1. Introduction
Conjugated fatty acids (CFA) are polyunsaturated fatty acids (PUFA) with two or more carbon-carbon double bonds that are separated by a carbon–carbon single bond. CFA usually include a mixture of cis (c) and trans (t) double bonds. Examples include rumenic acid, 9c,11t-18:2, an isomer of linoleic acid (L), 9c,12c-18:2, and α-eleostearic acid, 9c,11t,13t-18:3, an isomer of α-linolenic acid (Ln), 9c,12c,15c-18:3. Examples of CFA are known with conjugation across two, three, and four carbon–carbon double bonds. There are also examples of CFA having additional, non-conjugated carbon–carbon double bonds, such as rumelenic acid, 9c,11t,15c-18:3. Diconjugated FA are often referred to as conjugated linoleic acids (CLA), and triconjugated FA are often referred to as conjugated linolenic acids (CLNA). Following this pattern, we refer to tetraconjugated FA as conjugated stearidonic acids (CSDA), after stearidonic acid, 6c,9c,12c,15c-18:4.
In comparison with their corresponding non-conjugated isomers, CFA exhibit differences in their physical, chemical, and biological properties. For example, they exhibit characteristic UV absorption bands [
1], have greater susceptibility to oxidation [
2,
3,
4,
5] and isomerization [
4,
6,
7], and there is evidence that they have anti-carcinogenic and other health-related properties [
8]. CLA occur mainly in animal sources, and there is dietary exposure through consumption of meat and dairy foods [
9]. However, CLA are usually present as only a small percentage of the total FA content in such foods. CLNA and CSDA are found in plant sources, and they can be present as a high percentage of total FA content. For example, punicic acid, 9
c,11
t,13
c-18:3, can occur in over 70% of the total FA content in the oil of pomegranate (
Punica granatum) seeds [
10,
11]. Mostly, however, CLNA and CSDA occur in plant sources that are not commonly used as food. Pomegranate is an exception, though consumption of punicic acid depends upon ingestion of the seeds and not just the arils. There have been some efforts at transgenic incorporation of CFA into food sources, though with limited success [
12].
The susceptibility of CFA to oxidation and isomerization is an important consideration in their chemical analysis. In the preparation of the fatty acid methyl esters (FAME) of CFA, a common approach for analysis is gas chromatography (GC), where the choices of methylating reagent, temperature, and time of reaction are relevant factors. Basic reagents such as NaOCH
3 and tetramethylguanidine (TMG) are recognized as preserving the endogenous distributions of CFA when present as triacylglycerols (TAG) and in some other forms, but they are not effective in converting FA to FAME in all types of lipids [
7,
13,
14]. Acidic reagents such as methanolic HCl, H
2SO
4, and BF
3 are more broadly applicable for conversion of FA to FAME, but they are known to cause isomerization of CFA to a degree that depends on the temperature and time of the reaction [
6,
7,
13,
14,
15]. Extensive isomerization, including migration of double bonds, can occur under sufficiently harsh conditions. Under milder conditions, geometric isomerization may still occur, especially the conversion of
cis double bonds of CFA to the more stable
trans forms. At sufficiently short reaction times and low temperatures, isomerization may be avoided, but there is a tendency toward incomplete conversion of FA to FAME. Another potential problem with the use of acidic methylating reagents for CLA analysis is the production of artifacts, particularly the formation of allylic methoxy species [
15].
Many plant sources of CLNA are known to contain a single, predominant CLNA species, as in the case of
Punica granatum and punicic acid. Tung oil from the seeds of the tung tree,
Vernicia fordii, is an example of a plant source having two prominent CLNA isomers, α-eleostearic acid (9
c,11
t, 13
t-18:3) and β-eleostearic acid (9
t,11
t,13
t-18:3) [
16]. Several published reports have noted the presence of minor amounts of additional CLNA isomers in samples with a single, predominant isomer [
10,
11,
16]. Seven naturally occurring CLNA are well known in plant sources, and their configurations and common names are given in
Table 1. There is a recent report of four additional CLNA (10
t,12
t,14
t-18:3, 11
t,13
t,15
c-18:3, 11
t,13
t,15
t-18:3, and 8
c,10
t,12
t-18:3) occurring at low levels in melon and other fruit seed samples [
17]. FA with conjugation across the 5, 7, 9 positions have been reported for some red seaweed species [
18]. To our knowledge, there have been no reports of naturally occurring CLNA species of configuration
ccc,
cct,
tcc, or
tct. Two naturally occurring CSDA isomers, α-parinaric acid and β-parinaric acid, are well known [
13], and these are also listed in
Table 1.
In this work, we report the FA composition, excluding oxygenated FA, of seven CFA-containing seed oil samples. Lipids were extracted from seeds of
Jacaranda mimosifolia,
Punica granatum,
Momordica charantia,
Impatiens balsamina,
Calendula officinalis, and
Catalpa ovata. A sample of tung oil was also purchased and analyzed. FA in seeds are predominately in the form of triacylglycerols (TAG) stored in the seed oil bodies [
19]. Oil samples were treated with a methanolic base for the conversion of FA to FAME and analyzed by GC-FID and GC-MS using positive ion chemical ionization with methane as the reagent gas (PCI-CH
4). PCI-CH
4 is useful in analysis of FAME because it provides characteristic ions indicative of molecular weight, although it provides limited structural information and is of limited help in distinguishing between isomers [
20]. Ions characteristic of molecular weights observed for FAME by PCI-CH
4 included [M + H]
+, [M − H]
+, [M + 29]
+, [M + 41]
+, [M + H − 32]
+, and [M + H − 50]
+, though which of these were prominent varied. For a given FAME and its isomers, three to five of these ions were typically prominent, depending on the FAME. For example, for the 18:0 FAME and most other saturated FAME, [M + H]
+, [M − H]
+, and [M + 29]
+ were especially prominent ions. For the FAME of 18:1 isomers, [M + H]
+, [M + 29]
+, [M + H − 32]
+, and [M + H − 50]
+ were prominent.
Samples were run on two GC columns with differences in FAME retention properties that were helpful in confirming the identities of some peaks. One was a column with a poly(ethylene glycol) stationary phase. Such columns separate FAME according to carbon number and degree of unsaturation but do not separate monoenoic
cis and
trans isomers [
21]. The other was a column with a more polar biscyanopropyl stationary phase that is effective at separation of monoenoic
cis and
trans isomers [
21]. Relative performance with regard to isomers of PUFA is more complex, but there are definite retention differences on the two columns.
Analyzing the seven seed oil samples together in a single study was beneficial in that the major CFA in each sample being known, they effectively served as standards that were helpful in identifying minor CFA peaks in the other samples. Similarly, the identification or partial identification of certain other peaks was helped by comparison among the samples, as later described for specific cases.
2. Materials and Methods
Samples. Tung oil was obtained from Sigma-Aldrich (St. Louis, MO, USA). Catalpa ovata, Calendula officinalis, Punica granatum, and Jacaranda mimosifolia seeds were obtained from Sheffield’s Seed Co., Inc. (Locke, NY, USA). Momordica charantia (hybrid Gori) seeds were obtained from Seeds of India (Marlboro, NJ, USA). Impatiens balsamina seeds were obtained from Outsidepride.com (Independence, OR, USA).
Reagents and Solvents. Optima™ Grade Methanol and certified A.C.S. grade petroleum ether were from Fisher Scientific (Fair Lawn, NJ, USA). Chloroform and isooctane were HPLC grade from Sigma-Aldrich (St. Louis, MO, USA). Deionized water was from an in-house Millipore (Bedford, MA, USA) Milli-Q Water Purification System.
Sodium hydroxide and potassium hydroxide (Aldrich Chemical Company, Milwaukee, WI, USA), sodium chloride (Sigma Chemical Company, St. Louis, MO, USA), potassium bicarbonate and sodium nitrite (Sigma-Aldrich), and potassium chloride (Fisher Scientific) were all reagent grade. Nitric acid was J.T. Baker Ultrex. 3 N methanolic HCl and 0.5 N methanolic base (sodium metal in methanol/NaOCH3) were from Supelco (Bellefonte, PA, USA).
Extraction of Lipids from Seed Samples. Seeds were used as received and were ground in an electric grinder (Smart Grind, Black & Decker, Towson, MD, USA). Lipid extractions were based on the method of Folch et al. [
22] Ground seed samples (250 mg to 1 g) were placed in 25 mL or 50 mL glass screw top tubes with Teflon-lined caps. Extraction with 2:1 chloroform:methanol was conducted in two stages and with a combined solvent to sample ratio (mL/g) of 30:1 rather than the single stage 20:1 ratio specified by Folch et al. [
22] In the first stage, half the total solvent amount was added to the tube containing the seed sample, along with a magnetic stir bar. Samples were stirred, with periodic vortexing, for 20 min. Samples were then centrifuged at 1000 rpm for 2 min, and the supernatant was collected using a Pasteur pipet. The supernatant was washed with a 0.2× volume of 0.9% KCl. Phase separation was aided by centrifugation (1000 rpm for 2 min), and the lower chloroform layer was collected. The extraction and washing procedure was then repeated on the sample but with a stirring and vortexing period of 10 min. The two washed chloroform portions were combined, and solvent was evaporated to constant mass at room temperature under a stream of nitrogen gas, leaving a final, oily residue. Lipid residue yields ranged from 14% (
Catalpa o. and
Calendula o.) to 24% (
Momordica c.). Residues were reconstituted with isooctane and stored in a −20 °C freezer when not used immediately.
Derivatization Procedures. Two base treatments and one acid treatment were used for conversion of FA to FAME. Base treatment 1 was used routinely. Base treatment 2 and the acid treatment were used in the specific cases later mentioned. Unless otherwise noted, sample sizes were 20 to 25 mg of residue or oil.
Base Treatment 1 [
23]. 1 mL of 0.5N NaOCH
3 in methanol was added to each sample. Samples were vortexed and placed in a heating block at 50 °C for 15 min. After a brief cooling period, 1.5 mL of isooctane were added. Samples were vortexed, and then 4 mL of saturated aqueous NaCl solution were added. Samples were again vortexed, and the isooctane layer was collected using a Pasteur pipet. A second extraction using 1.5 mL of isooctane was then performed, and the resulting isooctane layer was combined with that from the first extraction. A washing step using 2 mL of 2% KHCO
3 was then performed, giving the final isooctane extract used for GC analysis.
Base Treatment 2 [
23]. 0.5 g of oil was mixed with 4 mL of petroleum ether and 1 mL of 2 M KOH in methanol. Samples were vortexed for 30 s and left to settle at room temperature for 30 min. The petroleum ether layer was collected using a Pasteur pipet.
Acid Treatment [
23]. 1.5 mL of methanol and 1 mL of 3N HCl in methanol were added to each sample. Samples were then vortexed and placed in a heating block at 50 °C for 1 h. Extraction with isooctane was performed as described for base treatment 1.
GC Standards. Multi-component FAME standards GLC 68B (16 FAME from C14 to C24) and GLC 463 (52 FAME from C4 to C24) were obtained from Nu-Check Prep (Elysian, MN, USA). CLA standards UC-59M and UC-61M were also obtained from Nu-Check Prep, as was methyl tricosanoate. Methyl pentacosanoate was obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Methyl hexacosanoate was obtained from Matreya LLC (Pleasant Gap, PA, USA). The methyl ester of 7c-18:1 was obtained from Larodan (Solna, Sweden). Standard L6031, containing the FAME of the eight geometrical isomers of 9,12,15-18:3, was obtained from Sigma-Aldrich. Standards were prepared as isooctane solutions at total FAME concentrations ranging from 0.2 mg/mL to 2 mg/mL.
In lieu of a commercial standard, a published procedure was adapted to produce the geometric isomers of linoleic acid (9
c,12
c-18:2) [
24]. A commercial corn oil sample with L at about 70% of FA content was first treated with methanolic KOH, as described above for base treatment 2, to convert FA to FAME. The recovered petroleum ether was evaporated under N
2 to yield the neat FAME oil (0.47 g recovered). To the FAME oil were added 2 drops of 6 M nitric acid and 0.2 mL of 2 M sodium nitrite. The sample was held at 40 °C for 10 min., after which 2 mL of deionized water and 2 mL heptane were added. After vortexing and settling, the heptane portion was recovered. A second extraction with 2 mL of heptane was performed, and the resulting heptane portion was combined with the first one. Further dilution with heptane was carried out to give an appropriate GC signal level. Additionally, using base treatment 2 for conversion of FA to FAME, a small amount of
trans-18:2 isomers was observed for the presumably deodorized corn oil as received. The observed distribution was 0.02%, 0.28%, 0.21%, 99.5% for the
tt,
ct,
tc, and
cc isomers, respectively. For the isomerized oil, the observed distribution was 7%, 20%, 20%, 53%.
Gas Chromatography and Mass Spectrometry. Two GC systems were used. One system was equipped with a single FID detector, and one with the effluent split between a mass selective detector (MSD) and an FID.
GC and GC-MS Apparatus
The GC-FID system was an Agilent Technologies (Santa Clara, CA, USA) 6890N GC with G2613A injector, split/splitless inlet, flame ionization detector (FID) and using OpenLabs CDS ChemStation Edition for GC Systems (Rev. C.01.05) software. The GC-MS instrument was an Agilent Technologies 5975C Inert XL EI/CI MSD with a 7890A GC System and G4513A injector, split/splitless inlet, G3180B two-way splitter with makeup gas, auxiliary FID, and using MassHunter GC/MS Acquisition software (version B.07.00 SP1.1549).
6890N GC-FID Method
The column for GC-FID was a Supelco (Bellefonte, PA, USA) SP-2560 (100 m × 0.25 mm × 0.2 µm film thickness) using hydrogen as the carrier gas with operation in constant flow mode at 1.3 mL/min. For the FID, the total hydrogen flow was set at 35 mL/min, air flow at 350 mL/min, and make-up gas (N2) flow at 25 mL/min. Experiments were conducted in split mode with a 100:1 split. The inlet temperature was 250 °C, and the FID temperature was 260 °C. The temperature program was as follows: initially 70 °C, then a gradient of 7.8 °C/min to 130 °C, followed by a gradient of 3.1 °C/min to 189 °C with a 2 min hold, then a gradient of 1.6 °C/min to 200 °C, and finally a gradient of 3.2 °C/min to 230 °C, with a 22 min hold. The run time was 67 min, and 1 µL was injected.
5975C/7890A GC-MSD-FID Methods
Method 1. The first column used for GC-MS/FID was also a Supelco SP-2560 (100 m × 0.25 mm × 0.2 µm film thickness) using helium as the carrier gas operated in constant flow mode at 1 mL/min. As above, the total hydrogen flow for the FID was set at 35 mL/min, air flow at 350 mL/min, and make-up gas (N2) flow at 25 mL/min. GC-MS/FID experiments were conducted in split mode with a 50:1 split. The inlet temperature was 250 °C, and the FID temperature was 260 °C. The temperature program started with an initial temperature of 70 °C, with a temperature gradient of 5 °C/min to 130 °C, then a gradient of 2 °C/min to 180 °C and a gradient of 1 °C/min to 196 °C, and finally a gradient of 2 °C/min to 230 °C with 30 min hold. The MSD was operated in a positive chemical ionization mode with methane reagent gas (PCI-CH4). The scan range was from m/z 50 to m/z 450 at 1.9 Hz.
Method 2. The second column used for GC-MS/FID was a Supelco Omegawax 250 (30 m × 0.25 mm × 0.25 µm film thickness) with helium carrier gas in constant flow mode at 1 mL/min. The inlet temperature was 250 °C, and the FID temperature was 270 °C. The FID again used hydrogen flow at 35 mL/min, air flow at 350 mL/min, and make-up gas (N2) flow at 25 mL/min. Operation was in split mode at 50:1 split, and 1 µL was injected. The temperature program had an initial temperature of 70 °C, a temperature gradient of 10 °C/min to 170 °C, and then a gradient of 1 °C/min to 250 °C, with 1 min hold. The run time was 91 min. The MSD was operated in PCICH4 mode, with scans from m/z 50 to m/z 450 at 0.9 Hz.
Calculations. FID peaks were manually integrated within the Agilent Chemstation data analysis software, and retention time versus peak area data were transferred to an Excel spreadsheet for further analysis. FAME mass percentages were obtained directly, and these were converted to mole percentages. Molar response factors based on the FID responses of the FAME of the GLC 68B standard were applied to the mole percentages. The corrected molar percentages were used for construction of
Table 2. The data in
Table 3,
Table 4,
Table 5,
Table 6 and
Table 7 are from the individual isomer percentages of the relevant total isomer peak area.
3. Results and Discussion
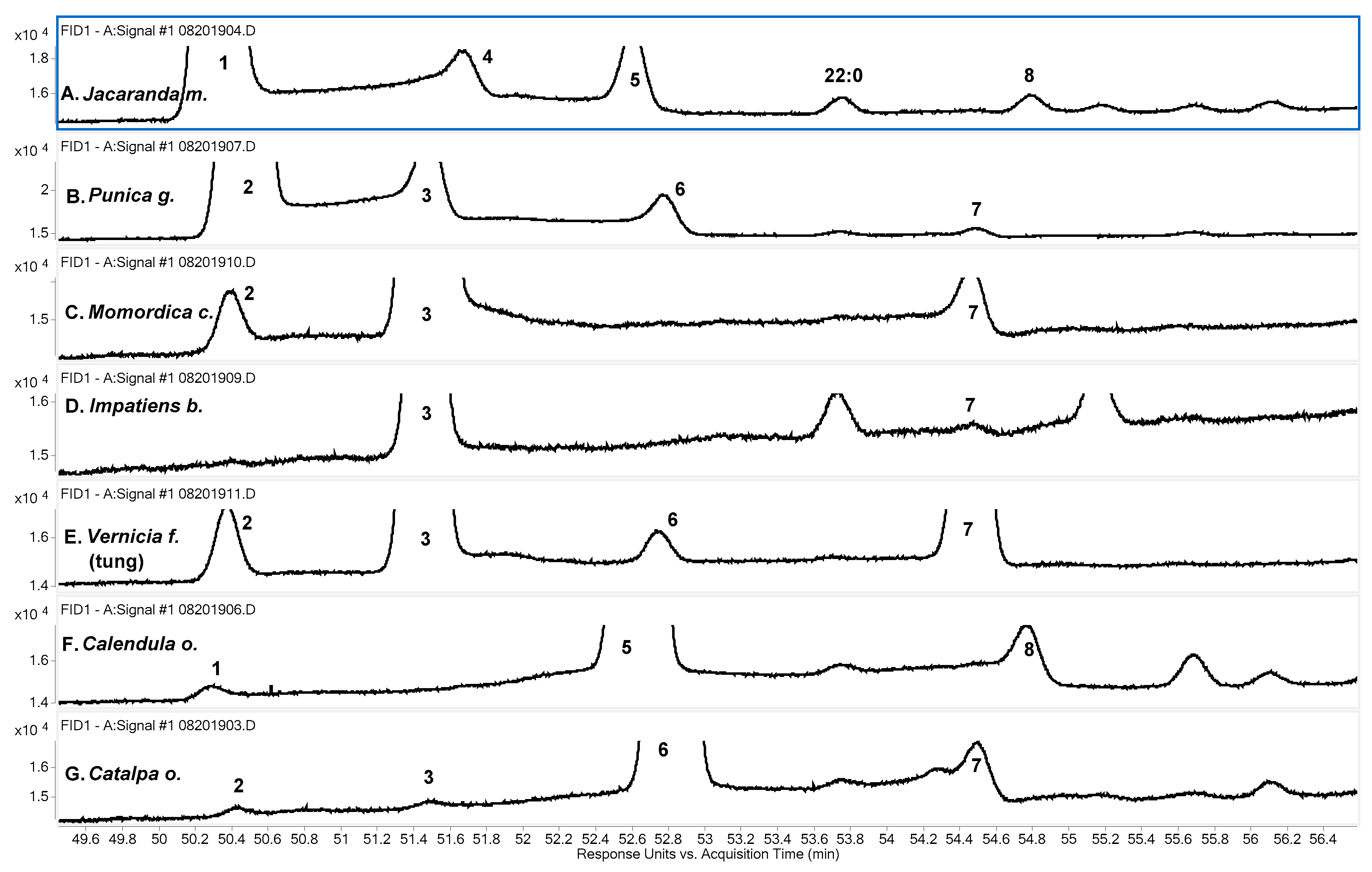
Figure 1 shows portions of FID chromatograms obtained using the SP-2560 column that include the major FAME peaks obtained for the seven CLNA-containing samples. From top to bottom, the samples are
Jacaranda m.,
Punica g.,
Momordica c.,
Impatiens b.,
Vernicia f. (tung oil),
Calendula o., and
Catalpa o. The order follows the retention time (RT) order of the most abundant CLNA FAME for each sample. The methyl ester of jacaric acid is the earliest eluting of these, at about 74 min. It is followed in order by the FAME of punicic acid, α-eleostearic acid (the major CLNA species found in
Momordica c. and
Impatiens b. seeds and in tung oil), α-calendic acid, and catalpic acid. Tung oil (
Figure 1E) has a second prominent CLNA species, β-eleostearic acid, with FAME RT a little longer than that of the catalpic acid FAME. Another CLNA, β-calendic acid, known to occur in
Calendula o. (16), has FAME RT a little past that of the FAME of β-eleostearic acid, but the peak is too small to see in
Figure 1F. Considering the
cis/
trans configurations of the three conjugated double bonds of each isomer, the retention order on the SP-2560 column can be described as
ctc <
ctt <
ttc <
ttt. The retention order of 8,10,12 versus 9,11,13 is not consistent. For
ctc and
ttc, the 8,10,12 isomer has an earlier RT than the 9,11,13 isomer, but the opposite is observed for the
ttt isomers.
For five of the seven samples, the dominant CLNA FAME is the most prominent peak in the FID chromatograms. For
Jacaranda m. (
Figure 1A), the peak for the linoleic acid FAME is slightly larger than that for jacaric acid. For
Impatiens b. (
Figure 1D), the α-linolenic acid FAME is the most prominent peak, and there is only a small peak for the α-eleostearic acid FAME. Impatiens species are known to contain the conjugated 18:4 FA isomers α-parinaric acid and β-parinaric acid (
Table 1). In
Figure 1D, the peak for the FAME of α-parinaric is seen at about 87 min. Six species of FA, including isomers, account for almost all the obvious peaks in the chromatograms in
Figure 1. These are: 16:0, 18:0, 18:1, 18:2, 18:3, and 18:4.
Table 2 provides a more detailed look at the FA distributions found for each sample and includes relative abundances for species found down to 0.01%. Relative quantitation was performed on FID chromatograms obtained using the SP-2560 column. Results obtained using the Omegawax column were examined as needed for help in resolving uncertainty about peak identification. Only FID peaks that were present at ≥0.01% and with identification supported by MS (peaks observed in extracted ion chromatograms (EIC) using the ions characteristic of molecular weight, established from the mass spectrum of known FAME, and with matching confirmed by examination of extracted peak mass spectra), or that matched the RT on both columns with a peak identified in another sample, were used in constructing the table. Isomers are grouped together in
Table 2, so, e.g., the entries for 18:3 include the sum of the area percentages for all 18:3 species found (Ln and all other non-conjugated 18:3 plus all CLNA).
A small amount of methyl laurate (12:0) was found for Punica g., but no FAME smaller than 14:0 was confirmed for any other sample. The FAME of the odd-carbon FA 15:0, 17:0, and 23:0 were found in small amounts for all samples, but 19:0, 21:0, and 25:0 were not uniformly detected. The FAME of palmitic acid (16:0) was prominent for all samples. The FAME of a single 16:1 isomer, palmitoleic acid (9c-16:1), was observed for all samples except Momordica c. No other 16:1 isomers were confirmed.
In total, the FAME of 18-carbon FA accounted for over 90% of the integrated signal for all samples. The lowest 18-carbon FA content was found for Impatiens b., at 91.85%. The others ranged from 94.77% (Calendula o.) to 97.52% (Momordica c.). The 18:3 isomers were, in total, the most abundant 18-carbon FA for all samples except for Jacaranda m., for which the total 18:2 isomers were more abundant. For Catalpa o., the total of 18:2 isomers was only slightly less than the 18:3 total, and for Impatiens b., the total of 18:4 isomers was only slightly less than the 18:3 total. Momordica c. was found to contain 30% of stearic acid (S; 18:0), which was much higher than for any of the other samples. Total 18:1 isomers ranged from 1.71% for Momordica c. to 15.78% for Impatiens b. The total 18:4 content for Impatiens b. was found to be 27.5% of its total FA content. The presence of 18:4 was not confirmed for any other sample. The isomer distributions for 18:1, 18:2, 18:3, and 18:4 are discussed in more detail below. The total of FAME found for FA greater than 18 carbons ranged from about 0.5% for Impatiens b. to about 1.3% for Jacaranda m. and tung oil. Of these longer FA, only two unsaturated species were observed: 20:1 (11c-20:1 and smaller amounts of two other isomers), found in all samples, and 20:2 (a single isomer, 11c,14c-20:2), found in all except Momordica c. FAME of 20:0, 22:0, and 24:0 were found in all samples. Small amounts of the FAME for 26:0 were found for Punica g., Jacaranda m., Catalpa o., and Impatiens b. Fame of any FA larger than 26:0 were not confirmed for any sample.
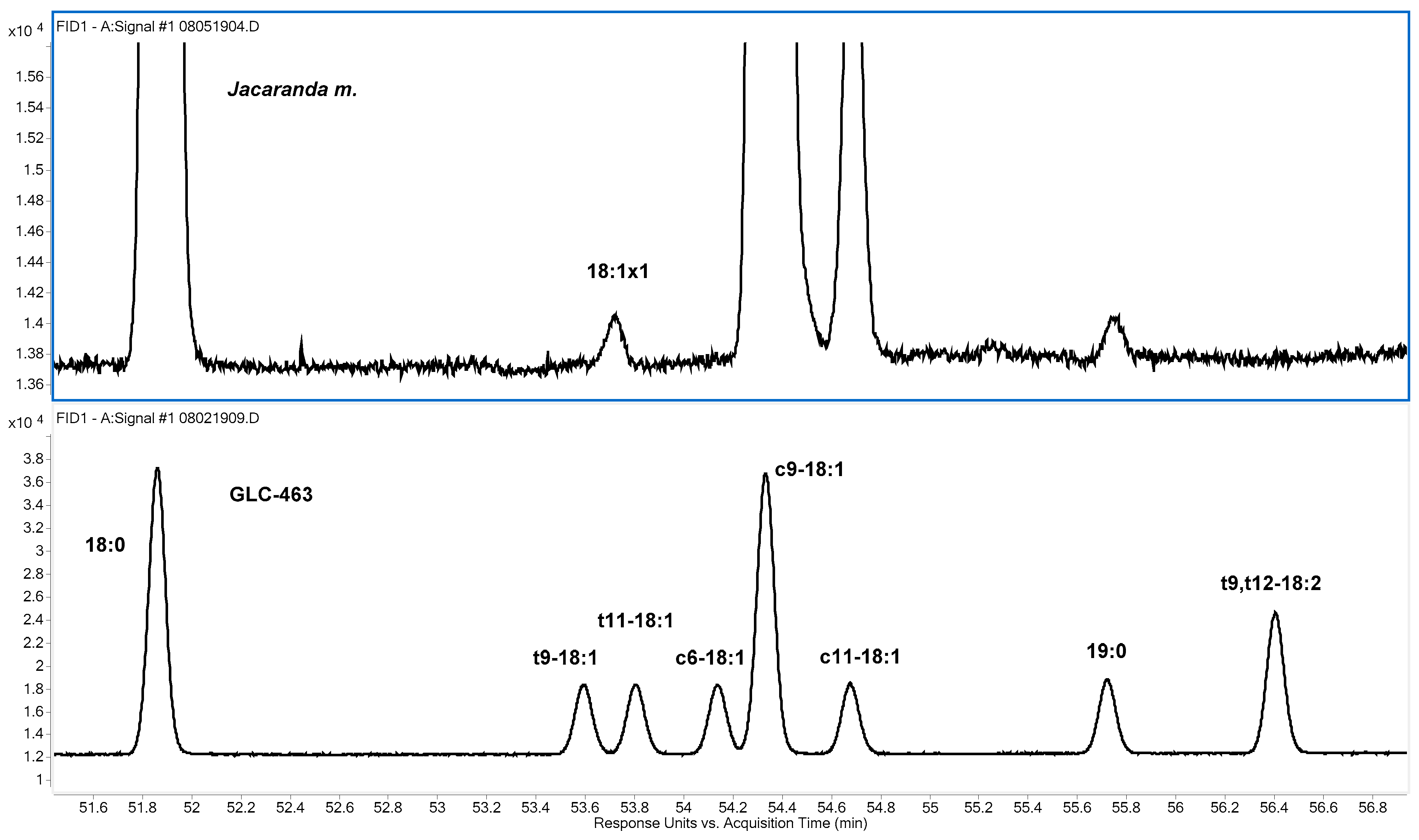
3.1. 18:1. Isomers
As indicated in
Table 3, oleic acid (9
c-18:1) was by far the most abundant 18:1 isomer found for all samples, and
cis-vaccenic acid (11
c-18:1) was the second most abundant. A small amount of a third isomer, designated 18:1x1, was also found for all samples. Three other 18:1 isomers, 9
t, 11
t, and 6
c, are listed in
Table 3 because they are present in the GLC 463 standard, but none of the three were confirmed for any of the samples. On the SP-2560 column, the 18:1x1 isomer eluted at a position between the 9
t- and 11
t-18:1 isomers, as shown in
Figure 2. With 100 m 100% cyanopropyl polysiloxane columns, it is possible to separate most of the trans-18:1 isomers, and the position of 18:1x1 in
Figure 2 is consistent with what was observed for 10
t-18:1 by Kramer, et al. [
25,
26]. The 18:1x1 peak was not observed with the Omegawax column, presumably due to overlap with 9
c- or 11
c-18:1.
3.2. 18:2. Isomers
Table 4 gives the 18:2 isomer distribution as a percentage of the total integrated 18:2 FID signal for each sample. Linoleic acid was the major isomer for all, but several of the samples had other prominent 18:2 isomers. A total of seven 18:2 isomers were found. The commercial standards used contained L (in both GLC 68B and GLC 463) and 9
t,12
t-18:2 (in GLC 463). The isomerized corn oil sample described above established the RT of the two other 9,12-18:2 isomers. The elution order of the geometric L isomers on highly polar cyanopropyl columns such as SP-2560 (USP G5 phases) is known to be
tt <
ct <
tc <
cc [
24,
27].
Six 18:2 isomers were observed in
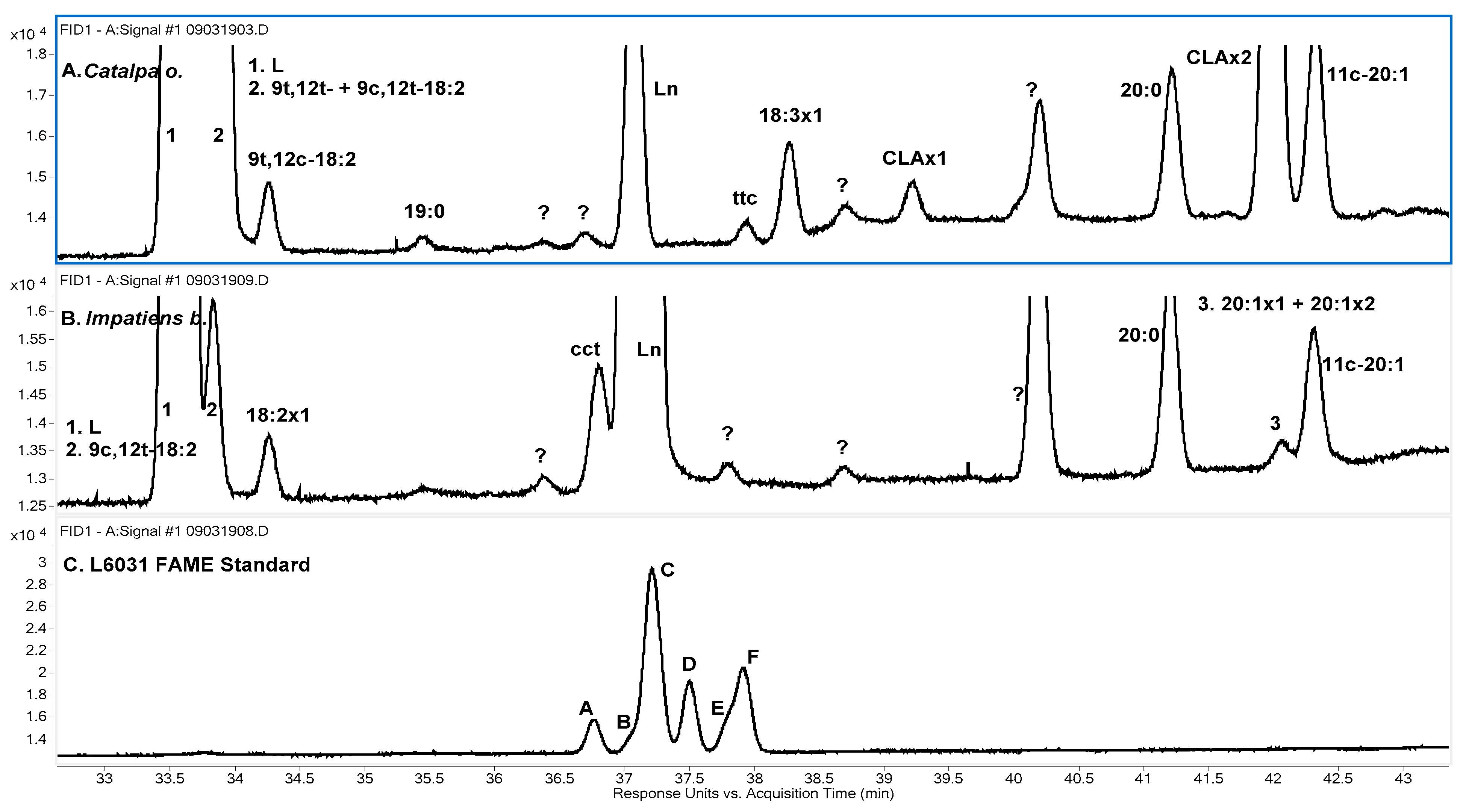
Catalpa o., and
Figure 3A shows the relevant portion of its FID chromatogram on the SP-2560 column. About 27% of its 18:2 content was found to be 9
t,12
t. This isomer was not observed for any of the other samples. Two 18:2 isomers of
Catalpa o. matched the retention times of the
ct- and
tc-9,12-18:2 isomers of the isomerized corn oil sample. The matches were also observed on the Omegawax column, and it is assumed these isomers are 9
c,12
t and 9
t,12
c. The 9
c,12
t isomer was found in all samples and was present at about 10% of total 18:2 signal for
Momordica c. and tung oil. The 9
t,12
c isomer was found only in
Catalpa o.
Another isomer, designated 18:2x1, was found only in
Impatiens b. It eluted a short time after L on the SP-2560 column (
Figure 3B). On the Omegawax column it also eluted shortly after L (
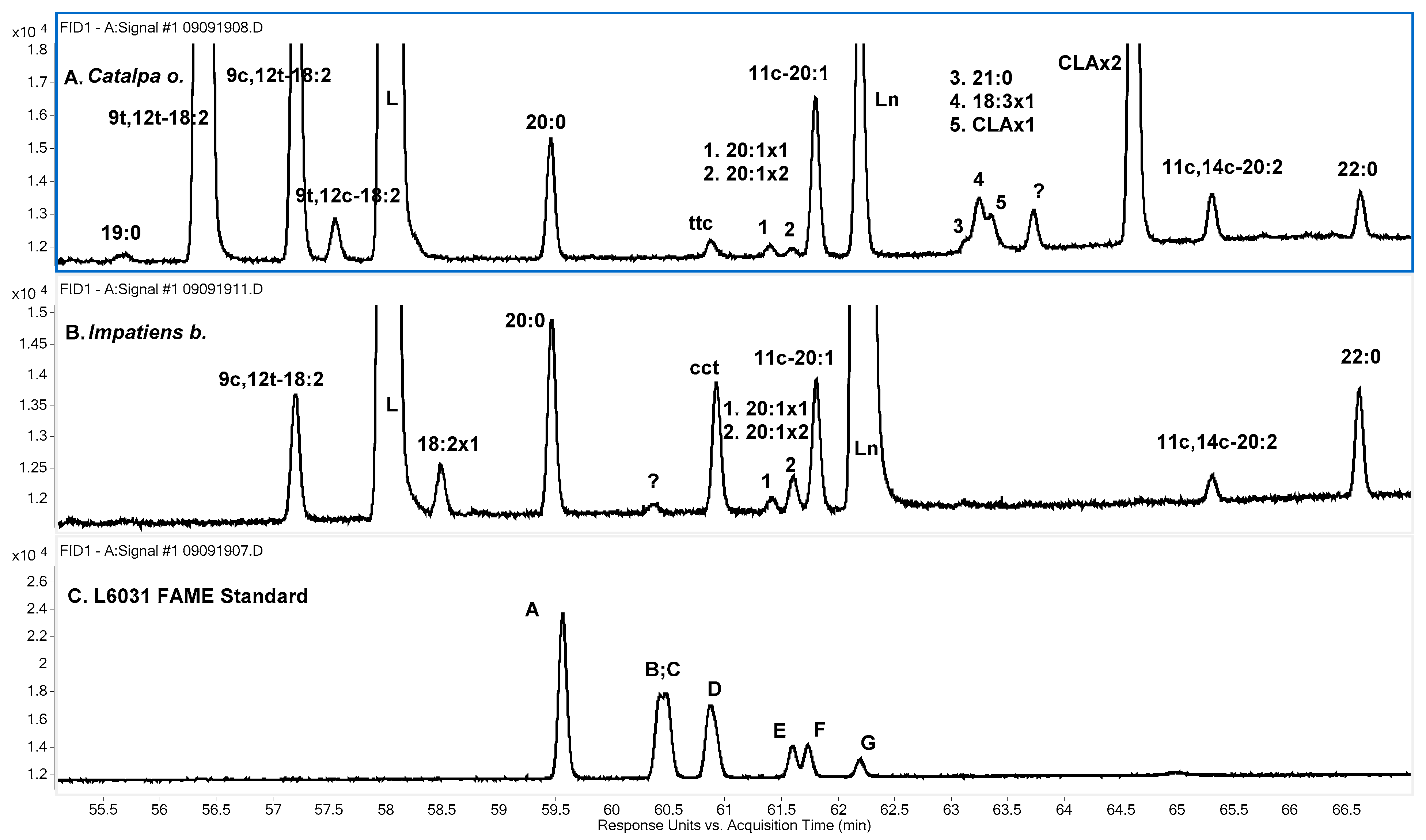
Figure 4B), although the overall elution order of the 18:2 isomers was different on the two columns. For
Catalpa o. on the Omegawax column (
Figure 4A), 9
t,12
t and 9
c,12
t elute just after L and cannot be distinguished from each other by RT. The coelution of these two isomers on the Omegwax column was confirmed using the isomerized corn oil sample. The position of 9
c,12
t can be clearly seen for
Impatiens b., for which 9
t,12
t is not present, in
Figure 4B. 9
t,12
c elutes after 9
c,12
t on both columns (
Figure 3A and
Figure 4A), and it has the same RT as 18:2x1 of
Impatiens b. on the Omegawax column (
Figure 4A,B). However, the RT of the four 9,12-18:2 geometric isomers having all been established, 18:2x1 must be an 18:2 positional isomer.
The other two unknown 18:2 isomers elute after α-Ln on both columns, suggesting that they are conjugated 18:2 species. They are given the designations CLAx1 and CLAx2 in
Table 4 and
Figure 3 and
Figure 4. Both CLA isomers were found in
Jacaranda m. and
Catalpa o., but their presence was not confirmed in any of the other samples. The elution on the SP-2560 column of CLAx1 near 21:0 suggests it is a
ct- or
tc-CLA isomer, and the later elution of CLAx2 in the region of 20:2 isomer elution suggests it is a
tt-CLA isomer [
13]. RT matching with Nu-Check UC-59M and UC-61M standards indicates that CLAx1 is the 10
t,12
c isomer and that CLAx2 is 8
t,10
t, 9
t,11
t, or 10
t,12
t. The latter three isomers elute at the same RT under the conditions used, just after the 11
t,13
t isomer, similar to what was reported by Cruz-Hernandez et al. [
13].
3.3. 18:3. Isomers
Table 5 gives the 18:3 isomer distributions and includes conjugated and non-conjugated species. The GLC 68B standard contains only one 18:3 isomer, α-Ln, and the GLC 463 standard contains two 18:3 isomers, α-Ln and 6
c,9
c,12
c-18:3 (γ-Ln). All samples contain α-Ln, but
Impatiens b. is the only sample for which it is a large percentage of the total 18:3. No γ-Ln was observed for any sample.
A few of the samples contained small amounts of other non-conjugated 18:3 isomers. A total of three were found. Two of the three are identified as geometric isomers of 9,12,15-18:3 by RT matching with peaks observed for the L6031 standard. The order of elution for the eight geometric 9,12,15-18:3 isomers on highly polar cyanopropyl columns can be stated as
ttt <
ctt <
tct <
ttc +
cct <
ctc <
tcc <
ccc [
24,
27,
28,
29]. The
ctt and
tct isomers are typically close in RT and may even be coincident, depending on the particular column and temperature program. It is similar for the
ttc and
cct isomers, but they are more difficult to separate. Considering the known isomer relative concentrations, peaks observed for the L6031 standard on the SP-2560 column in this work are consistent with the order given above (
Figure 3C). The resolution of the L6031 peaks is not as good on the Omegawax column (
Figure 4C), and the time range over which they elute is about half that of the SP-2560 column. Additionally, the elution order on the Omegawax column is not the same as on the SP-2560 column and does not follow any obvious pattern with respect to the
cis/
trans configuration of the double bonds.
A peak observed for
Catalpa o., labeled
ttc in
Figure 3A, is a RT match to the fourth peak, labeled D, of L6031, a coelution of the
ttc and
cct 9,13,15-18:3 isomers, on the SP-2560 column (
Figure 3C). There is a peak with similar RT, labeled
cct, for
Impatiens b. (
Figure 3B); however, its RT is about 3 s past the apex of L6031 D, while the RT of the
Catalpa o. peak is virtually identical to the apex of L6031 D. The L6031 peak is slightly skewed, suggesting that the less abundant
cct isomer has a slightly longer RT than the more abundant
ttc component and the tentative identifications of the
Catalpa o. peak as the
ttc isomer and the
Impatiens b. peak as the
cct isomer.
Results for the Omegawax column verify that the
Catalpa o. and
Impatiens b. peaks are from different isomers. The
Catalpa o. peak, labeled
ttc in
Figure 4A, matches the latest eluting isomer peak of L6031, labeled F in
Figure 4C, which is again, by consideration of peak areas, a mixture of a (1
c, 2
t) isomer and a (2
c, 1
t) isomer. The
Impatiens b. peak, labeled
cct in
Figure 4B, matches the first eluting L6031 isomer, labeled A in
Figure 4C. The latter is a (2
c, 1
t) isomer, and, considering the SP-2560 results, it is the
cct isomer.
From the SP-2560 results and the identification of the cct isomer in Impatiens b., it follows that the Catalpa o. peak under consideration, and likewise the (1c, 2t) component of L6031 peak F on the Omegawax column, is the ttc isomer. The elution order of the α-Ln geometric isomers on the Omegawax column can thus be partially described as A (cct) < B (ccc) < C (ttt + ctt or tct) < D (ctt or tct) < E (ctc or tcc) < F (ttc + ctc or tcc).
An unknown 18:3 isomer, labeled 18:3x1 in
Table 5 and
Figure 3A and
Figure 4A, was only found in tung oil and in
Catalpa o. It elutes later than any of the L6031 peaks on both columns, so the positions of its double bonds must be other than 9,12.15.
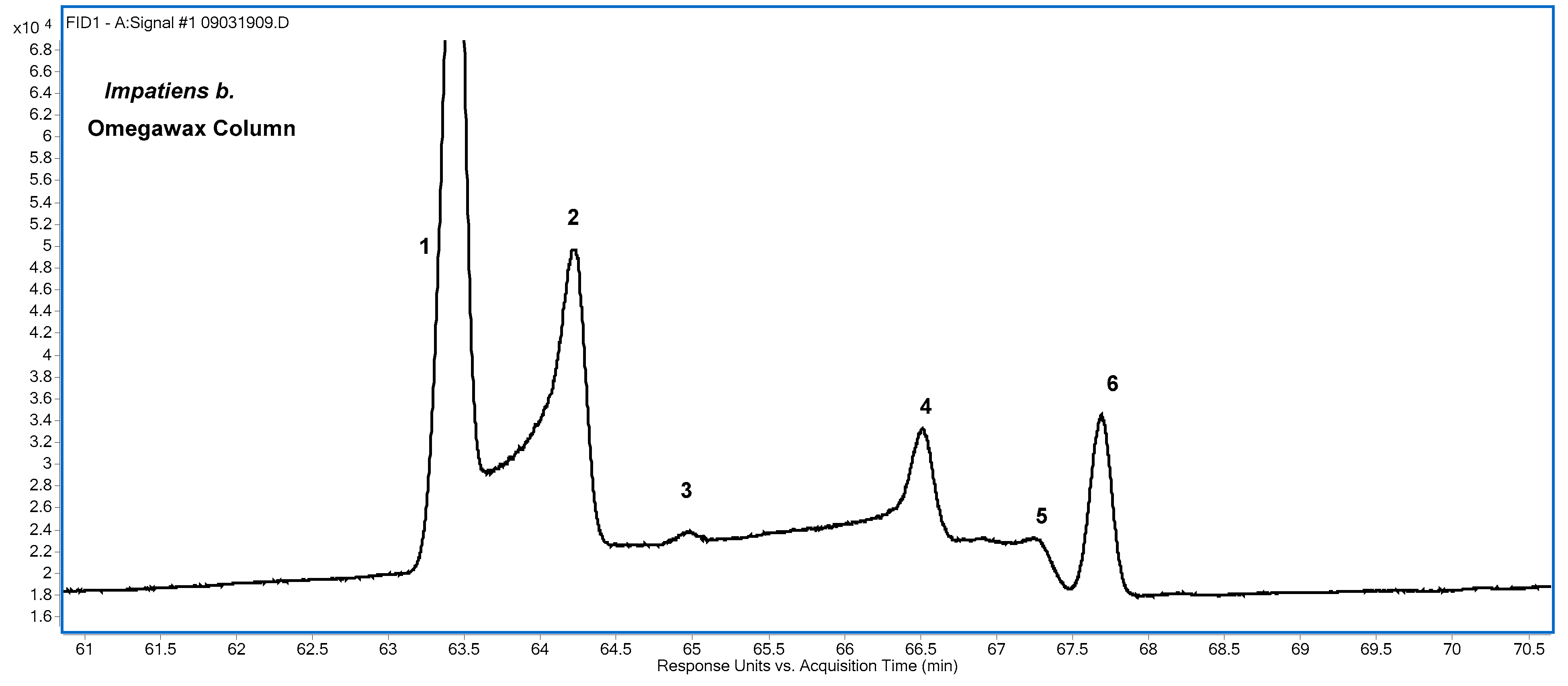
The elution order of the CLNA isomers was the same for both columns, but the separation of the isomers, and especially of the FAME of punicic acid and α-eleostearic acid, was better on the Omegawax column,
Figure 5. Overlap of the FAME of punicic acid and α-eleostearic acid on the SP-2560 column hid the presence of the smaller of the two isomers for
Punica g.,
Momordica c.,
Impatiens b., and tung oil. For
Catalpa o., for which both isomers are minor components, they could be distinguished but were only partially separated, with a resolution of approximately 0.5.
Figure 5 shows the CLNA regions of the FID chromatograms of the seven samples obtained using the Omegawax column. The samples used to obtain
Figure 5 were diluted by between 2:1 and 10:1 from normal analysis levels to give better RT matching between the largest CLNA peak in each sample with the much smaller corresponding peaks in other samples. The abundance of the dominant CLNA species for most of the samples, e.g., punicic acid for
Punica g. and α-eleostearic acid for tung oil, resulted in peak fronting at the sample concentrations used for determination of the FA profiles presented in this work. However, such concentrations were necessary to obtain peak area and RT estimates of minor components, and some peaks, which are assigned a percentage in
Table 5, are only faintly seen or are not observed in the chromatograms of
Figure 5.
It can be seen in
Figure 5 that the FAME of 22:0, behenic acid, elutes within the region of CLNA FAME elution. Another potential interfering FAME on the Omegawax column is that of all-
cis-5,8,11,14,17-20:5 (eicosapentaenoic acid (EPA)), and the FAME of erucic acid (22:1) elutes just after the CLNA region. On the SP-2560 column, potential interfering FAME include those of EPA, nervonic acid (24:1), and 22:3. The FAME of lignoceric acid (24:0) elutes just before the CLNA region on the SP-2560 column. However, no evidence of the FAME of EPA, 22:1, 24:1, or 22:3 was found for any of the samples.
Four of the samples (
Punica g.,
Momordica c., tung oil, and
Catalpa o.) contained all four of the 9,11,13-18:3 isomers listed in
Table 1. Three of the 9,11,13-18:3 isomers were found in the
Impatiens b. sample. Catalpic acid was not detected in
Impatiens b., which has a much lower total CLNA level than the other samples. None of the 8,10,12-18:3 isomers could be verified for any of these five samples.
Jacaranda m. contained the three 8,10,12-18:3 isomers listed in
Table 1 and gave a fourth CLNA peak that does not match the RT of any of the isomers in
Table 1. As is apparent in
Figure 5 and as mentioned earlier, the well-known CLNA isomers elute according to the pattern
ctc <
ctt <
ttc <
ttt, and this order holds on both columns used in this work. The FAME of the two
ctc isomers, jacaric acid and punicic acid, have a ΔRT of about 0.13 min in
Figure 5 (peaks 1 and 2); the two
ttc isomers (α-calendic and catalpic) have a ΔRT of about 0.19 min (peaks 5 and 6); and the two
ttt isomers (β-eleostearic and β-calendic, peaks 7 and 8) have a ΔRT of about 0.28 min. The separation of
ctc and
ctt (peaks 1 vs. 4 and 2 vs. 3) is about 1 min in
Figure 5; the separation of
ctt and
ttc is about 1 min (peaks 4 vs. 5 and 3 vs. 6); and the separation of
ttc and
ttt is about 1.7 min (peaks 5 vs. 8 and 6 vs. 7). Since it is close in RT to α-eleostearic acid (ΔRT about 0.2 min on both columns), it is proposed that the unidentified CLNA peak (peak 4) for
Jacaranda m. is from the 8,10,12 analog to α-eleostearic acid, 8c,10t,12t-18:3.
Calendula o. likewise contained the three 8,10,12-18:3 isomers listed in
Table 1 and a small amount of the probable 8
c,10
t,12
t-18:3 isomer. No 9,11,13-18:3 isomers could be verified for
Jacaranda m. or
Ca lendula o.
The probable presence of the 8
c,10
t,12
t-18:3 isomer in
Jacaranda m. and
Calendula o. is in agreement with the observation of Wang, et al. [
17] of the natural occurrence of this isomer, which had not previously been reported.
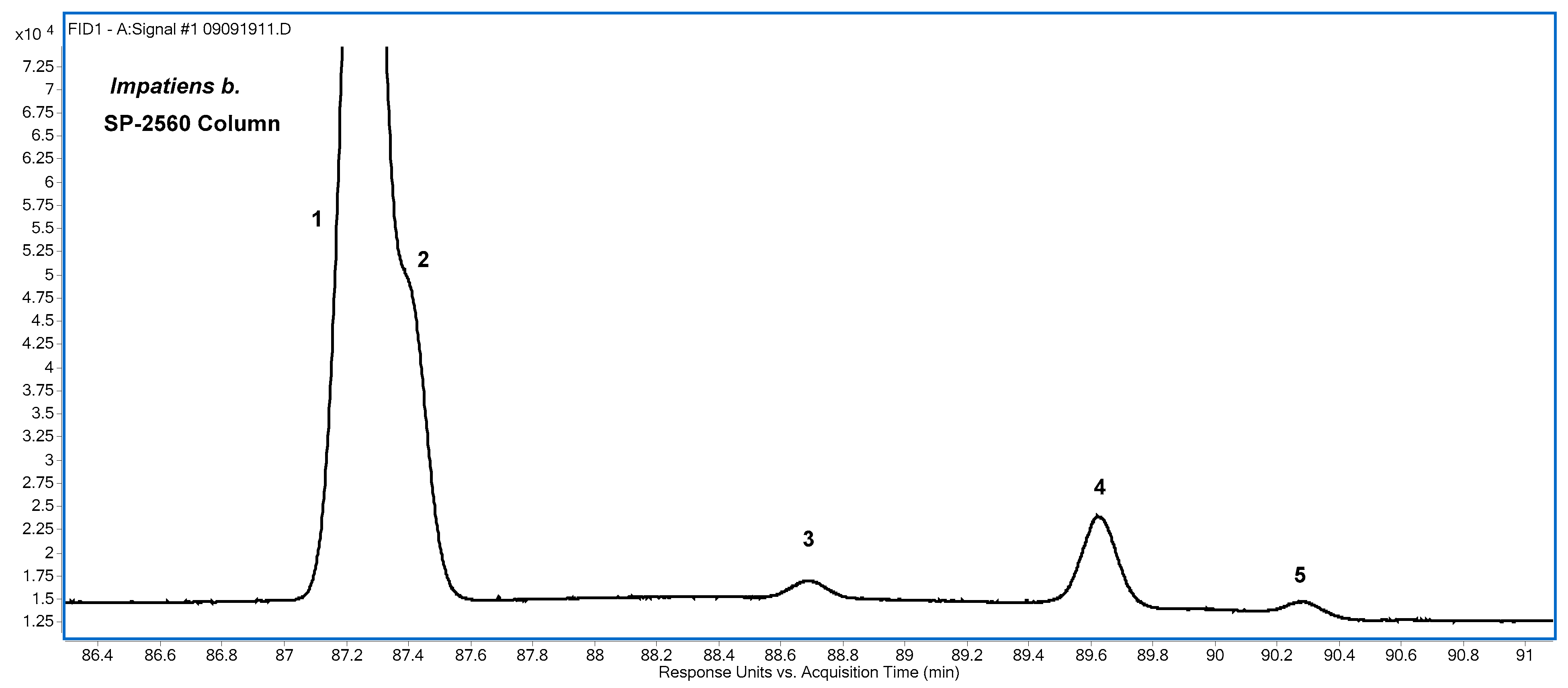
3.4. 18:4. Isomers
CSDA isomers are the predominant CFA species found in
Impatiens b. The isomer distribution based on integration of the 18:4 FAME FID peaks is given in
Table 6, and the CSDA region of an FID chromatogram of Impatiens b. obtained using the SP-2560 column is shown in
Figure 6. Hamberg (29) identified four 9,11,13,15-18:4 isomers from an
Impatiens b. sample and reported the following relative abundances determined by GC with a methyl silicone column: α-parinaric acid (74%); 9
c,11
t,13
t,15
t-18:4 (18%); 9
t,11
t,13
t,15
c-18:4 (4%); and β-parinaric acid (4%). Using capillary electrophoresis (CE) and UV detection, Ohman et al. (30) reported the same four CSDA isomers in a sample of
Impatiens b. They did not report relative abundances, but a sample electropherogram suggests an approximate distribution of:
cttc (70%),
cttt (15%),
tttc (10%) and
tttt (5%). The relative abundance estimates in
Table 6 were obtained using the SP-2560 column.
The first peak in
Figure 6 has two incompletely resolved components, introducing some uncertainty into the peak area assignments; but based on relative areas, they are assigned as the FAME of α-parinaric acid and its
cttt isomer. The two peaks are better separated on the Omegawax column (
Figure 7), but the underlying ‘hump’ makes quantitative estimates problematic. The hump presumably consists primarily of unresolved FAME of CSDA, and such an interpretation is supported by examination of the PCI-CH
4 mass spectra of various portions of it. Although not obvious in
Figure 6, a similar hump is observed using the SP-2560 column, but it is smaller. Integration of the entire hump areas of
Figure 6 and
Figure 7, including peaks, and then subtracting the peak areas indicates that about 10% of the total CSDA FID signal is unresolved on the SP-2560 column, and about 55% is unresolved on the Omegawax column.
Hamberg [
30] cautioned that thermal
cis-to-
trans isomerization can occur during GLC analysis of CSDA and estimated that it occurred to the extent of about 5% to 8% for a given CSDA isomer under the conditions used. Such isomerization occurring in the GC inlet would result in errors in estimates of endogenous CDSA isomers and possibly produce non-endogenous isomers. Isomerization occurring on-column would seem likely to cause peak broadening. The inlet temperatures used in the SP-2560 method and the Omegawax method in this work were both 250 °C. The oven temperatures at the time of elution were similar: about 230 °C for the SP-2560 method and about 225 °C for the Omegawax method. The time of exposure to elevated temperatures can also influence the degree of isomerization of conjugated PUFA [
7], but here the RT of CSDA FAME is longer on the SP-2560 column (about 88 min) than on the Omegawax column (about 65 min). It appears that the larger hump area on the Omegawax column has more to do with its poorer resolution of the CSDA FAME than with thermal isomerization occurring during GC analysis.
Following Hamberg [
30] and Ohman et al. [
31], the third largest peak in
Figure 6, peak 4, is assigned as the FAME of the
tttc isomer of α-parinaric acid. The two smallest peaks, peaks 3 and 5, are each about 1% of the CSDA FAME FID area. A significant increase in the area of peak 5 upon use of the acid treatment for the conversion of FA to FAME suggests that it is the FAME of the
tttt isomer, β-parinaric acid. Peak 3 appears to be a fifth CSDA isomer. It exhibits a small peak in the extracted ion chromatogram (EIC) obtained using ions characteristic of the known CSDA isomers, but, unlike in the FID chromatogram, its EIC peak is smaller than that observed for β-parinaric acid, and the extracted peak mass spectrum is of poor quality due to the weak signal level. Peak 3 is therefore not included as part of the isomer distribution given in
Table 6, pending further investigation. Apart from this possible fifth CSDA isomer, the major differences among the CSDA abundance estimates of this work, Hamberg [
30] and Ohman et al. [
31] are for the
tttc and
tttt isomers. Sample-to-sample differences may account for some portion of the differences, but treatment differences, including extraction, derivatization, and instrumental, may also have an influence.
3.5. 20:1. Isomers
Three 20:1 isomers were observed at the same retention times in all seven samples, consistent with these being the same three isomers in all seven samples. The distributions are shown in
Table 7. The GLC-463 standard contains the 5
c-, 8
c-, and 11
c-20:1 isomers. On the SP-2560 column, the 20:1 isomers elute in the same region as the non-conjugated 18:3 isomers (
Figure 3). On the Omegawax column, they elute after 20:0 and in the same region as
tt-CLA isomers (CLAx2 in
Figure 4A). The major isomer found for all samples was 11
c-20:1. On the SP-2560 column, the two less abundant 20:1 isomers of the samples have RT close to that of the 8
c-20:1 isomer of GLC-463. One isomer elutes slightly before 8
c-20:1, and the other elutes slightly after it. Following Kramer et al. [
32], the later eluting isomer, designated 20:1x2 in
Figure 3 and
Table 7, is probably 9
c-20:1, although 14
t-20:1 cannot be excluded. The earlier eluting isomer, designated 20:1x1, is probably one of 13
t, 14
t-, 6
c-, or 7
c-20:1. On the Omegawax column, the two less abundant 20:1 isomers elute together, just before 11
c-20:1 and very close to the
tt-CLA isomer found for
Jacaranda m. and
Catalpa o., designated as CLAx2 in
Figure 4A.
4. Conclusions
The FA distributions of seven CFA-containing seed oil samples were determined, including isomer distributions for 18:1, 18:2, 18:3, 18:4, and 20:1 FA. Quantification was performed, after conversion of FA to FAME by base-catalyzed methylation, using FID data, but the use of MS with PCI-CH4 was helpful in establishing FAME molecular weights. The use of two columns with different retention characteristics, i.e., one with a poly(ethylene glycol) stationary phase and one with a bis-cyanopropyl stationary phase, was also helpful in peak identification. Highly polar cyanopropyl columns are considered essential for analysis of milk and meat samples containing multiple CLA species (13, 14), and they are also superior to less polar columns for analysis of samples containing CLNA and CSDA. However, complimentary information obtained using a poly(ethylene glycol) column was crucial in peak identification in several instances in this work. Contrary to what might be expected, separation of CLNA and CSDA isomers was better on the Omegawax column than on the SP-2560 column, although peak definition and hence quantitative estimates were superior with the SP-2560 column.
In addition to the major CFA components, which are well known for these samples, all seven samples were found to contain small amounts of additional CFA. Four samples, Punica g., Momordica c., Vernicia f. (tung), and Catalpa o., were found to contain all four of the known, naturally occurring 9,11,13-CLNA isomers: punicic acid, α-eleostearic acid, catalpic acid, and β-eleostearic acid. Three of these were observed in the Impatiens b. sample, which contains lower levels of CLNA than the other 9,11,13-CLNA-containing samples, but catalpic acid was not detected. Jacaranda m. and Calendula o. were found to contain the three well-known, naturally occurring 8,10,12-CLNA isomers: jacaric acid, α-calendic acid, and β-calendic acid, plus a fourth isomer, 8c,10t,12t-18:3, which, until recently, had not been observed to occur naturally. There is evidence of a fifth CSDA isomer in Impatiens b., for which four CSDA isomers have been previously reported (29, 30), but additional work is needed for confirmation. The reported distributions account for 99% or more of the total FID peak areas observed for the samples. Additional work is required to identify the peaks not accounted for, which may be due to oxygenated FA or species other than FA.