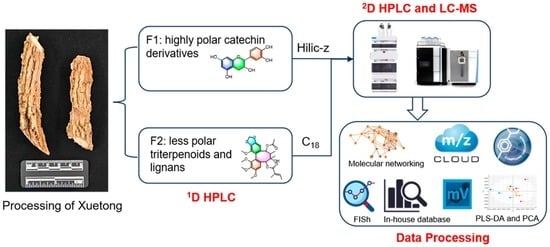
Targeted Offline Two-Dimensional HPLC and UHPLC-Orbitrap-MS Combined with Molecular Networking Reveal the Effect of Processing on Chemical Constituents of Xuetong (the Stem of Kadsura heteroclita)
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Processing and Sample Solution Preparation
2.3. Targeted Offline 2D HPLC Separation
2.4. 2D UHPLC-MS Analysis
2.5. Data Processing and Molecular Networking
3. Results and Discussion
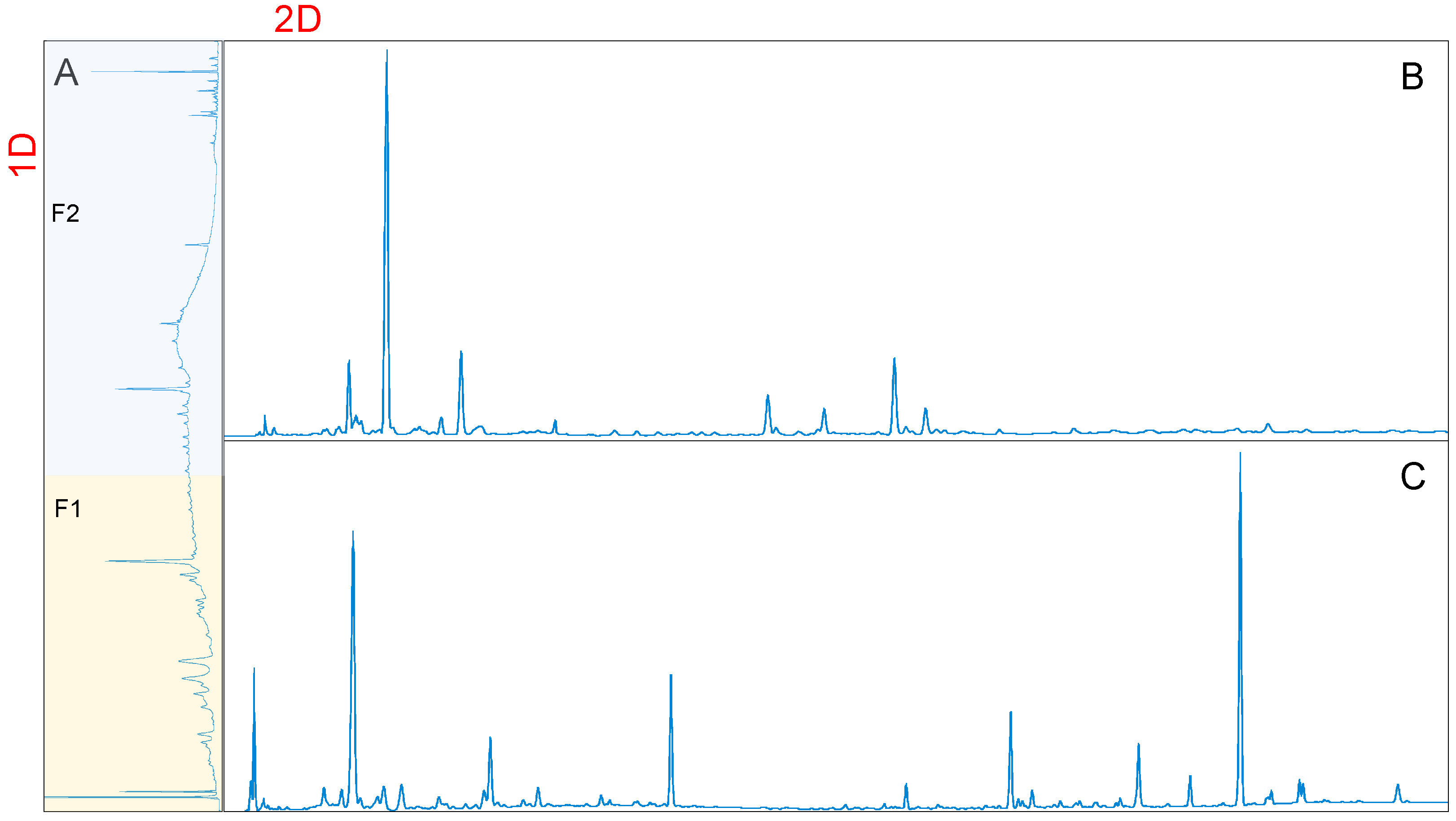
3.1. Optimization of the Targeted Offline 2D Chromatographic Conditions
3.2. Identification of the Major Compounds by Targeted 2D HPLC Method
3.3. Compound Identification by 2D UHPLC-MS
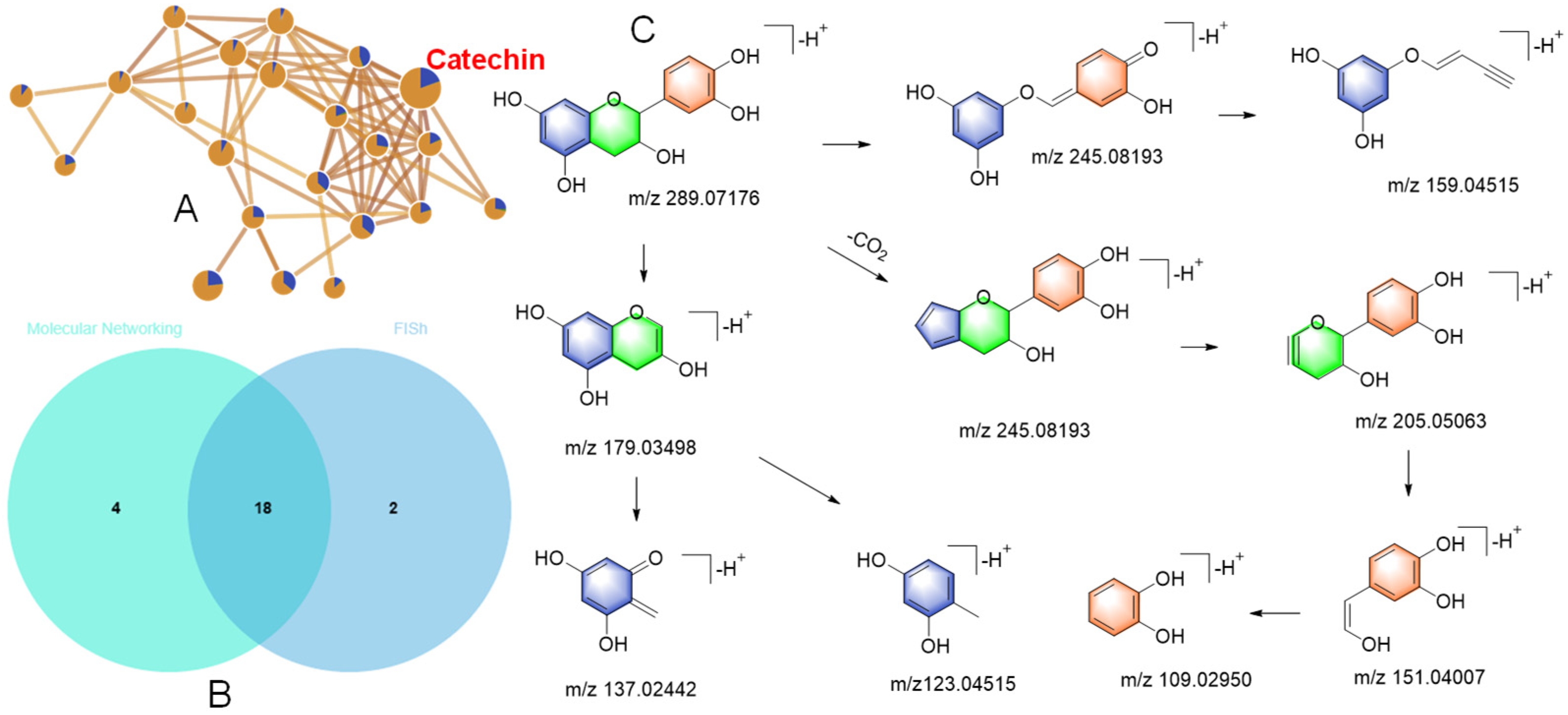
3.3.1. Identification of Catechin Derivatives and Phenols in F1
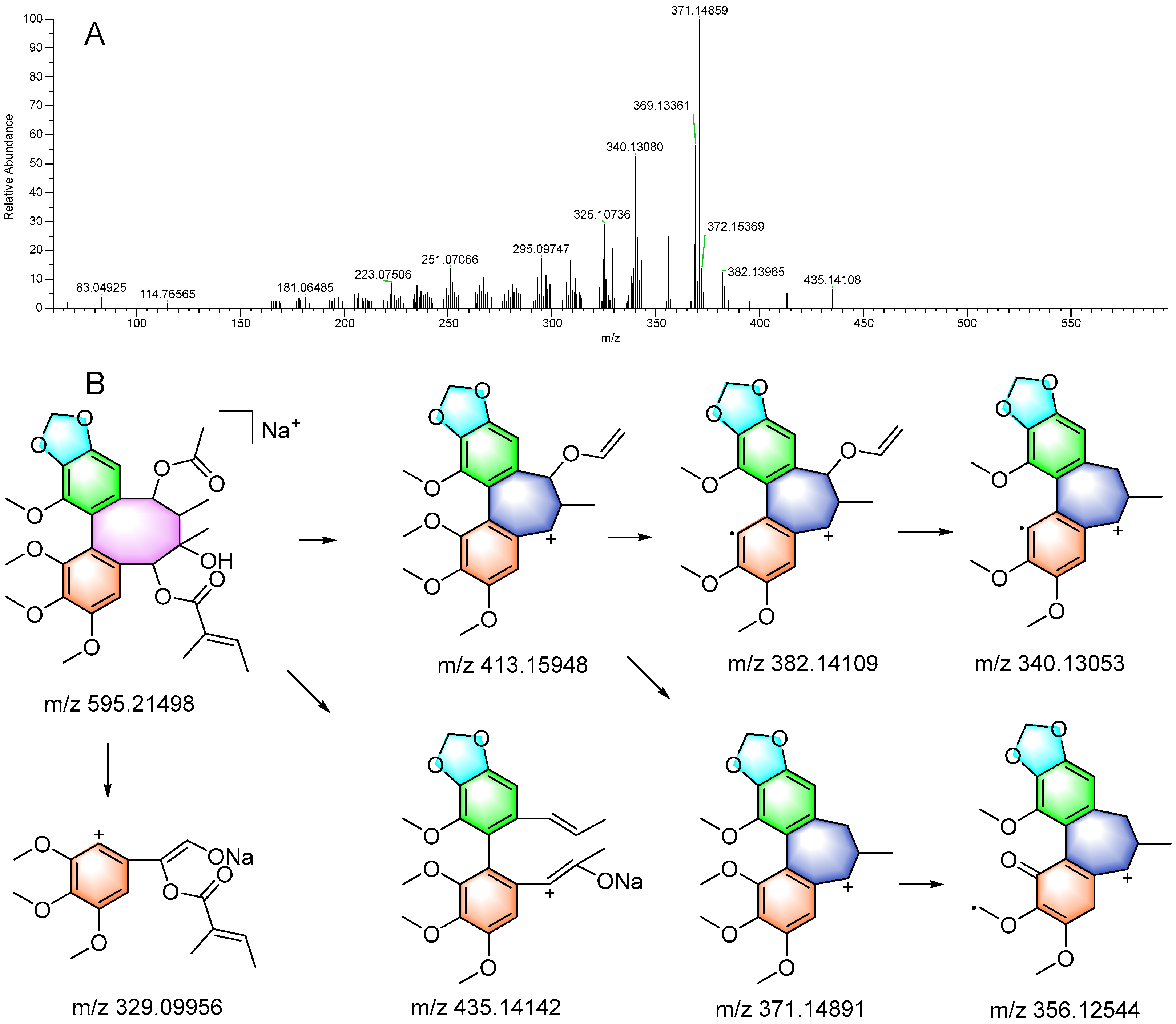
3.3.2. Identification of Lignan and Triterpenoid Derivatives in F2
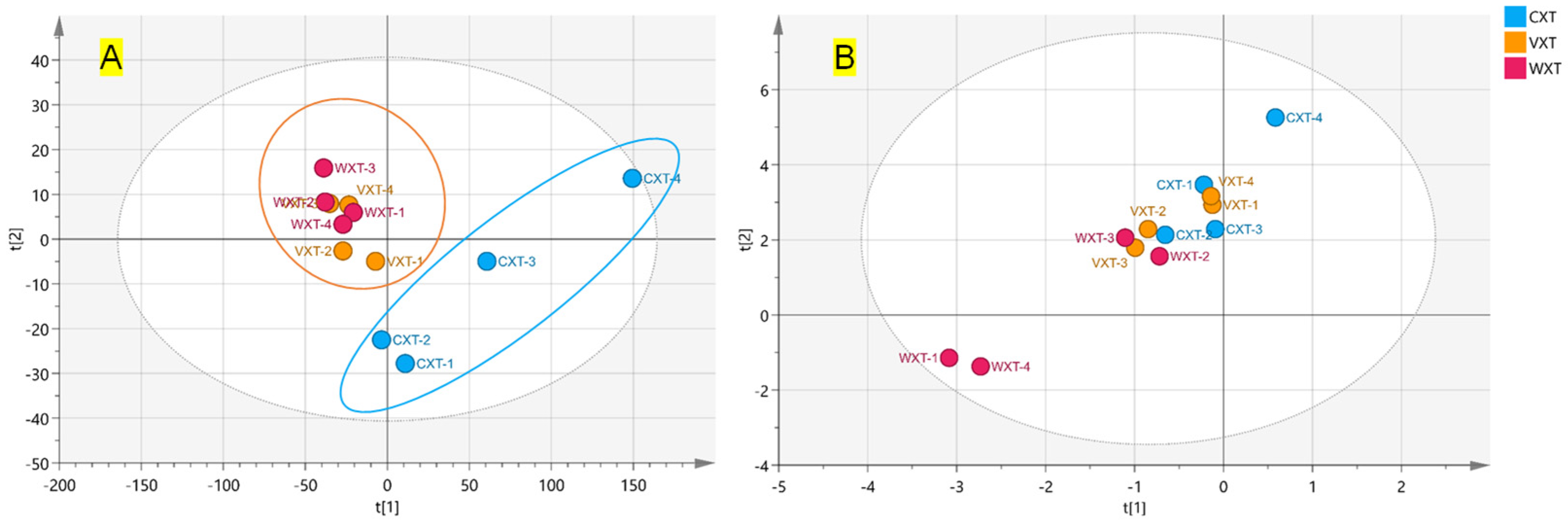
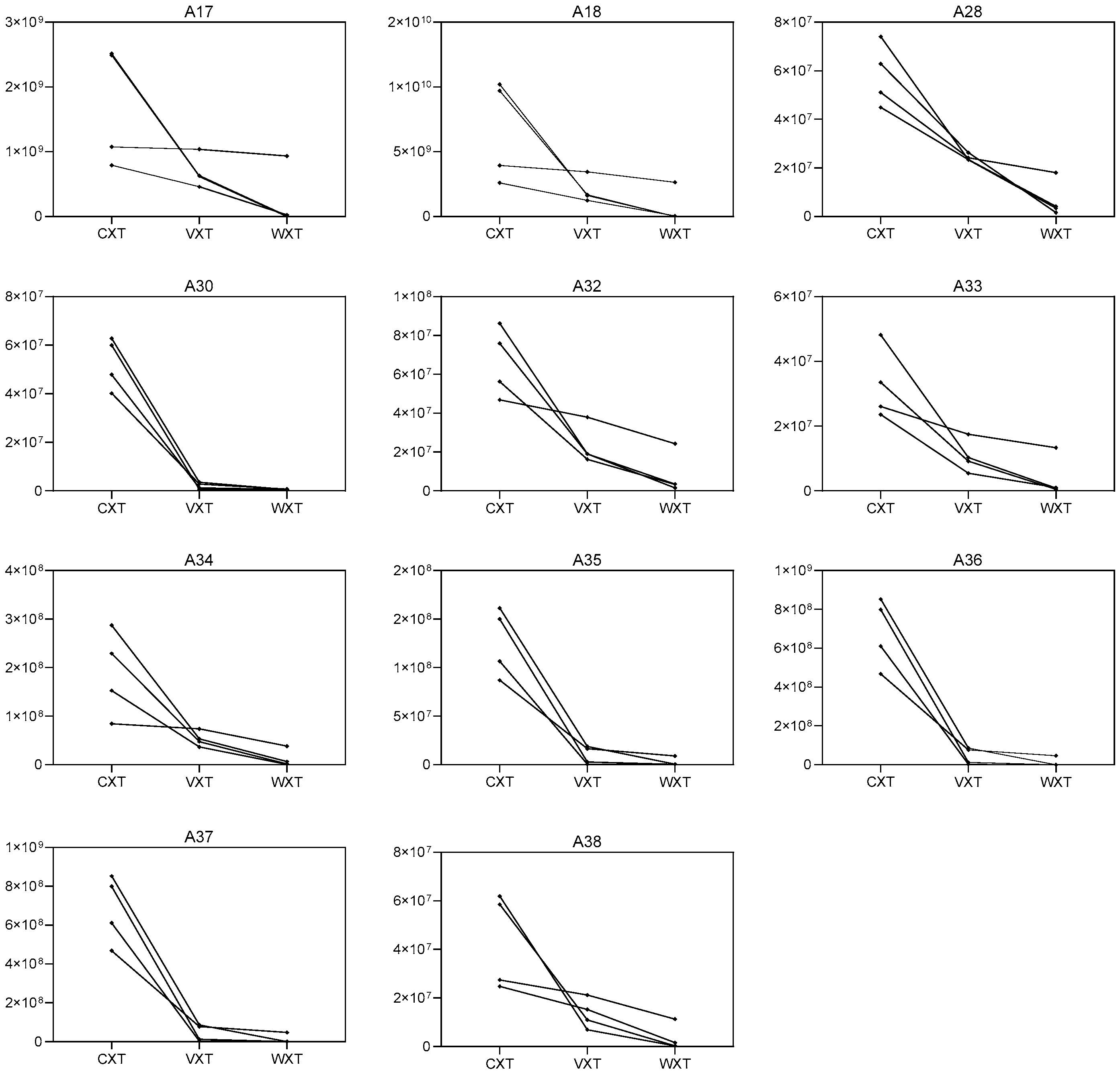
3.4. Effect of Processing on the Constituents of Xuetong
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hou, A.; Lv, J.; Zhang, S.; Zhang, J.; Yang, L.; Jiang, H.; Kuang, H. Salt processing: A unique and classic technology for Chinese medicine processing. Front. Pharmacol. 2023, 14, 116047. [Google Scholar] [CrossRef]
- Li, R.; Zhang, Q.; Liu, J.; He, L.; Huang, Q.; Peng, W.; Wu, C. Processing methods and mechanisms for alkaloid-rich Chinese herbal medicines: A review. J. Integr. Med. 2021, 19, 89–103. [Google Scholar] [CrossRef]
- Chen, L.; Verpoorte, R.; Yen, H.; Peng, W.; Cheng, Y.; Chao, J.; Pao, L. Effects of processing adjuvants on traditional Chinese herbs. J. Food Drug Anal. 2018, 26, S96–S114. [Google Scholar] [CrossRef]
- Chen, Z.; Ye, S.; Zhu, R. The extraordinary transformation of traditional Chinese medicine: Processing with liquid excipients. Pharm. Biol. 2020, 58, 561–573. [Google Scholar] [CrossRef]
- Zhan, Z.; Yang, L.; Zhang, Q.; Jialuo, C.; Deng, G.; Ouyang, L.; Wu, P. Present situation and emerging problems of processing Chinese materia medica. China J. Tradit. Chin. Med. Pharm. 2018, 33, 3233–3238. [Google Scholar]
- Wang, M.; Jiang, S.; Yuan, H.; Zafar, S.; Hussain, N.; Jian, Y.; Li, B.; Gong, L.; Peng, C.; Liu, C.; et al. A review of the phytochemistry and pharmacology of Kadsura heteroclita, an important plant in Tujia ethnomedicine. J. Ethnopharmacol. 2021, 268, 113567. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lin, Y.; Zeng, R.; Li, X.; Zhang, T.; Tasneem, S.; Chen, C.; Qiu, Y.; Li, B.; Liao, J.; et al. Analgesic and anti-inflammatory effects and molecular mechanisms of Kadsura heteroclita stems, an anti-arthritic Chinese Tujia ethnomedicinal herb. J. Ethnopharmacol. 2019, 238, 111902. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zeng, R.; Lin, Y.; Li, X.; Tasneem, S.; Yang, Z.; Qiu, Y.; Li, B.; Wang, Y.; Cai, X.; et al. Kadsura heteroclita stem suppresses the onset and progression of adjuvant-induced arthritis in rats. Phytomedicine 2019, 58, 152876. [Google Scholar] [CrossRef]
- Yu, H.; Fan, J.; Shehla, N.; Qiu, Y.; Lin, Y.; Wang, Z.; Cao, L.; Li, B.; Daniyal, M.; Qin, Y.; et al. Biomimetic hybrid membrane-coated xuetongsu assisted with laser irradiation for efficient rheumatoid arthritis therapy. ACS Nano 2022, 16, 502–521. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yang, Y.; Hussain, N.; Jian, Y.; Li, B.; Qiu, Y.; Yu, H.; Wang, H.; Wang, W. Dibenzocyclooctadiene lignans from the family Schisandraceae: A review of phytochemistry, structure-activity relationship, and hepatoprotective effects. Pharmacol. Res. 2023, 195, 106872. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, Y.; Shen, A.; Fu, D.; Liu, D.; Liu, Y.; Liang, X. Offline two-dimensional normal-phase × reversed-phase liquid chromatography coupled with high-resolution mass spectrometry for comprehensive analysis of chemical constituents in Euphorbia kansui. J. Chromatogr. 2023, 1693, 463897. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, C.; Li, W.; Wang, H.; Hu, Y.; Yang, W.; Jia, L.; Wang, X.; Gao, X.; Guo, D. Offline two-dimensional liquid chromatography coupled with ion mobility-quadrupole time-of-flight mass spectrometry enabling four-dimensional separation and characterization of the multicomponents from white ginseng and red ginseng. J. Pharm. Anal. 2020, 10, 597–609. [Google Scholar] [CrossRef]
- Messaili, S.; Colas, C.; Fougère, L.; Destandau, E. Combination of molecular network and centrifugal partition chromatography fractionation for targeting and identifying Artemisia annua L. antioxidant compounds. J. Chromatogr. 2020, 1615, 460785. [Google Scholar] [CrossRef]
- Qin, G.-F.; Zhang, X.; Zhu, F.; Huo, Z.-Q.; Yao, Q.-Q.; Feng, Q.; Liu, Z.; Zhang, G.-M.; Yao, J.-C.; Liang, H.-B. MS/MS-based molecular networking: An efficient approach for natural products dereplication. Molecules 2023, 28, 157. [Google Scholar] [CrossRef]
- Yu, J.S.; Seo, H.; Kim, G.B.; Hong, J.; Yoo, H.H. MS-based molecular networking of designer drugs as an approach for the detection of unknown derivatives for forensic and doping applications: A case of NBOMe derivatives. Anal. Chem. 2019, 91, 5483–5488. [Google Scholar] [CrossRef] [PubMed]
- Parailloux, M.; Godin, S.; Lobinski, R. Nontargeted screening for flavonoids in Salicornia plant by reversed-phase liquid chromatography-electrospray orbitrap data-dependent MS2/MS3. Molecules 2023, 28, 3022. [Google Scholar] [CrossRef]
- Yu, Y.; Yao, C.; Guo, D. Insight into chemical basis of traditional Chinese medicine based on the state-of-the-art techniques of liquid chromatography−mass spectrometry. Acta Pharm. Sin. B 2021, 11, 1469–1492. [Google Scholar] [CrossRef]
- Yu, Y.; Yao, C.; Wei, W.; Li, H.; Huang, Y.; Yao, S.; Qu, H.; Chen, Q.; Mei, Q.; Wu, W.; et al. Integration of offline two-dimensional chromatography and mass defect filtering-based precursor ion list data acquisition for targeted characterization of diterpenoid alkaloids in the lateral roots of Aconitum carmichaelii. J. Chromatogr. 2022, 1684, 463554. [Google Scholar] [CrossRef]
- Han, C.; Chen, J.; Chen, B.; Sen-Chun Lee, F.; Wang, X. Fingerprint chromatogram analysis of Pseudostellaria heterophylla (Miq.) Pax root by high performance liquid chromatography. J. Sep. Sci. 2006, 29, 2197–2202. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, Z.; Yang, M.; Liu, R.; Wang, W.; Liu, P.; Guo, D. Structural determination of seven new triterpenoids from Kadsura heteroclita by NMR techniques. Magn. Reson. Chem. 2007, 45, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Baskiyar, S.; Ren, C.; Heck, K.L.; Hall, A.M.; Gulfam, M.; Packer, S.; Seals, C.D.; Calderón, A.I. Bioactive natural products identification using automation of molecular networking software. J. Chem. Inf. Model. 2022, 62, 6378–6385. [Google Scholar] [CrossRef] [PubMed]
- Caldas, L.A.; Zied, D.C.; Sartorelli, P. Dereplication of extracts from nutraceutical mushrooms Pleurotus using molecular network approach. Food Chem. 2022, 370, 131019. [Google Scholar] [CrossRef] [PubMed]


| Peaks | Peak Area | Retention Time | ||||
|---|---|---|---|---|---|---|
| Precision | Stability | Repeatability | Precision | Stability | Repeatability | |
| X1 | 2.479 | 2.359 | 2.531 | 1.428 | 1.683 | 1.534 |
| X2 | 0.684 | 0.698 | 3.225 | 1.796 | 2.484 | 2.185 |
| X3 | 5.372 | 2.391 | 2.789 | 1.970 | 1.767 | 2.002 |
| X4 | 3.229 | 4.313 | 4.843 | 1.160 | 1.263 | 1.694 |
| X5 | 1.225 | 1.097 | 5.006 | 1.006 | 1.188 | 1.620 |
| X6 | 1.138 | 0.598 | 5.821 | 0.826 | 1.060 | 1.386 |
| X7 | 2.573 | 2.941 | 5.603 | 0.683 | 1.145 | 1.395 |
| Y3 | 1.134 | 1.533 | 3.798 | 0.948 | 0.071 | 0.492 |
| Y7 | 2.035 | 1.313 | 4.155 | 0.326 | 0.034 | 0.321 |
| Y8 | 0.699 | 1.266 | 2.892 | 0.080 | 0.019 | 0.328 |
| Y9 | 4.531 | 1.265 | 4.543 | 0.037 | 0.010 | 0.072 |
| Y10 | 5.012 | 1.172 | 4.498 | 0.016 | 0.009 | 0.093 |
| Y13 | 1.159 | 1.025 | 6.592 | 0.015 | 0.007 | 0.050 |
| Y14 | 1.472 | 1.393 | 4.400 | 0.017 | 0.007 | 0.015 |
| Y15 | 1.526 | 1.306 | 2.493 | 0.019 | 0.005 | 0.050 |
| Y16 | 2.312 | 1.221 | 5.273 | 0.020 | 0.004 | 0.031 |
| Y18 | 6.521 | 3.594 | 4.681 | 0.034 | 0.005 | 0.089 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, H.; Su, W.; Liang, L.; Xie, Q.; Lyu, M.; Yu, H.; Li, B.; Wang, W. Targeted Offline Two-Dimensional HPLC and UHPLC-Orbitrap-MS Combined with Molecular Networking Reveal the Effect of Processing on Chemical Constituents of Xuetong (the Stem of Kadsura heteroclita). Separations 2024, 11, 87. https://doi.org/10.3390/separations11030087
Yuan H, Su W, Liang L, Xie Q, Lyu M, Yu H, Li B, Wang W. Targeted Offline Two-Dimensional HPLC and UHPLC-Orbitrap-MS Combined with Molecular Networking Reveal the Effect of Processing on Chemical Constituents of Xuetong (the Stem of Kadsura heteroclita). Separations. 2024; 11(3):87. https://doi.org/10.3390/separations11030087
Chicago/Turabian StyleYuan, Hanwen, Wei Su, Ling Liang, Qingling Xie, Mengying Lyu, Huanghe Yu, Bin Li, and Wei Wang. 2024. "Targeted Offline Two-Dimensional HPLC and UHPLC-Orbitrap-MS Combined with Molecular Networking Reveal the Effect of Processing on Chemical Constituents of Xuetong (the Stem of Kadsura heteroclita)" Separations 11, no. 3: 87. https://doi.org/10.3390/separations11030087
APA StyleYuan, H., Su, W., Liang, L., Xie, Q., Lyu, M., Yu, H., Li, B., & Wang, W. (2024). Targeted Offline Two-Dimensional HPLC and UHPLC-Orbitrap-MS Combined with Molecular Networking Reveal the Effect of Processing on Chemical Constituents of Xuetong (the Stem of Kadsura heteroclita). Separations, 11(3), 87. https://doi.org/10.3390/separations11030087