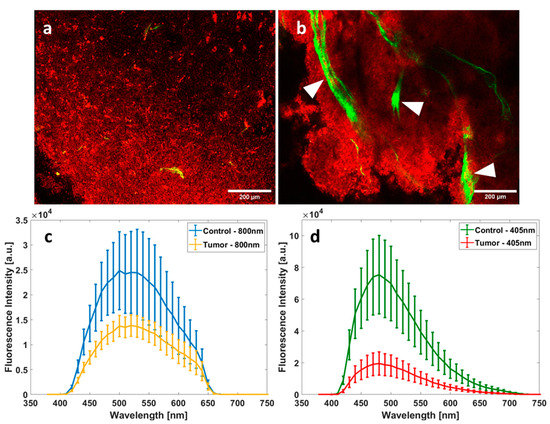
Figure 2.
Spectral and imaging acquisitions of control and tumor samples under one- and two-photon excitation. Fluorescence and SHG microscopic images from the multimodal microscope set-up under 890 nm excitation for control (a) and for GBM (b); Scale bars: 200 µm; red pixels refers to fluorescence signal while green pixels refers to SHG signal; white arrows highlight blood vessels. Comparison between the mean fluorescence emission of control tissues and GBM tissues using 800 nm as excitation wavelength (c) and 405 nm as excitation wavelength (d); a.u: arbitrary unit.
The presence of these large blood vessels, dense tissue structures, and collagen fibers in these images, which are sources of absorption and light scattering, explains the higher scattering and absorption coefficient depicted in Figure 1.
On the other hand, Figure 2c shows the mean fluorescence intensity spectra generated using 800 nm as the excitation wavelength for control and tumor tissues while Figure 2d shows the mean fluorescence spectra of both tissue types using 405 as the excitation wavelength. Differences in spectral shapes and in the intensity of the emitted fluorescence could be easily noticed, comparing 405 nm and 800 nm spectra of the two tissue types.
Using one-photon excitation (405 nm), the probability of a molecule absorbing a photon is higher than that using two-photon excitation (800 nm), so tissues have a higher intensity fluorescence at 405 nm (Figure 2d) than at 800 nm (Figure 2c). However, two-photon excitation is able to efficiently excite some molecule types (here, lipopigments and porphyrins emitting between 500 and 680 nm) that one-photon excitation cannot; that is why we observe a larger spectrum at 800 nm. Regardless of the excitation wavelength, tumorous tissues present a lower fluorescence intensity than control tissues. Going into the details, one can notice that tumor tissues generally show two peaks using an 800 nm excitation wavelength, centered at 540 and 630 nm, respectively, while control tissues on the other hand show a peak centered around 520 nm and another smaller one at around 630 nm at the same excitation.
3.3. FLIM
Figure 3 illustrates the fluorescence lifetime imaging histogram represented using the phasor approach applied to flavins (FAD) and nicotinamide adenine dinucleotide (NADH), respectively (a,c), and the corresponding repartition of the long lifetime contribution for each of the tissue types (b,d). In fact, every pixel generated from the fluorescence lifetime images is displayed as a coordinate on the phasor plot and, using the Fourier transform, the G and S coordinates are calculated [31,32]. The semicircle present in Figure 3a,c represents the universal circle into which pixels with single exponential decays fall. Looking at Figure 3a, which represents the phasor applied to FAD, one can detect two histograms that comply within the semicircle. The phasor falls within the line that passes through these two histograms maxima with average fluorescence lifetime values of 2 and 0.7 ns, respectively. From that, one can extract the alpha coefficient (which represents the long lifetime intensity fraction α) for each of the samples in question. This repartition is observed in Figure 3b,d for both tissue types and FAD and NADH. For FAD, the control group shows α coefficients strictly larger than 0.8 and the tumor group shows α coefficients strictly smaller than 0.8 with a p-value = 4.87 × 10−8 < 0.05, while for NADH there are fewer differences between the control group and tumor, but it is still significant with a p-value = 7.2 × 10−3 < 0.05.
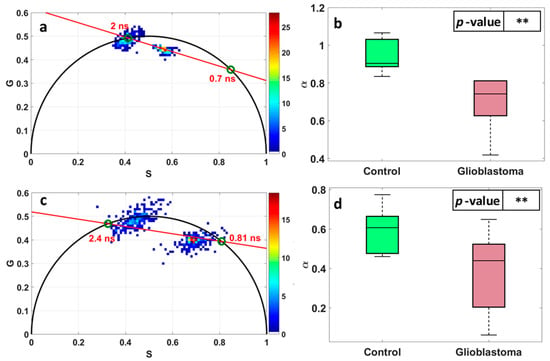
Figure 3.
Fluorescence lifetime imaging (FLIM) histogram represented using the phasor approach applied to FAD (a) and NADH (c). S and G are the coordinates of each phasor count, calculated through equations 1 and 2. Repartition of the long lifetime contribution α of FAD (b) and NADH (d) for each tissue type. **: indicates that p-value < 0.05.
4. Discussion
In this work, we aim to complete our previous study performed in the visible spectral range [1]. So, we worked on the near-infrared range, where the scattering, the absorption, and the anisotropy coefficients were measured using the integrating sphere technique. In addition, spectral measurements were acquired using one- and two-photon excitation. Lastly, we studied the fluorescence lifetime of two molecules using two-photon excitation: NADH and FAD. As mentioned previously, there is a large decrease in both the scattering and absorption coefficients when shifting from the visible to the near-infrared domain regardless of the type. In fact, if we take a look at the therapeutic window of tissues, we see that the scattering and the absorption coefficients of hemoglobin (either oxy or deoxy) attain their minimum between 600 and 900 nm. Because hemoglobin is a major constituent of brain tissues and the main actor in optical properties, when its optical parameters decrease in this excitation wavelength domain, the overall tissue parameters will also decrease. On the other hand, the contrary is observed for the anisotropy coefficient and this is mainly due to slightly more isotropic scattering in the visible domain, which renders this coefficient smaller under a 405 nm excitation [33].
As with past results obtained in the visible range, a discriminative criterion can be found in the near-infrared domain, especially when looking at the absorption coefficient, whereby a discrimination threshold can be defined: a tissue with a µa value higher than 0.25 mm−1 is considered tumorous and less than 0.25 mm−1 is considered healthy. This is simply explained by the fact that in tumorous tissues there is more vascularization, which leads to higher absorption [1,33]. Additionally, the higher the malignancy of the tumor, the more the chromatin is condensed and so the higher the absorption is. These results are generally in accordance with what was previously published in the literature in this excitation wavelength domain, noting that the tissue preparation and measurements techniques are completely different in some of the cases [1,33,34].
This trend is less identified when taking the scattering coefficient into account, whereby when looking at the scattering boxplot at 800 nm, no major differentiation can be made. Moreover, μs values for the different tissue types show a big discrepancy in general. This gap in the results might be affected by different parameters: the difference in the grades of tumor leading to changes in vascularization and chromatin condensation and the fact that GBM-infiltrated extracted tissues could contain healthy regions overlapped with tumorous regions due to the infiltrating character of GBM tumors. The sample cuts may also alter the results, whereby not all samples can be exactly cut at 200 μm, thus leading to some regions being thicker than others. This particularly alters the optical coefficients that tend to be region- and sample-specific as reported in the literature [33].
However, if we take the average of each type into account, we can have a threshold relying on the average of the scattering coefficient value of 17.1 mm−1 for the control group and 20.2 mm−1 for the tumor group, which is by itself discriminative. Additionally, and as previously mentioned, the g values can also act as discriminatory features between different tissue types either in the visible or in the near-infrared domains. As such, and in synchrony with the literature, tumorous tissues usually have g values in the range of 0.91–0.92 as opposed to 0.87–0.89 for the healthy control tissues under a near-infrared excitation [34,35]. This suggests that, no matter what excitation wavelength is being used, one is almost always able to discriminate between healthy and tumorous tissue simply by studying the absorption, scattering, and anisotropy coefficients.
To supplement the optical coefficients study, we decided to study the endogenous fluorescence emission of the two tissue types. This kind of study has shown its reliability and its efficiency to discriminate healthy from tumor tissues and to track the metabolic state of tissues [28,36]. The presence of several distinctive features (such as different auto-fluorescence levels that correspond to cell cytoplasm or even mitochondria) permitted this correlation. Previous studies have demonstrated the choice of the excitation wavelength to be used [28]. It is clear that, when using one-photon excitation (at 405 nm), one is able to attain a much higher fluorescence intensity than by using two-photon microscopy (excitation at 800 nm), regardless of the tissue type. By fact of numbers, the fluorescence intensity is approximately four times higher under one-photon excitation for the control tissue and approximately two times higher for the tumorous tissue. However, under two-photon excitation, where more molecule types can fluoresce compared to one-photon excitation, a broader spectral range than under one-photon excitation can be clearly observed by comparing the spectral shapes, especially in the 550–650 nm spectral range, corresponding to the lipopigments and porphyrins emission range. According to the literature, one can detect in this wavelength range multiple molecules, mainly lipopigments and porphyrins [37]. This shows that, by using two-photon excitation, not only are we using a lower laser power to excite our tissues and collect fluorescence, we are also detecting an additional number of molecules unperceived using one-photon excitation. Therefore, this observation, supported by other observations reported in the literature [38] where one- and two-photon excitation was used to highlight that the presence of albumin affects the fluorescence emission of ASP+, shows the importance of characterizing endogenous molecules under both one- and two-photon excitation in order to understand and reveal their emission behavior when the excitation wavelength/technique changes.
Furthermore, the study of fluorescence spectra confirms that the fluorescence emission is significantly reduced in tumorous tissues as compared to control tissues, regardless of the excitation wavelength [39]. The reason behind this difference in auto-fluorescence is a major debatable topic [40]. The main reason ascribed to this difference is the decrease of lipid levels (lipopigments emission). This decrease is linked to the sensitivity of lipopigments to the metabolic state of the tissue [41], even if it is not tumoral, and that they are affected by several metabolic processes such as oxidative and glycolytic metabolism [29,42,43]. Other explanations, such as tumor necrosis and hyperfusion, have also been reported in the literature as possible explanations for this phenomenon [44,45].
To obtain more information about the metabolic state of tissues, a fluorescence lifetime study was added to this work. Two molecules were considered—the NADH and the FAD. These molecules are involved in several important enzymatic reactions in metabolism as an electron carrier in redox reactions. They have two lifetime components, a long and a short one depending on their chemical states as free or protein-bound. Our lifetime results are globally in synchrony with what has been previously presented in the literature [27,46], where the protein-bound NADH corresponds to the long lifetime in the phasor plot, while the protein-bound FAD corresponds to the short lifetime in the phasor plot [46]. Based on that, and as clearly seen in the phasor plots previously presented, one can discern between two separate histograms in the case of either FAD or NADH. This suggests that one group might manifest a higher concentration of protein-bound FAD while the other might manifest a higher concentration of protein-bound NADH. Luckily, the use of the phasor FLIM method makes it possible to track this behavior by studying the corresponding repartition of the long lifetime contribution for each of the tissues in question. Our results indicate that the control group generally has higher alpha values either in the case of FAD or NADH; meaning that the control group generally matches with the long lifetime values of these fluorophores. Knowing that, one can deduce that the control group reflects a higher concentration of protein-bound NADH and a lower concentration of protein-bound FAD, whereas the opposite applies for the tumor group. These results are consistent with the available literature. However, some groups suggest a slightly higher protein-bound FAD in control tissues [27,32]. This decrease in protein-bound NADH in tumor tissues is explained by the fact that, under hypoxic conditions, cells will favor glycolysis over oxidative phosphorylation in order to produce energy [47]. In parallel, cancerous cells always favor this means of energy production regardless of the conditions [46], meaning that this decrease in protein-bound NADH in tumor cells corresponds to the change in the NADH binding sites that are themselves driven by the change in the metabolic pathway [46]. Similarly, the increase in protein-bound FAD in tumor tissues could be due to decreased levels of NAD+ in these tissues [48]. Here again, using another approach, we were able to accurately discriminate between control and tumor tissues.
5. Conclusions
This study provides new insights into the differentiation between healthy and tumorous tissues. In fact, our continuous quest for discriminative parameters between different brain tissue types led us to a complete multimodal study that focuses mainly on tissue built-in parameters. In fact, and as this work shows, the study of different tissue optical properties, combined with spectral and lifetime analysis, has the promising potential to provide reliable parameters for the discrimination between healthy and tumorous tissues not only in the visible, but also in the near-infrared, domains. However, the sample size used in this study does not allow us to consider that the defined discrimination threshold values of the optical coefficients in this work are absolute or definitive. Therefore, the obtained results should be verified and completed in future works with a larger sample cohort and adding qualitative imaging analysis. Indeed, this work is part of a larger project that consists of building a large multimodal tissue database, in collaboration with Paris Psychiatrie et Neuroscience Hospital. This database consists of collecting multiple optical signatures of different types of healthy and tumor brain tissues using multimodal analysis in order to discriminate each tissue type [1,21,36]. Thus, this work is a preliminary study with promising preliminary results that fills in part of the large multimodal database that we are building.
Therefore, the multimodality is of crucial importance, especially when applied in vivo, as some of these parameters might be altered during real-time surgery (such as blood contamination), leading to a decrease in the collected fluorescent signal. In addition, this multimodality increases the chances of accurately discriminating between not only healthy and solid tumorous tissues but also between healthy and infiltrating brain tumors.
Author Contributions
H.M. worked on the data analysis and on writing the manuscript. E.K.M. collected the data and worked on the data analysis. B.D. and P.V. provided the samples. D.A.H. designed the research project, the protocols for the experiments, supervised the work, and worked on the writing of the text. She provided necessary funds for all the experiments and measurements. All authors reviewed the manuscript and approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Funding
This work is financially supported by ITMO Cancer AVIESAN (Alliance Nationale pour les Sciences de la Vie et de la Santé, National Alliance for Life Sciences and Health) within the framework of the Cancer Plan for MEVO and IMOP projects, by CNRS with the “Défi instrumental” grant, by ligue nationale contre le cancer (LNCC), by the P2IO LabEx and the Institut National de Physique Nucléaire et de Physique des Particules (IN2P3). This work was conducted in the PIMPA Platform partly funded by the French program “Investissement d’Avenir” run by the “Agence Nationale pour la Recherche” (grant “Infrastructure d’avenir en Biologie Santé—ANR—11-INBS-0006”).
Institutional Review Board Statement
This study was conducted and approved by the Institutional Review Board of GHU Paris Psychiatrie et Neuroscience Hospital—University Paris Descartes (CPP Ile de France 3, S.C.3227). All methods were performed in accordance with the relevant guidelines and regulations of this protocol.
Informed Consent Statement
Written informed consent was obtained from all patients involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to their confidentiality as they were acquired from samples of human patients following the approval of the GHU Paris Psychiatrie et Neuroscience Hospital—University Paris Descartes Review Board (CPP Ile de France 3, S.C.3227).
Acknowledgments
We would like to thank Bertrand Devaux, former chief head of the neurosurgery department at the GHU Paris Psychiatrie et Neurosciences Hospital center, the Delegation for Clinical Research and Innovation (DRCI) and the Biological Resources Center (CRB) of GHU Paris Psychiatrie et Neurosciences Hospital center for providing the samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Poulon, F.; Mehidine, H.; Juchaux, M.; Varlet, P.; Devaux, B.; Pallud, J.; Haidar, D.A. Optical properties, spectral, and lifetime measurements of central nervous system tumors in humans. Sci. Rep. 2017, 7, 13995. [Google Scholar] [CrossRef] [PubMed]
- Mehidine, H.; Chalumeau, A.; Poulon, F.; Jamme, F.; Varlet, P.; Devaux, B.; Refregiers, M.; Haidar, D.A. Optical Signatures Derived From Deep UV to NIR Excitation Discriminates Healthy Samples From Low and High Grades Glioma. Sci. Rep. 2019, 9, 8786. [Google Scholar] [CrossRef] [PubMed]
- Harter, D.H.; Wilson, T.A.; Karajannis, M.A. Glioblastoma multiforme: State of the art and future therapeutics. Surg. Neurol. Int. 2014, 5, 64. [Google Scholar] [CrossRef] [PubMed]
- Lara-Velazquez, M.; Al-Kharboosh, R.; Jeanneret, S.; Vazquez-Ramos, C.; Mahato, D.; Tavanaiepour, D.; Rahmathulla, G.; Quinones-Hinojosa, A. Advances in Brain Tumor Surgery for Glioblastoma in Adults. Brain Sci. 2017, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Valdés, P.A.; Roberts, D.W.; Lu, F.-K.; Golby, A. Optical technologies for intraoperative neurosurgical guidance. Neurosurg. Focus 2016, 40, E8. [Google Scholar] [CrossRef]
- Walter, S.; Susanne, S.; Simon, W.; Herbert, S.; Clemens, F.; Claudia, G.; Alwin, E.G.; Rainer, K.; Hans, J.R. Intraoperative Detection of Malignant Gliomas by 5-Aminolevulinic Acid-induced Porphyrin Fluorescence. Neurosurgery 1998, 42, 518–526. [Google Scholar] [CrossRef]
- Alston, L.; Mahieu-Williame, L.; Hebert, M.; Kantapareddy, P.; Meyronet, D.; Rousseau, D.; Guyotat, J.; Montcel, B. Spectral complexity of 5-ALA induced PpIX fluorescence in guided surgery: A clinical study towards the discrimination of healthy tissue and margin boundaries in high and low grade gliomas. Biomed. Opt. Express 2019, 10, 2478–2492. [Google Scholar] [CrossRef]
- Stummer, W.; Pichlmeier, U.; Meinel, T.; Wiestler, O.D.; Zanella, F.; Reulen, H.-J.; ALA-Glioma Study Group. Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: A randomised controlled multicentre phase III trial. Lancet Oncol. 2006, 7, 392–401. [Google Scholar] [CrossRef]
- Stummer, W.; Tonn, J.-C.; Mehdorn, H.M.; Nestler, U.; Franz, K.; Goetz, C.; Bink, A.; Pichlmeier, U. Counterbalancing risks and gains from extended resections in malignant glioma surgery: A supplemental analysis from the randomized 5-aminolevulinic acid glioma resection study. J. Neurosurg. 2011, 114, 613–623. [Google Scholar] [CrossRef]
- Croce, A.; Bottiroli, G. Autofluorescence spectroscopy and imaging: A tool for biomedical research and diagnosis. Eur. J. Histochem. 2014, 58, 2461. [Google Scholar] [CrossRef]
- Shrirao, A.B.; Schloss, R.S.; Fritz, Z.; Shrirao, M.V.; Rosen, R.; Yarmush, M.L. Autofluorescence of blood and its application in biomedical and clinical research. Biotechnol. Bioeng. 2021, 118, 4550–4576. [Google Scholar] [CrossRef]
- Costanzo, V.; D’Apolito, L.; Sardella, D.; Iervolino, A.; La Manna, G.; Capasso, G.; Frische, S.; Trepiccione, F. Single nephron glomerular filtration rate measured by linescan multiphoton microscopy compared to conventional micropuncture. Pflügers Arch.-Eur. J. Physiol. 2022, 474, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, S.; Pieper, M.; Hüttmann, G.; Bölke, T.; Wollenberg, B.; Marsh, L.M.; Garn, H.; König, P. Autofluorescence multiphoton microscopy for visualization of tissue morphology and cellular dynamics in murine and human airways. Lab. Investig. 2016, 96, 918–931. [Google Scholar] [CrossRef]
- Haidar, D.A.; Leh, B.; Zanello, M.; Siebert, R. Spectral and lifetime domain measurements of rat brain tumors. Biomed. Opt. Express 2015, 6, 1219–1233. [Google Scholar] [CrossRef]
- Marcu, L.; Jo, J.A.; Butte, P.V.; Yong, W.H.; Pikul, B.K.; Black, K.L.; Thompson, R.C. Fluorescence Lifetime Spectroscopy of Glioblastoma Multiforme. Photochem. Photobiol. 2007, 80, 98–103. [Google Scholar] [CrossRef]
- Palmer, S.; Litvinova, K.; Dunaev, A.; Yubo, J.; McGloin, D.; Nabi, G. Optical redox ratio and endogenous porphyrins in the detection of urinary bladder cancer: A patient biopsy analysis. J. Biophotonics 2016, 10, 1062–1073. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Chen, Y.; Chen, J.; Chen, H.; Xu, Y.; Zhu, X.; Zhuo, S.; Shi, Z.; Chen, J. Optical diagnosis of gallbladder cancers via two-photon excited fluorescence imaging of unstained histological sections. Lasers Med. Sci. 2014, 30, 225–233. [Google Scholar] [CrossRef]
- Costanzo, V.; Costanzo, M. Intravital Imaging with Two-Photon Microscopy: A Look into the Kidney. Photonics 2022, 9, 294. [Google Scholar] [CrossRef]
- Wu, X.; Chen, G.; Lu, J.; Zhu, W.; Qiu, J.; Chen, J.; Xie, S.; Zhuo, S.; Yan, J. Label-Free Detection of Breast Masses Using Multiphoton Microscopy. PLoS ONE 2013, 8, e65933. [Google Scholar] [CrossRef]
- Butte, P.V.; Fang, Q.; Jo, J.A.; Yong, W.H.; Pikul, B.K.; Black, K.L.; Marcu, L. Intraoperative delineation of primary brain tumors using time-resolved fluorescence spectroscopy. J. Biomed. Opt. 2010, 15, 027008. [Google Scholar] [CrossRef]
- Poulon, F.; Pallud, J.; Varlet, P.; Zanello, M.; Chretien, F.; Dezamis, E.; Abi-Lahoud, G.; Nataf, F.; Turak, B.; Devaux, B.; et al. Real-time Brain Tumor imaging with endogenous fluorophores: A diagnosis proof-of-concept study on fresh human samples. Sci. Rep. 2018, 8, 14888. [Google Scholar] [CrossRef] [PubMed]
- William, H.Y.; Yong, W.H.; Butte, P.V.; Pikul, B.K.; Jo, J.A.; Fang, Q.; Papaioannou, T.; Black, K.; Marcu, L. Distinction of brain tissue, low grade and high grade glioma with time-resolved fluorescence spectroscopy. Front. Biosci. 2006, 11, 1255–1263. [Google Scholar] [CrossRef]
- Alfano, R.; Tang, G.; Pradhan, A.; Lam, W.; Choy, D.; Opher, E. Fluorescence spectra from cancerous and normal human breast and lung tissues. IEEE J. Quantum Electron. 1987, 23, 1806–1811. [Google Scholar] [CrossRef]
- Das, B.B.; Glassman, W.S.; Alfano, R.R.; Cleary, J.; Prudente, R.; Celmer, E.J.; Lubicz, S. UV-fluorescence spectroscopic technique in the diagnosis of breast, ovarian, uterus, and cervix cancer. In Laser-Tissue Interaction II; SPIE: Los Angeles, CA, USA, 1991; pp. 368–373. [Google Scholar] [CrossRef]
- Papayan, G.; Petrishchev, N.; Galagudza, M. Autofluorescence spectroscopy for NADH and flavoproteins redox state monitoring in the isolated rat heart subjected to ischemia-reperfusion. Photodiagnosis Photodyn. Ther. 2014, 11, 400–408. [Google Scholar] [CrossRef]
- Toms, S.A.; Lin, W.-C.; Weil, R.J.; Johnson, M.D.; Jansen, E.D.; Mahadevan-Jansen, A. Intraoperative Optical Spectroscopy Identifies Infiltrating Glioma Margins with High Sensitivity. Neurosurgery 2005, 57, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Skala, M.C.; Riching, K.M.; Gendron-Fitzpatrick, A.; Eickhoff, J.; Eliceiri, K.W.; White, J.G.; Ramanujam, N. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. USA 2007, 104, 19494–19499. [Google Scholar] [CrossRef] [PubMed]
- Zanello, M.; Poulon, F.; Pallud, J.; Varlet, P.; Hamzeh, H.; Lahoud, G.A.; Andreiuolo, F.; Ibrahim, A.; Pages, M.; Chretien, F.; et al. Multimodal optical analysis discriminates freshly extracted human sample of gliomas, metastases and meningiomas from their appropriate controls. Sci. Rep. 2017, 7, 41724. [Google Scholar] [CrossRef]
- Mehidine, H.; Refregiers, M.; Jamme, F.; Varlet, P.; Juchaux, M.; Devaux, B.; Haidar, D.A. Molecular changes tracking through multiscale fluorescence microscopy differentiate Meningioma grades and non-tumoral brain tissues. Sci. Rep. 2021, 11, 3816. [Google Scholar] [CrossRef]
- Mehidine, H.; Devaux, B.; Varlet, P.; Haidar, D.A. Comparative Study Between a Customized Bimodal Endoscope and a Benchtop Microscope for Quantitative Tissue Diagnosis. Front. Oncol. 2022, 12, 881331. [Google Scholar] [CrossRef]
- Wright, B.K.; Andrews, L.M.; Jones, M.R.; Stringari, C.; Digman, M.A.; Gratton, E. Phasor-flim analysis of NADH distribution and localization in the nucleus of live progenitor myoblast cells. Microsc. Res. Technol. 2012, 75, 1717–1722. [Google Scholar] [CrossRef]
- Trinh, A.L.; Chen, H.; Chen, Y.; Hu, Y.; Li, Z.; Siegel, E.R.; Linskey, M.E.; Wang, P.H.; Digman, M.A.; Zhou, Y.-H. Tracking Functional Tumor Cell Subpopulations of Malignant Glioma by Phasor Fluorescence Lifetime Imaging Microscopy of NADH. Cancers 2017, 9, 168. [Google Scholar] [CrossRef] [PubMed]
- Jacques, S.L. Optical properties of biological tissues: A review. Phys. Med. Biol. 2013, 58, R37–R61. [Google Scholar] [CrossRef] [PubMed]
- Yaroslavsky, A.N.; Schulze, P.C.; Yaroslavsky, I.V.; Schober, R.; Ulrich, F.; Schwarzmaier, H.-J. Optical properties of selected native and coagulated human brain tissues in vitro in the visible and near infrared spectral range. Phys. Med. Biol. 2002, 47, 2059–2073. [Google Scholar] [CrossRef] [PubMed]
- Roggan, A.; Minet, O.; Schroeder, C.; Mueller, G.J. Determination of optical tissue properties with double integrating sphere technique and Monte Carlo simulations. In Cell and Biotissue Optics: Applications in Laser Diagnostics and Therapy; SPIE: Los Angeles, CA, USA, 1994; Volume 2100, pp. 42–57. [Google Scholar] [CrossRef]
- Zanello, M.; Poulon, F.; Varlet, P.; Chretien, F.; Andreiuolo, F.; Pages, M.; Ibrahim, A.; Pallud, J.; Dezamis, E.; Abi-Lahoud, G.; et al. Multimodal optical analysis of meningioma and comparison with histopathology. J. Biophotonics 2016, 10, 253–263. [Google Scholar] [CrossRef]
- Marcu, L.; French, P.M.W.; Elson, D.S. (Eds.) Fluorescence Lifetime Spectroscopy and Imaging: Principles and Applications in Biomedical Diagnostics; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group: Abingdon, UK, 2014. [Google Scholar]
- Engbjerg, J.S.; Costanzo, V.; Sardella, D.; Bordoni, L.; Jakobsen, S.; D’Apolito, L.; Frøkiær, J.; Trepiccione, F.; Capasso, G.; Frische, S. The Probe for Renal Organic Cation Secretion (4-Dimethylaminostyryl)-N-Methylpyridinium (ASP+)) Shows Amplified Fluorescence by Binding to Albumin and Is Accumulated In Vivo. Mol. Imaging 2022, 2022, 7908357. [Google Scholar] [CrossRef]
- Croce, A.C.; Fiorani, S.; Locatelli, D.; Nano, R.; Ceroni, M.; Tancioni, F.; Giombelli, E.; Benericetti, E.; Bottiroli, G. Diagnostic Potential of Autofluorescence for an Assisted Intraoperative Delineation of Glioblastoma Resection Margins. Photochem. Photobiol. 2003, 77, 309–318. [Google Scholar] [CrossRef]
- Gogvadze, V.; Orrenius, S.; Zhivotovsky, B. Mitochondria in cancer cells: What is so special about them? Trends Cell Biol. 2008, 18, 165–173. [Google Scholar] [CrossRef]
- Mehidine, H.; Sibai, M.; Poulon, F.; Pallud, J.; Varlet, P.; Zanello, M.; Devaux, B.; Haidar, D.A. Multimodal imaging to explore endogenous fluorescence of fresh and fixed human healthy and tumor brain tissues. J. Biophotonics 2018, 12, e201800178. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Koppenol, W.H.; Bounds, P.L.; Dang, C.V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 2011, 11, 325–337. [Google Scholar] [CrossRef]
- Wang, H.-W.; Gukassyan, V.; Chen, C.-T.; Wei, Y.-H.; Guo, H.-W.; Yu, J.-S.; Kao, F.-J. Differentiation of apoptosis from necrosis by dynamic changes of reduced nicotinamide adenine dinucleotide fluorescence lifetime in live cells. J. Biomed. Opt. 2008, 13, 054011. [Google Scholar] [CrossRef] [PubMed]
- Baraghis, E.; Devor, A.; Fang, Q.; Srinivasan, V.J.; Wu, W.; Lesage, F.; Ayata, C.; Kasischke, K.A.; Boas, D.A.; Sakadžić, S. Two-photon microscopy of cortical NADH fluorescence intensity changes: Correcting contamination from the hemodynamic response. J. Biomed. Opt. 2011, 16, 106003. [Google Scholar] [CrossRef] [PubMed]
- Kantelhardt, S.R.; Leppert, J.; Krajewski, J.; Petkus, N.; Reusche, E.; Tronnier, V.M.; Hüttmann, G.; Giese, A. Imaging of brain and brain tumor specimens by time-resolved multiphoton excitation microscopy ex vivo1. Neuro-Oncology 2007, 9, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, T.; Yuan, H.; Viglianti, B.L.; Peltz, C.; Asopa, S.; Vujaskovic, Z.; Dewhirst, M.W. Spatial Heterogeneity and Oxygen Dependence of Glucose Consumption in R3230Ac and Fibrosarcomas of the Fischer 344 Rat. Cancer Res. 2005, 65, 5163–5171. [Google Scholar] [CrossRef]
- Maeda-Yorita, K.; Aki, K. Effect of Nicotinamide Adenine Dinucleotide on the Oxidation-Reduction Potentials of Lipoamide Dehydrogenase from Pig Heart. J. Biochem. 1984, 96, 683–690. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).