Self-Regulated Symmetry Breaking Model for Stem Cell Differentiation
Abstract
:1. Introduction
2. Mean-Field Approximation Model for Self-Tuned Symmetry Breaking
2.1. Landau’s Potential Energy for Stem Cell Populations
2.2. Feedback Mechanism for Noise Regulation
3. Results and Discussion
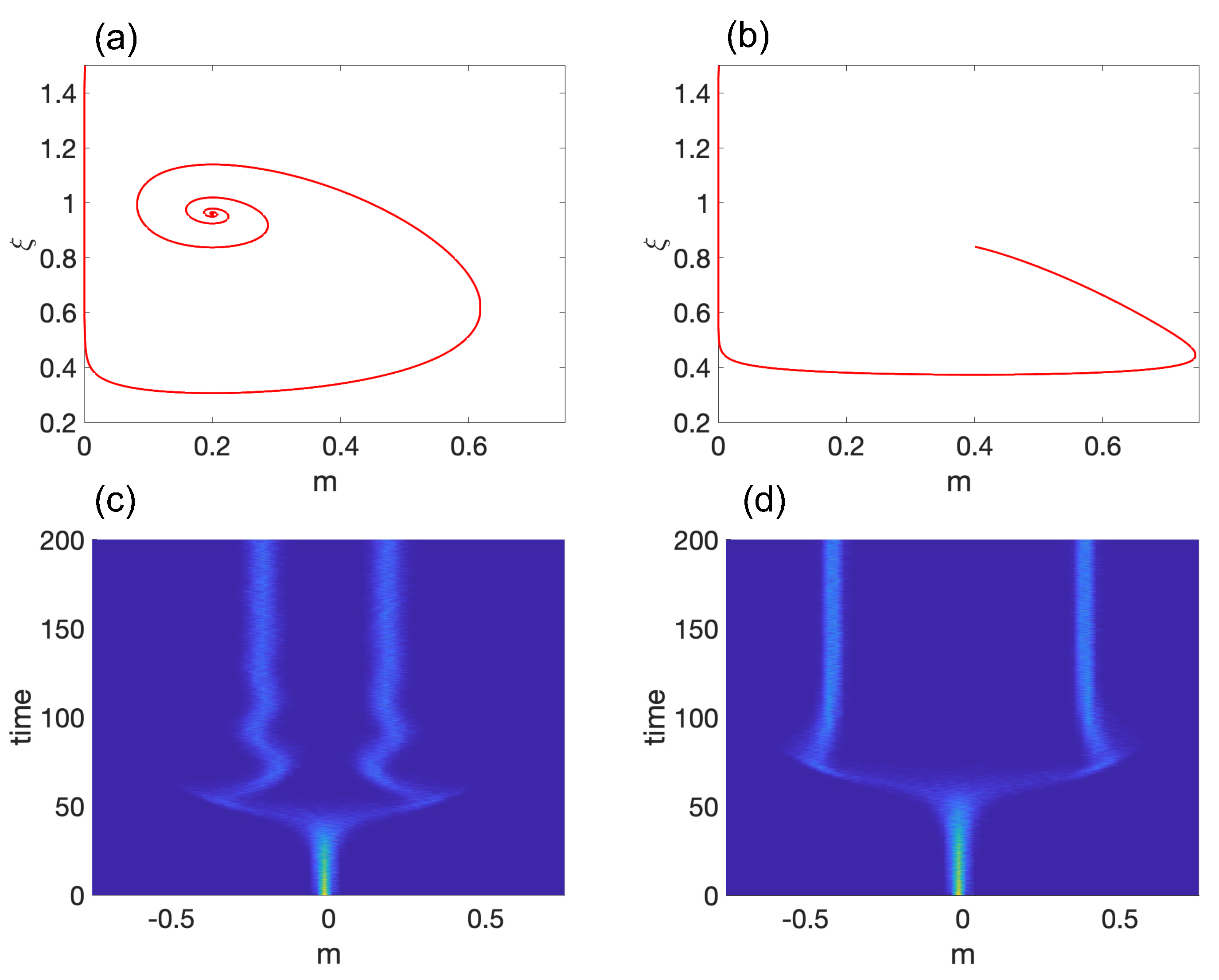
3.1. Model 1
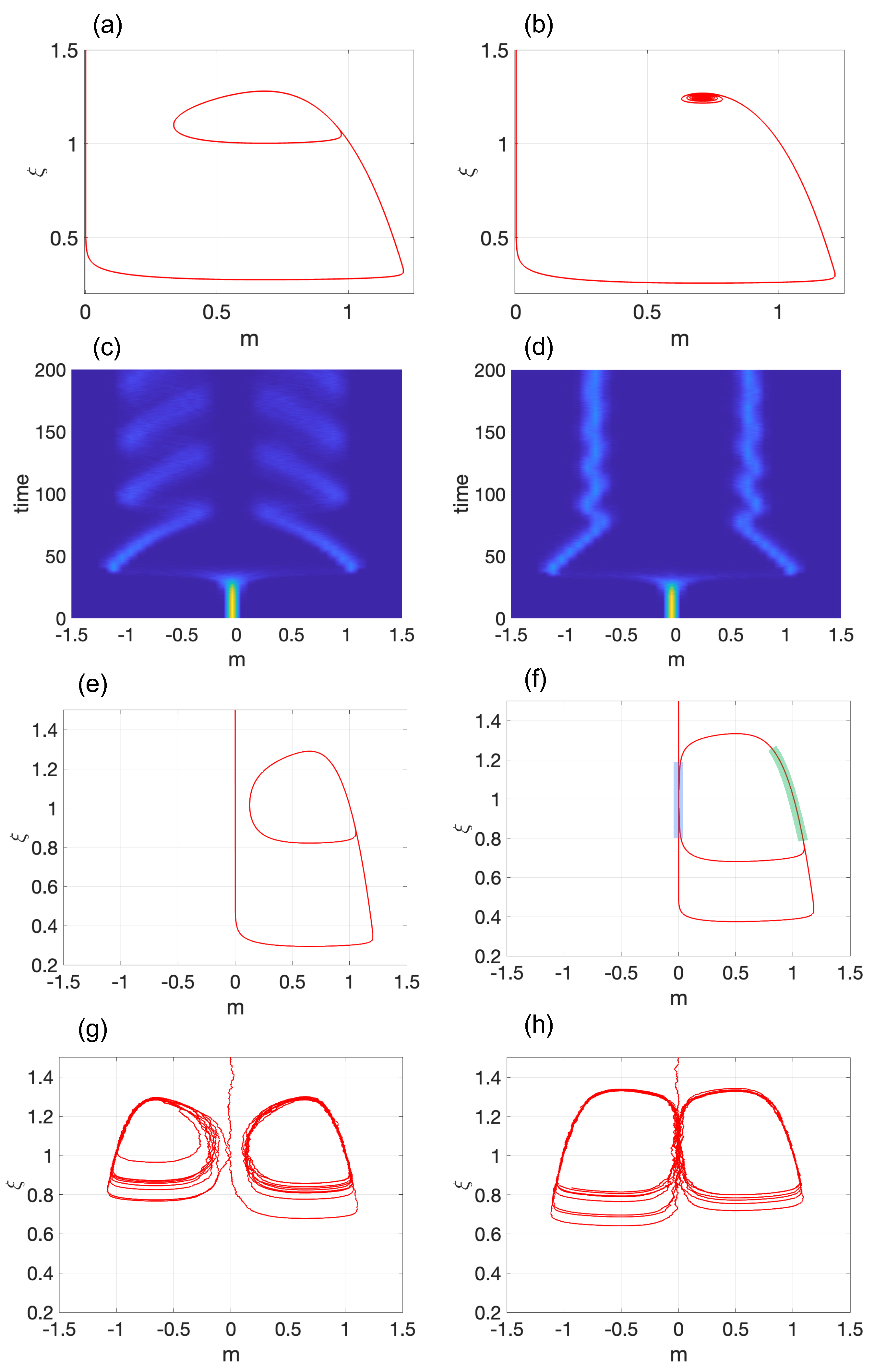
3.2. Model 2
3.3. Model 3
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kærn, M.; Elston, T.C.; Blake, W.J.; Collins, J.J. Stochasticity in gene expression: From theories to phenotypes. Nat. Rev. Genet. 2005, 6, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; van Oudenaarden, A. Nature, Nurture, or Chance: Stochastic Gene Expression and Its Consequences. Cell 2008, 135, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C.H. (Ed.) Organisers and Genes; Cambridge University Press: Cambridge, UK, 1940. [Google Scholar]
- Huang, S. Reprogramming cell fates: Reconciling rarity with robustness. BioEssays 2009, 31, 546–560. [Google Scholar] [CrossRef]
- Foster, D.V.; Foster, J.G.; Huang, S.; Kauffman, S.A. A model of sequential branching in hierarchical cell fate determination. J. Theor. Biol. 2009, 260, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Cross, M.A.; Enver, T. The lineage commitment of haemopoietic progenitor cells. Curr. Opin. Genet. Dev. 1997, 7, 609–613. [Google Scholar] [CrossRef]
- Hu, M.; Krause, D.; Greaves, M.; Sharkis, S.; Dexter, M.; Heyworth, C.; Enver, T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997, 11, 774–785. [Google Scholar] [CrossRef]
- Parmentier, R.; Racine, L.; Moussy, A.; Chantalat, S.; Sudharshan, R.; Gao, N.P.; Stockholm, D.; Corre, G.; Fourel, G.; Deleuze, J.F.; et al. Global genome decompaction leads to stochastic activation of gene expression as a first step toward fate commitment in human hematopoietic cells. PLoS Biol. 2022, 20, e3001849. [Google Scholar] [CrossRef]
- Zimatore, G.; Tsuchiya, M.; Hashimoto, M.; Bioeng, A.; Kasperski, A.; Giuliani, A. Self-organization of whole-gene expression through coordinated chromatin structural transition. Biophys. Rev. 2021, 2, 031303. [Google Scholar] [CrossRef]
- Arias, A.M.; Hayward, P. Filtering transcriptional noise during development: Concepts and mechanisms. Nat. Rev. Genet. 2006, 7, 34–44. [Google Scholar] [CrossRef]
- Pujadas, E.; Feinberg, A.P. Regulated Noise in the Epigenetic Landscape of Development and Disease. Cell 2012, 148, 1123–1131. [Google Scholar] [CrossRef]
- Szutorisz, H.; Georgiou, A.; Tora, L.; Dillon, N. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell 2006, 127, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, C.; Kaneko, K. A dynamical-systems view of stem cell biology. Science 2012, 338, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Macarthur, B.D.; Lemischka, I.R. Statistical Mechanics of Pluripotency. Cell 2013, 154, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ojalvo, J.; Martinez Arias, A. Towards a statistical mechanics of cell fate decisions. Curr. Opin. Genet. Dev. 2012, 22, 619–626. [Google Scholar] [CrossRef]
- Silva, J.; Smith, A. Capturing Pluripotency. Cell 2008, 132, 532–536. [Google Scholar] [CrossRef]
- Stanoev, A.; Schröter, C.; Koseska, A. Robustness and timing of cellular differentiation through population-based symmetry breaking. Development 2021, 148, dev197608. [Google Scholar] [CrossRef]
- Przybyla, L.; Voldman, J. Probing embryonic stem cell autocrine and paracrine signaling using microfluidics. Annu. Rev. Anal. Chem. 2012, 5, 293. [Google Scholar] [CrossRef]
- Saiz, N.; Mora-Bitrià, L.; Rahman, S.; George, H.; Herder, J.P.; García-Ojalvo, J.; Hadjantonakis, A.K. Growth factor-mediated coupling between lineage size and cell fate choice underlies robustness of mammalian development. eLife 2020, 9, 1–38. [Google Scholar] [CrossRef]
- Hayashi, K.; Lopes, S.M.d.S.; Tang, F.; Surani, M.A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 2008, 3, 391–401. [Google Scholar] [CrossRef]
- Goldenfeld, N. Lectures on Phase Transitions and the Renormalization Group; CRC Press: Boca Raton, FL, USA, 2018; p. 2554. [Google Scholar]
- Bargaje, R.; Trachana, K.; Shelton, M.N.; McGinnis, C.S.; Zhou, J.X.; Chadick, C.; Cook, S.; Cavanaugh, C.; Huang, S.; Hood, L. Cell population structure prior to bifurcation predicts efficiency of directed differentiation in human induced pluripotent cells. Proc. Natl. Acad. Sci. USA 2017, 114, 2271–2276. [Google Scholar] [CrossRef]
- Ferrell, J.E. Bistability, bifurcations, and Waddington’s epigenetic landscape. Curr. Biol. 2012, 22, R458. [Google Scholar] [CrossRef] [PubMed]
- Moris, N.; Pina, C.; Arias, A.M. Transition states and cell fate decisions in epigenetic landscapes. Nat. Rev. Genet. 2016, 17, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, C.; Kaneko, K. Theory of Robustness of Irreversible Differentiation in a Stem Cell System: Chaos Hypothesis. J. Theor. Biol. 2001, 209, 395–416. [Google Scholar] [CrossRef]
- Huang, S.; Guo, Y.P.; May, G.; Enver, T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev. Biol. 2007, 305, 695–713. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Furusawa, C.; Kaneko, K. Oscillatory Protein Expression Dynamics Endows Stem Cells with Robust Differentiation Potential. PLoS ONE 2011, 6, e27232. [Google Scholar] [CrossRef]
- Koseska, A.; Ullner, E.; Volkov, E.; Kurths, J.; García-Ojalvo, J. Cooperative differentiation through clustering in multicellular populations. J. Theor. Biol. 2010, 263, 189–202. [Google Scholar] [CrossRef]
- Ullner, E.; Zaikin, A.; Volkov, E.I.; García-Ojalvo, J. Multistability and clustering in a population of synthetic genetic oscillators via phase-repulsive cell-to-cell communication. Phys. Rev. Lett. 2007, 99, 148103. [Google Scholar] [CrossRef]
- Taga, M.E.; Bassler, B.L. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 2003, 100, 14549–14554. [Google Scholar] [CrossRef]
- Danino, T.; Mondragón-Palomino, O.; Tsimring, L.; Hasty, J. A synchronized quorum of genetic clocks. Nature 2010, 463, 326–330. [Google Scholar] [CrossRef]
- You, L.; Cox, R.S.; Weiss, R.; Arnold, F.H. Programmed population control by cell–cell communication and regulated killing. Nature 2004, 428, 868–871. [Google Scholar] [CrossRef]
- Lander, A.D.; Gokoffski, K.K.; Wan, F.Y.; Nie, Q.; Calof, A.L. Cell lineages and the logic of proliferative control. PLoS Biol. 2009, 7, e1000015. [Google Scholar] [CrossRef] [PubMed]
- Kirouac, D.C.; Madlambayan, G.J.; Yu, M.; Sykes, E.A.; Ito, C.; Zandstra, P.W. Cell–cell interaction networks regulate blood stem and progenitor cell fate. Mol. Syst. Biol. 2009, 5, 293. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, S.A. Metabolic stability and epigenesis in randomly constructed genetic nets. J. Theor. Biol. 1969, 22, 437–467. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, S. The large scale structure and dynamics of gene control circuits: An ensemble approach. J. Theor. Biol. 1974, 44, 167–190. [Google Scholar] [CrossRef]
- Bornholdt, S.; Kauffman, S. Ensembles, dynamics, and cell types: Revisiting the statistical mechanics perspective on cellular regulation. J. Theor. Biol. 2019, 467, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Shmulevich, I.; Gluhovsky, I.; Hashimoto, R.F.; Dougherty, E.R.; Zhang, W. Steady-state analysis of genetic regulatory networks modelled by probabilistic Boolean networks. Comp. Funct. Genom. 2003, 4, 601–608. [Google Scholar] [CrossRef]
- Kang, C.; Mcelroy, M.; Voulgarakis, N.K. Emergent Criticality in Coupled Boolean Networks. Entropy 2023, 25, 235. [Google Scholar] [CrossRef]
- Cowan, B. Topics in Statistical Mechanics; Imperial College Press: London, UK; World Scientific Publishing Co.: Singapore, 2005. [Google Scholar]
- Falk, J.; Mendler, M.; Drossel, B. A minimal model of burst-noise induced bistability. PLoS ONE 2017, 12, e0176410. [Google Scholar] [CrossRef]
- Assaf, M.; Roberts, E.; Luthey-Schulten, Z.; Goldenfeld, N. Extrinsic noise driven phenotype switching in a self-regulating gene. Phys. Rev. Lett. 2013, 111, 058102. [Google Scholar] [CrossRef]
- Samoilov, M.; Plyasunov, S.; Arkin, A.P. Stochastic amplification and signaling in enzymatic futile cycles through noise-induced bistability with oscillations. Proc. Natl. Acad. Sci. USA 2005, 102, 2310–2315. [Google Scholar] [CrossRef]
- Bishop, L.M.; Qian, H. Stochastic bistability and bifurcation in a mesoscopic signaling system with autocatalytic kinase. Biophys. J. 2010, 98, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Strogatz, S. Nonlinear Dynamics and Chaos: With Applications to Physics, Biology, Chemistry, and Engineering; a Member of the Perseus Books Group; Westview Press: Boulder, CO, USA, 2015. [Google Scholar]
- Wiggins, S. Introduction to Applied Nonlinear Dynamical Systems and Chaos; Springer: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Cheng, Y.C.; Qian, H. Stochastic Limit-Cycle Oscillations of a Nonlinear System Under Random Perturbations. J. Stat. Phys. 2021, 182, 1–33. [Google Scholar] [CrossRef]
- Di Santo, S.; Burioni, R.; Vezzani, A.; Muñoz, M.A. Self-Organized Bistability Associated with First-Order Phase Transitions. Phys. Rev. Lett. 2016, 116, 240601. [Google Scholar] [CrossRef]
- Buendiá, V.; Di Santo, S.; Villegas, P.; Burioni, R.; Munõz, M.A. Self-organized bistability and its possible relevance for brain dynamics. Phys. Rev. Res. 2020, 2, 013318. [Google Scholar] [CrossRef]
- Tsuchiya, M.; Giuliani, A.; Hashimoto, M.; Erenpreisa, J.; Yoshikawa, K. Self-Organizing Global Gene Expression Regulated through Criticality: Mechanism of the Cell-Fate Change. PLoS ONE 2016, 11, e0167912. [Google Scholar] [CrossRef]
- Giuliani, A.; Tsuchiya, M.; Yoshikawa, K. Self-Organization of Genome Expression from Embryo to Terminal Cell Fate: Single-Cell Statistical Mechanics of Biological Regulation. Entropy 2018, 20, 13. [Google Scholar] [CrossRef]
- Tsuchiya, M.; Hashimoto, M.; Takenaka, Y.; Motoike, I.N.; Yoshikawa, K. Global Genetic Response in a Cancer Cell: Self-Organized Coherent Expression Dynamics. PLoS ONE 2014, 9, e97411. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, M.; Giuliani, A.; Hashimoto, M.; Erenpreisa, J.; Yoshikawa, K. Emergent Self-Organized Criticality in Gene Expression Dynamics: Temporal Development of Global Phase Transition Revealed in a Cancer Cell Line. PLoS ONE 2015, 10, e0128565. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McElroy, M.; Green, K.; Voulgarakis, N.K. Self-Regulated Symmetry Breaking Model for Stem Cell Differentiation. Entropy 2023, 25, 815. https://doi.org/10.3390/e25050815
McElroy M, Green K, Voulgarakis NK. Self-Regulated Symmetry Breaking Model for Stem Cell Differentiation. Entropy. 2023; 25(5):815. https://doi.org/10.3390/e25050815
Chicago/Turabian StyleMcElroy, Madelynn, Kaylie Green, and Nikolaos K. Voulgarakis. 2023. "Self-Regulated Symmetry Breaking Model for Stem Cell Differentiation" Entropy 25, no. 5: 815. https://doi.org/10.3390/e25050815
APA StyleMcElroy, M., Green, K., & Voulgarakis, N. K. (2023). Self-Regulated Symmetry Breaking Model for Stem Cell Differentiation. Entropy, 25(5), 815. https://doi.org/10.3390/e25050815






