Artemisinin DNA Base Interaction Studies in Presence of Fe(II): LC/TOF MS Separation of Reaction Products
Abstract
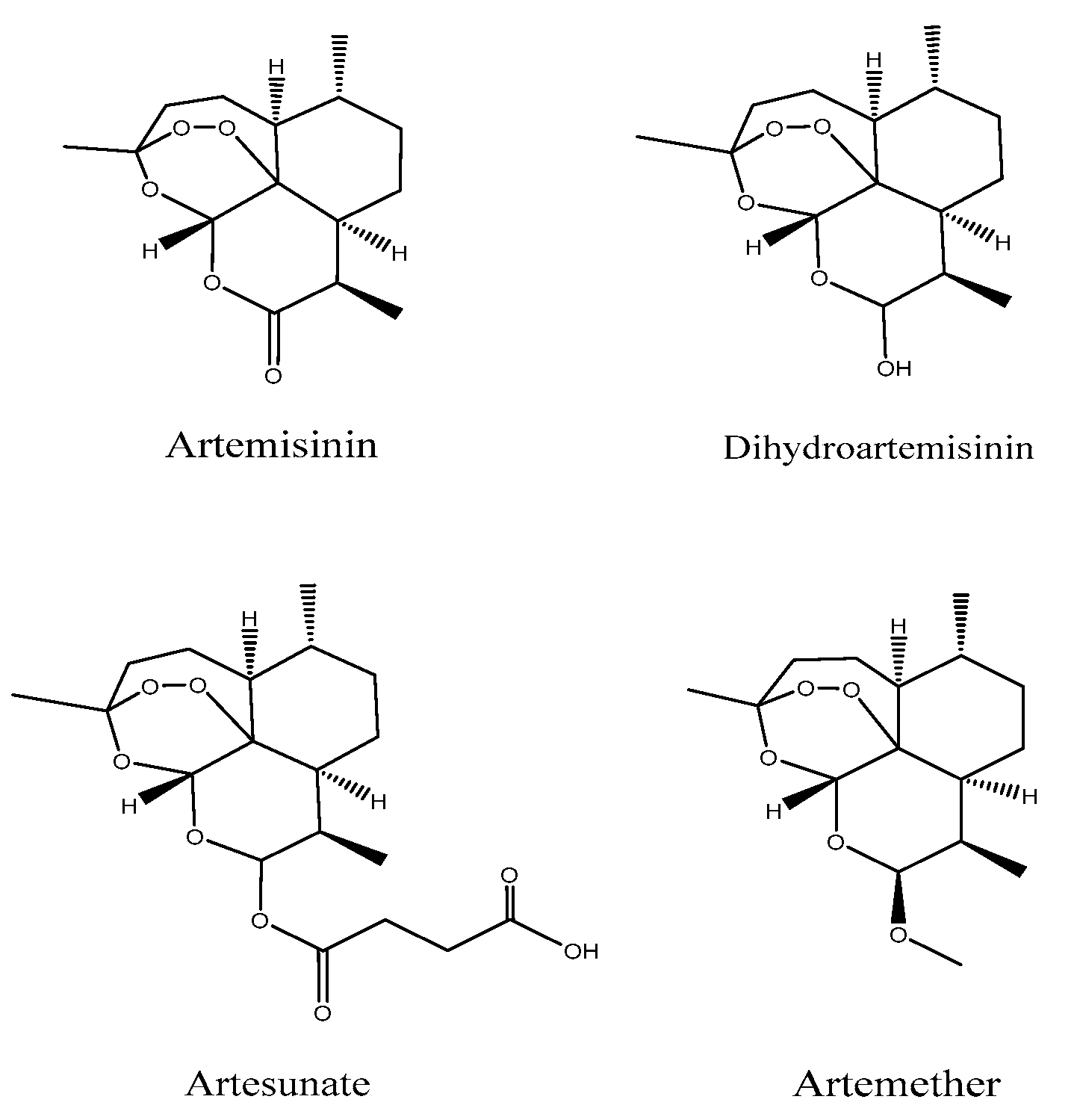
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Chromatographic Procedure
2.3. Mass Spectrometer Procedure
2.4. Sample Preparation
ART-DNA Reactions
2.5. Optimization of the Reaction Conditions
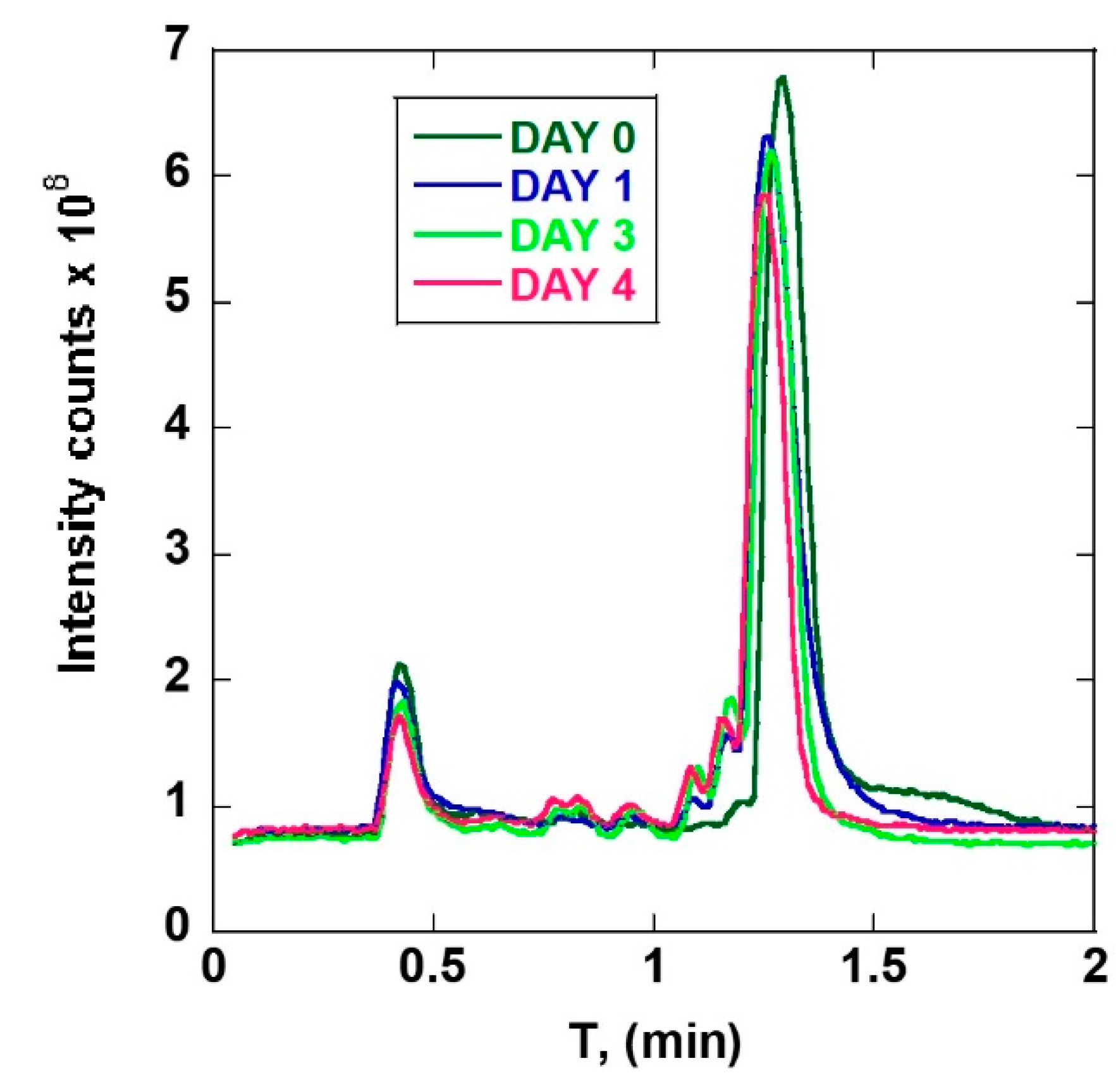
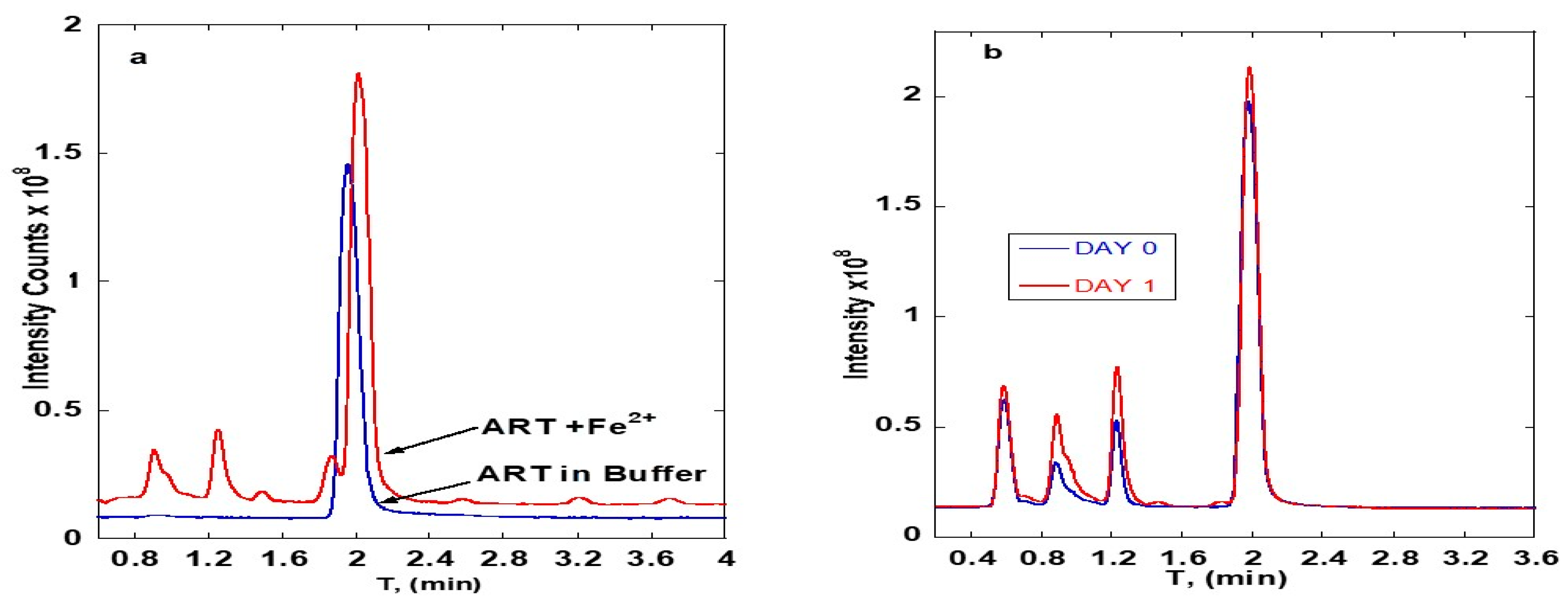
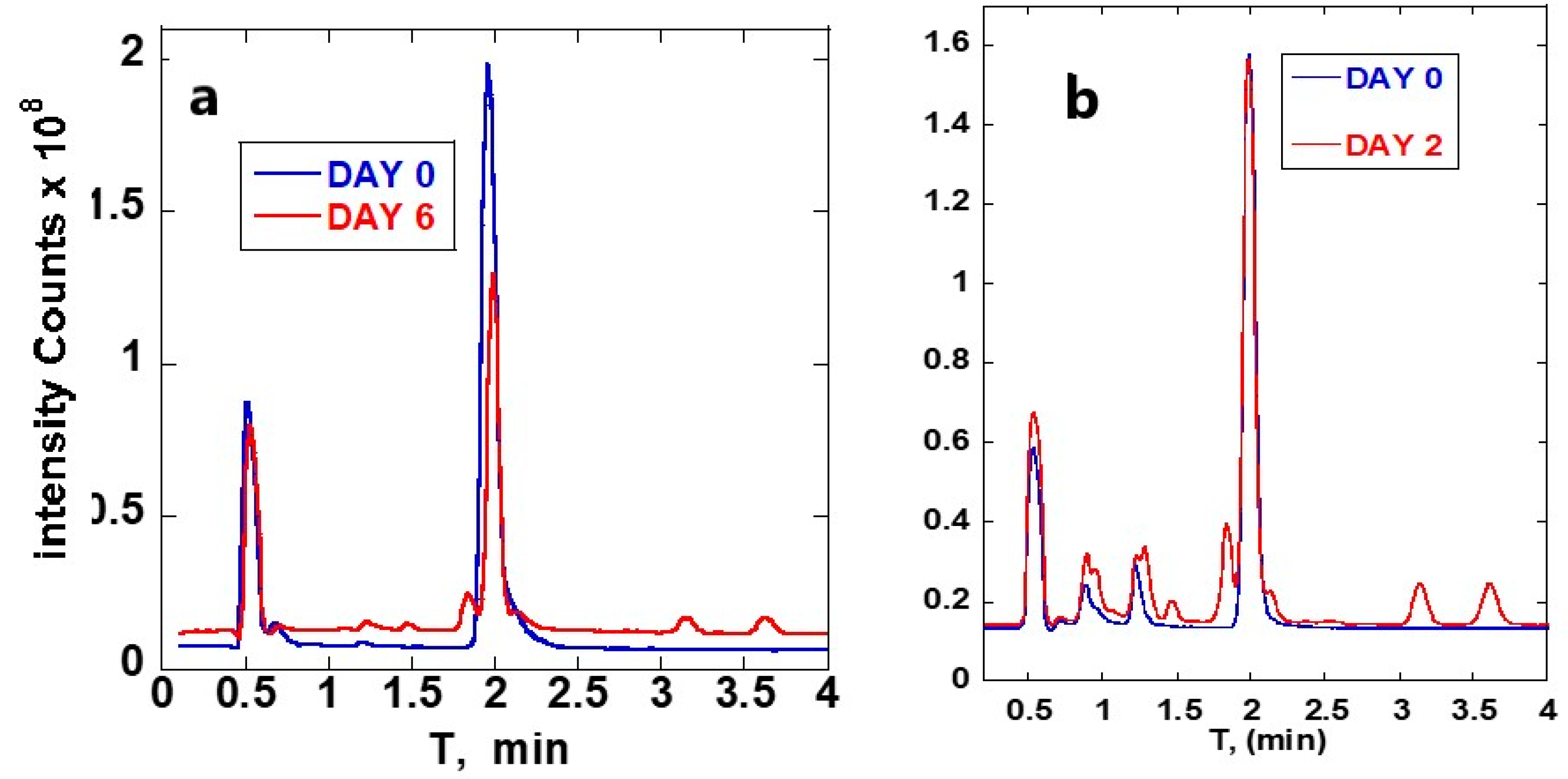
3. Results
3.1. ART-Deoxyadenosine
3.2. ART-Deoxycytidine
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ivanescu, B.; Miron, A.; Corciova, A. Sesquiterpene Lactones fromArtemisiaGenus: Biological Activities and Methods of Analysis. J. Anal. Methods Chem. 2015, 2015, 247685. [Google Scholar] [CrossRef] [Green Version]
- Van Geldre, E.; Vergauwe, A.; Eeckhout, E.V.D. State of the art of the production of the antimalarial compound artemisinin in plants. Plant Mol. Biol. 1997, 33, 199–209. [Google Scholar] [CrossRef]
- Rudrapal, M.; Chetia, D. Endoperoxide antimalarials: Development, structural diversity and pharmacodynamic aspects with reference to 1,2,4-trioxane-based structural scaffold. Drug Des. Dev. Ther. 2016, 10, 3575–3590. [Google Scholar] [CrossRef] [Green Version]
- Woodrow, C.J.; Haynes, R.K.; Krishna, S. Artemisinins. Postgrad. Med. J. 2005, 81, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Meshnick, S.R.; Taylor, T.E.; Kamchonwongpaisan, S. Artemisinin and the antimalarial endoperoxides: From herbal remedy to targeted chemotherapy. Microbiol. Rev. 1996, 60, 301–315. [Google Scholar] [CrossRef]
- Chen, X.; He, L.-Y.; Lai, S.; He, Y. Dihydroartemisinin inhibits the migration of esophageal cancer cells by inducing autophagy. Oncol. Lett. 2020, 20, 94. [Google Scholar] [CrossRef] [PubMed]
- Augustin, Y.; Staines, H.M.; Krishna, S. Artemisinins as a novel anti-cancer therapy: Targeting a global cancer pandemic through drug repurposing. Pharmacol. Ther. 2020, 216, 107706. [Google Scholar] [CrossRef]
- Konstat-Korzenny, E.; Ascencio-Aragon, J.A.; Niezen-Lugo, S.; Vazquez-Lopez, R. Artemisinin and its synthetic derivatives as a possible therapy for cancer. Med. Sci. 2018, 6, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Huang, L.; Li, J.; Fan, Q.; Long, Y.; Li, Y.; Zhou, B. Artemisinin directly targets malarial mitochondria through its specific mitochondrial activation. PLoS ONE 2010, 5, e9582. [Google Scholar] [CrossRef] [Green Version]
- Waseem, Y.; Hasan, A.C.; Ahmed, F. Artemisinin: A promising adjunct for cancer therapy. Cureus 2018, 10, e3628. [Google Scholar] [CrossRef] [Green Version]
- Accardo, A.; Laurent, S.A.L.; Mazarguil, H.; Meyer, M.; Robert, A.; Meunier, B. Interaction of iron(II)-heme and artemisinin with a peptide mimic of Plasmodium falciparum HRP-II. J. Inorg. Biochem. 2007, 101, 1739–1747. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Yao, X.; Jiang, Q.; Wang, K.; Zhang, Y.; Peng, H.; Tang, J.; Yang, W. Dihydroartemisinin-loaded magnetic nanoparticles for enhanced chemodynamic therapy. Front. Pharmacol. 2020, 11, 226. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Li, Y.-J.; You, C.; Sun, K.; An, P.; Sun, C.; Wang, M.-X.; Zhu, X.; Sun, B.-W. Iron oxide nanocarrier-mediated combination therapy of cisplatin and artemisinin for combating drug resistance through highly increased toxic reactive oxygen species generation. ACS Appl. Bio Mater. 2018, 1, 270–280. [Google Scholar] [CrossRef]
- Sen, R.; Ganguly, S.; Saha, P.; Chatterjee, M. Efficacy of artemisinin in experimental visceral leishmaniasis. Int. J. Antimicrob. Agents 2010, 36, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Patel, Y.; Mistry, N.; Mehra, S. Repurposing artemisinin as an anti-mycobacterial agent in synergy with rifampicin. Tuberculosis 2019, 115, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Ortalli, M.; Varani, S.; Cimato, G.; Veronesi, R.; Quintavalla, A.; Lombardo, M.; Monari, M.; Trombini, C. Evaluation of the pharmacophoric role of the O–O bond in synthetic antileishmanial compounds: Comparison between 1,2-dioxanes and tetrahydropyrans. J. Med. Chem. 2020, 63, 13140–13158. [Google Scholar] [CrossRef]
- Geroldinger, G.; Tonner, M.; Quirgst, J.; Walter, M.; De Sarkar, S.; Machín, L.; Monzote, L.; Stolze, K.; Duvigneau, J.C.; Staniek, K.; et al. Activation of artemisinin and heme degradation in Leishmania tarentolae pro-mastigotes: A possible link. Biochem. Pharmacol. 2020, 173, 113737. [Google Scholar] [CrossRef]
- Chen, G.-Q.; Benthani, F.A.; Wu, J.; Liang, D.; Bian, Z.-X.; Jiang, X. Artemisinin compounds sensitize cancer cells to ferroptosis by regulating iron homeostasis. Cell Death Differ. 2020, 27, 242–254. [Google Scholar] [CrossRef]
- Alotaibi, S.H.; Momen, A.A. Antinticancer drugs’ deoxyribonucleic acid (DNA) interactions. In Biophysical Chemistry—Advanced Applications; Khalid, M.A.A., Ed.; InTech Open: London, UK, 2019. [Google Scholar] [CrossRef] [Green Version]
- Vandercruyssen, K.; D’Hondt, M.; Vergote, V.; Jansen, H.; Burvenich, C.; De Spiegeleer, B. LC–UV/MS quality analytics of pediatric artemether formulations. J. Pharm. Anal. 2013, 4, 37–52. [Google Scholar] [CrossRef]
- Naumczuk, B.; Bocian, W.; Sitkowski, J.; Kawęcki, R.; Kozerski, L. Solvent-dependent regioselectivity of 2′-deoxyadenosine alkylation by 9-aminomethyl derivatives of SN38. New J. Chem. 2019, 43, 18975–18978. [Google Scholar] [CrossRef]
- Liu, X. Progress in the mechanism and kinetics of Fenton reaction. MOJ Ecol. Environ. Sci. 2018, 3, 60. [Google Scholar] [CrossRef] [Green Version]
- Naumczuk, B.; Kawęcki, R.; Bocian, W.; Bednarek, E.; Sitkowski, J.; Kozerski, L. Regioselective alkylation reaction of the 2′-deoxyctidine with 9-aminomethyl derivatives of SN38. J. Mol. Struct. 2018, 1176, 298–302. [Google Scholar] [CrossRef]
- Koskinen, M.; Calebiro, D.; Hemminki, K. Styrene oxide-induced 2′-deoxycytidine adducts: Implications for the mutagenicity of styrene oxide. Chem. Biol. Interact. 2000, 126, 201–213. [Google Scholar] [CrossRef]

| Retention Time | Main Fragments (m/z) |
|---|---|
| 0.7 | 251, 337, 353, 399 |
| 0.8 | 237, 251, 307, 399 |
| 0.9 | 251, 307, 337, 353 |
| 1.1 | 251, 283, 305, 337, 353 and 587 |
| 1.2 | 209, 237, 251, 265, 337 and 353 |
| Retention Time | Main Fragments |
|---|---|
| 0.91 | 265, 283, 305, 314, and 587 |
| 0.97 | 203, 209, 237, 265, 277, 283, 305, 314, 321 and 358 |
| 1.2 | 223, 300, 305, 314, 328, 356 and 587 |
| 1.4 | 203, 209, 251, 297, 332, 337, 346, 360, and 388 |
| 1.8 | 209, 251, 265, 315, 332, 337, 346, 353 and 360 |
| 2.5 | 203, 209, 224, 237, 251, 265, 279, 297, 337, 346, 358, 360 and 399 |
| 3.1 | 209, 251, 346, 360 and 374 |
| 3.6 | 203, 209, 224, 251, 265, 297, 346, 351, 360 and 374 |
| Retention Time (min) | Main Fragments (m/z) |
|---|---|
| 0.8 | 252, 283, 305, 337, 367, 587, and 619 |
| 0.9 | 203, 209, 237, 252, 277, 283, 293, 305, 411, 531, and 547 |
| 1.2 | 223, 255, 279, 305, 314, 321, 356, 443 and 587 |
| 1.4 | 203, 217, 251, 252, 269, 291, 293, 297, 305, 319, 328, 337, 346, and 370 |
| 1.8 | 203, 209, 251, 277, 337, 346, 353 and 360 |
| Retention Time | Main Fragments (m/z) |
|---|---|
| 1.2 | 203, 209, 284, 291, 305, 323, 332 and 337 |
| 1.4 | 203, 224, 245, 251, 284, 291, 337, 346, 360, and 388 |
| 1.8 | 209, 251, 277, 337 and 346 |
| 3.1 | 209, 251, 277, 284, 307, 346, 351, 360 and 374 |
| 3.6 | 209, 251, 277, 297, 307, 351, 360 and 374 |
| Retention Time | Main Fragments (m/z) |
|---|---|
| 0.8 | 283, 305, 314, 337, 587, and 619 |
| 0.9 | 209, 237, 277, 283, 293, 305, and 531 |
| 1.2 | 223, 305, 314, 321, 356, 587 and 605 |
| 1.3 | 251, 291, 300 and 307 |
| 1.4 | 251, 291, 297, 307, 319, 337, 346, 353, 360, 374 and 388 |
| 1.8 | 251, 277, 297, 337, 346, and 353 |
| 2.1 | 209, 237, 265, 283, 305, 337, 346, 353 and 356 |
| 3.1 | 251, 346, 351, 360, 367 and 374 |
| 3.6 | 251, 277, 297, 307, 351, 360, 367 and 374. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oke, K.; Mugweru, A. Artemisinin DNA Base Interaction Studies in Presence of Fe(II): LC/TOF MS Separation of Reaction Products. Separations 2021, 8, 161. https://doi.org/10.3390/separations8090161
Oke K, Mugweru A. Artemisinin DNA Base Interaction Studies in Presence of Fe(II): LC/TOF MS Separation of Reaction Products. Separations. 2021; 8(9):161. https://doi.org/10.3390/separations8090161
Chicago/Turabian StyleOke, Kogila, and Amos Mugweru. 2021. "Artemisinin DNA Base Interaction Studies in Presence of Fe(II): LC/TOF MS Separation of Reaction Products" Separations 8, no. 9: 161. https://doi.org/10.3390/separations8090161
APA StyleOke, K., & Mugweru, A. (2021). Artemisinin DNA Base Interaction Studies in Presence of Fe(II): LC/TOF MS Separation of Reaction Products. Separations, 8(9), 161. https://doi.org/10.3390/separations8090161





