The Development of a High-Throughput Homonuclear Decoupling HSQC NMR Platform for the Determination of 10 Sex Hormones in Animal-Source Food and Medicines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Samples and Standards Preparation
2.3. NMR Parameters and Experiments
2.4. Method Validation
3. Results and Discussion
3.1. Method Development
3.2. Method Validation Results
3.3. Quantification of 10 Sex Hormones in Animal-Source Products
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lösel, R.; Wehling, M. Nongenomic actions of steroid hormones. Nat. Rev. Mol. Cell Biol. 2003, 4, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Wehling, M. Specific, nongenomic actions of steroid hormones. Annu. Rev. Physiol. 1997, 59, 365–393. [Google Scholar] [CrossRef]
- Chiang, Y.R.; Wei, S.T.S.; Wang, P.H.; Wu, P.H.; Yu, C.P. Microbial degradation of steroid sex hormones: Implications for environmental and ecological studies. Microb. Biotechnol. 2020, 13, 926–949. [Google Scholar] [CrossRef] [PubMed]
- Ojoghoro, J.O.; Scrimshaw, M.D.; Sumpter, J.P. Steroid hormones in the aquatic environment. Sci. Total Environ. 2021, 792, 148306. [Google Scholar] [CrossRef]
- Luzio, A.; Monteiro, S.M.; Rocha, E.; Fontaínhas-Fernandes, A.A.; Coimbra, A.M. Development and recovery of histopathological alterations in the gonads of zebrafish (Danio rerio) after single and combined exposure to endocrine disruptors (17α-ethinylestradiol and fadrozole). Aquat. Toxicol. 2016, 175, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.D.; Han, J.L. Determination of steroid hormone residues in farmed fish using high-resolution orbital ion trap mass spectrometry. Anal. Methods 2022, 14, 4146–4152. [Google Scholar] [CrossRef] [PubMed]
- Lemos, L.S.; Olsen, A.; Smith, A.; Chandler, T.E.; Larson, S.; Hunt, K.; Torres, L.G. Assessment of fecal steroid and thyroid hormone metabolites in eastern North Pacific gray whales. Conserv. Physiol. 2020, 8, coaa110. [Google Scholar] [CrossRef]
- Evangelista, S.; Vazakidou, P.; Koekkoek, J.; Heinzelmann, M.T.; Lichtensteiger, W.; Schlumpf, M.; Tresguerres, J.A.F.; Linillos-Pradillo, B.; van Duursen, M.B.M.; Lamoree, M.H.; et al. High throughput LC-MS/MS method for steroid hormone analysis in rat liver and plasma-unraveling methodological challenges. Talanta 2024, 266, 124981. [Google Scholar] [CrossRef]
- Li, Y.; Yan, Z.; Li, X.; Yin, X.; Li, K. UPLC–TOF–MS method for simultaneous quantification of steroid hormones in tissue homogenates of zebrafish with solid-phase extraction. Molecules 2021, 26, 6213. [Google Scholar] [CrossRef]
- Ross, A.R.S.; Liao, X.; Brown, T.M. Simultaneous determination of steroid hormones and pharmaceuticals in killer whale (Orcinus orca) faecal samples by liquid chromatography tandem mass spectrometry. Conserv. Physiol. 2023, 11, coad081. [Google Scholar] [CrossRef]
- Bechshoft, T.; Wright, A.J.; Styrishave, B.; Houser, D. Measuring and validating concentrations of steroid hormones in the skin of bottlenose dolphins (Tursiops truncatus). Conserv. Physiol. 2020, 8, coaa032. [Google Scholar] [CrossRef] [PubMed]
- Praveen, A.; Prasad, D.; Mishra, S.; Nagarajan, S.; Chaudhari, S.R. Facile NMR approach for profiling curcuminoids present in turmeric. Food Chem. 2021, 341, 128646. [Google Scholar] [CrossRef] [PubMed]
- Li, F.F.; Guo, C.C.; Zhang, Y.L.; Wang, N.; Shi, Y.; Wang, S.Q. A screening method based on 1D CSSF-TOCSY for the identification and quantification of 11 illegal adulterants in herbal medicines. Microchem. J. 2020, 153, 104495. [Google Scholar] [CrossRef]
- Kakeshpour, T.; Bax, A. Simultaneous Quantification of H2O2 and Organic Hydroperoxides by 1H NMR Spectroscopy. Anal. Chem. 2022, 94, 5729–5733. [Google Scholar] [CrossRef]
- Haddad, L.; Renou, S.; Remaud, G.S.; Rizk, T.; Bejjani, J.; Akoka, S. A precise and rapid isotopomic analysis of small quantities of cholesterol at natural abundance by optimized 1H-13C 2D NMR. Anal. Bioanal. Chem. 2021, 413, 1521–1532. [Google Scholar] [CrossRef]
- Fels, L.; Ruf, F.; Bunzel, M. Quantification of Isomaltulose in Food Products by Using Heteronuclear Single Quantum Coherence NMR-Experiments. Front. Nutr. 2022, 9, 928102. [Google Scholar] [CrossRef]
- Wu, Y.S.; Li, B.X.; Long, Y.Y. Rapid quantitative 1H-13C two-dimensional NMR with high precision. RSC Adv. 2022, 12, 5349–5356. [Google Scholar] [CrossRef]
- Boerkamp, V.J.P.; Merkx, D.W.H.; Wang, J.; Vincken, J.P.; Hennebelle, M.; van Duynhoven, J.P.M. Quantitative assessment of epoxide formation in oil and mayonnaise by 1H-13C HSQC NMR spectroscopy. Food Chem. 2022, 390, 133145. [Google Scholar] [CrossRef]
- Bourmaud, C.L.; Bertella, S.; Rico, A.B.; Karlen, S.D.; Ralph, J.; Luterbacher, J. Quantification of native lignin structural features with gel-phase 2D-HSQC0 reveals lignin structural changes during extraction. Angew. Chem. Int. Ed. 2024, 136, 202404442. [Google Scholar] [CrossRef]
- Xiao, H.H.; Lv, J.; Mok, D.; Yao, X.S.; Wong, M.S.; Cooper, R. NMR Applications for Botanical Mixtures: The Use of HSQC Data to Determine Lignan Content in Sambucus williamsii. J. Nat. Prod. 2019, 82, 1733–1740. [Google Scholar] [CrossRef]
- Çiçek, S.S.; Esposito, T.; Girreser, U. Prediction of the sweetening effect of Siraitia grosvenorii (luo han guo) fruits by two-dimensional quantitative NMR. Food Chem. 2021, 335, 127622. [Google Scholar] [CrossRef] [PubMed]
- Girreser, U.; Ugolini, T.; Çiçek, S.S. Quality control of Aloe vera (Aloe barbadensis) and Aloe ferox using band-selective quantitative heteronuclear single quantum correlation spectroscopy (bs-qHSQC). Talanta 2019, 205, 120109. [Google Scholar] [CrossRef] [PubMed]
- Kontogianni, V.G.; Exarchou, V.; Troganis, A.; Gerothanassis, I.P. Rapid and novel discrimination and quantification of oleanolic and ursolic acids in complex plant extracts using two-dimensional nuclear magnetic resonance spectroscopy-comparison with HPLC methods. Anal. Chim. Acta 2009, 635, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, Y.; Xu, Y.; Ma, S.; Yang, H.; Liu, Y.; Wu, X. Quality Evaluation of Tripterygium Glycoside Tablets Based on Quantitative Band-Selective 2D 1H–13C HSQC and 1H NMR Fingerprinting. ACS Omega 2024, 9, 27321–27328. [Google Scholar] [CrossRef]
- Sutcharitchan, C.; Miao, S.; Li, W.; Liu, J.; Zhou, H.; Ma, Y.; Ji, S.; Cui, Y. High performance liquid chromatography-tandem mass spectrometry method for residue determination of 39 plant growth regulators in root and rhizome Chinese herbs. Food Chem. 2020, 322, 126766. [Google Scholar] [CrossRef]
- Martín-Gómez, B.; Valverde, S.; Bernal, J.; Ares, A.M. Development and validation of an analytical methodology based on solvent microextraction and UHPLC-MS/MS for determining bisphenols in honeys from different botanical origins. Food Chem. 2024, 450, 139358. [Google Scholar] [CrossRef]
- Zhao, Q.; Liu, Y.; Ma, H.; Qiao, Y.; Chao, J.; Hou, X.; Wang, Y.; Wang, Y. Combination of pure shift NMR and chemical shift selective filters for analysis of Fischer-Tropsch waste-water. Anal. Chim. Acta 2020, 1110, 131–140. [Google Scholar] [CrossRef]
- Weitzel, A.; Samol, C.; Oefner, P.J.; Gronwald, W. Robust metabolite quantification from j-compensated 2d 1H–13C HSQC experiments. Metabolites 2020, 10, 449. [Google Scholar] [CrossRef]
- Yan, Y.; Li, M.; Wei, Y.; Jia, F.; Zheng, Y.; Tao, G.; Xiong, F. Oyster-derived dipeptides RI, IR, and VR promote testosterone synthesis by reducing oxidative stress in TM3 cells. Food Sci. Nutr. 2023, 11, 6470–6482. [Google Scholar] [CrossRef]
- Banihani, S.A. Ameliorative effects of propolis upon reproductive toxicity in males. Clin. Exp. Reprod. Med. 2023, 50, 12–18. [Google Scholar] [CrossRef]
- Xu, H.; Pu, J.; Teng, Y.; Zhu, Q.; Guo, L.; Zhao, J.; Ding, H.; Fang, Y.; Ma, X.; Liu, H.; et al. Melatonin Inhibits Testosterone Synthesis in Rooster Leydig Cells by Targeting CXCL14 through miR-7481-3p. Int. J. Mol. Sci. 2023, 24, 16552. [Google Scholar] [CrossRef]

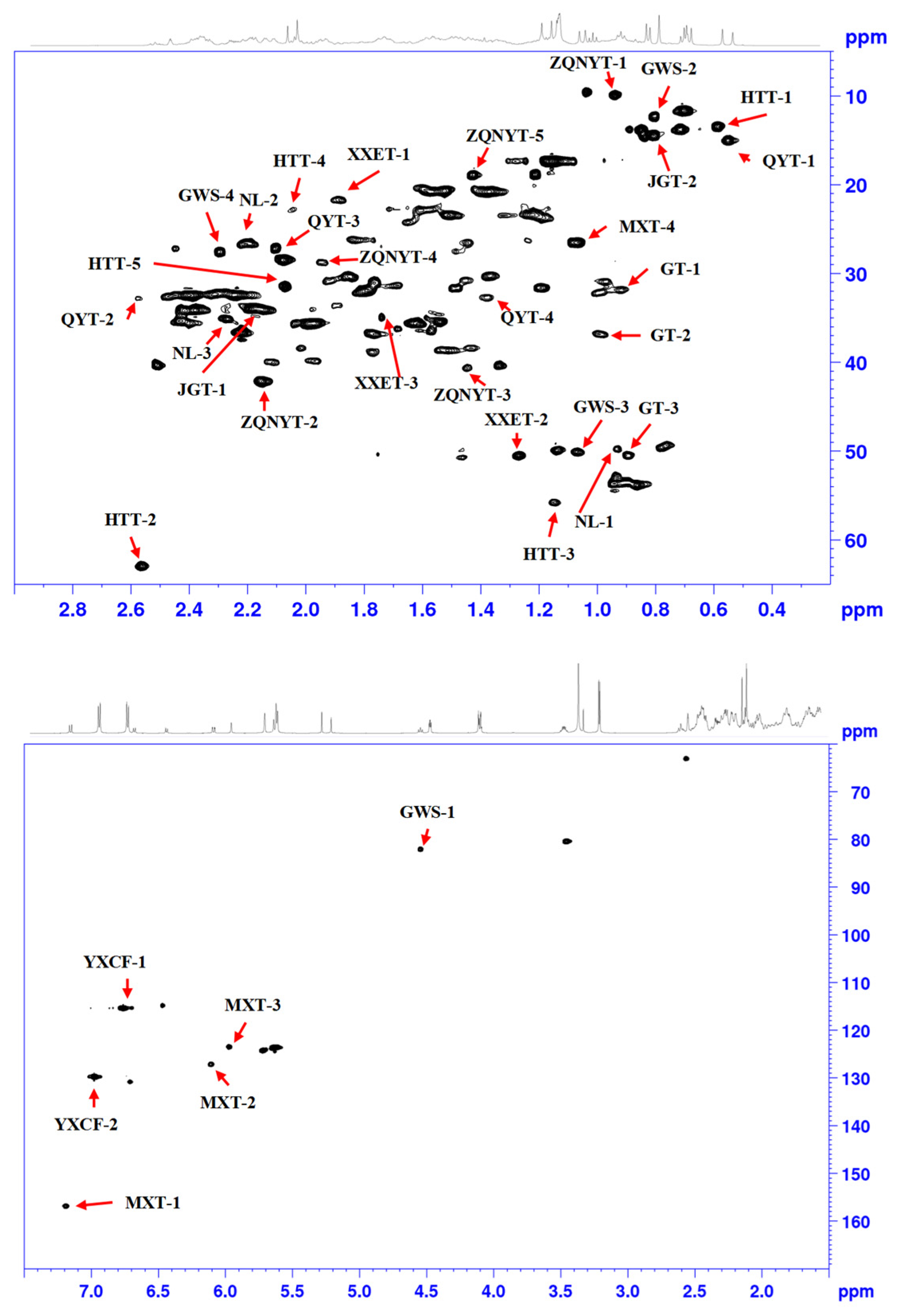
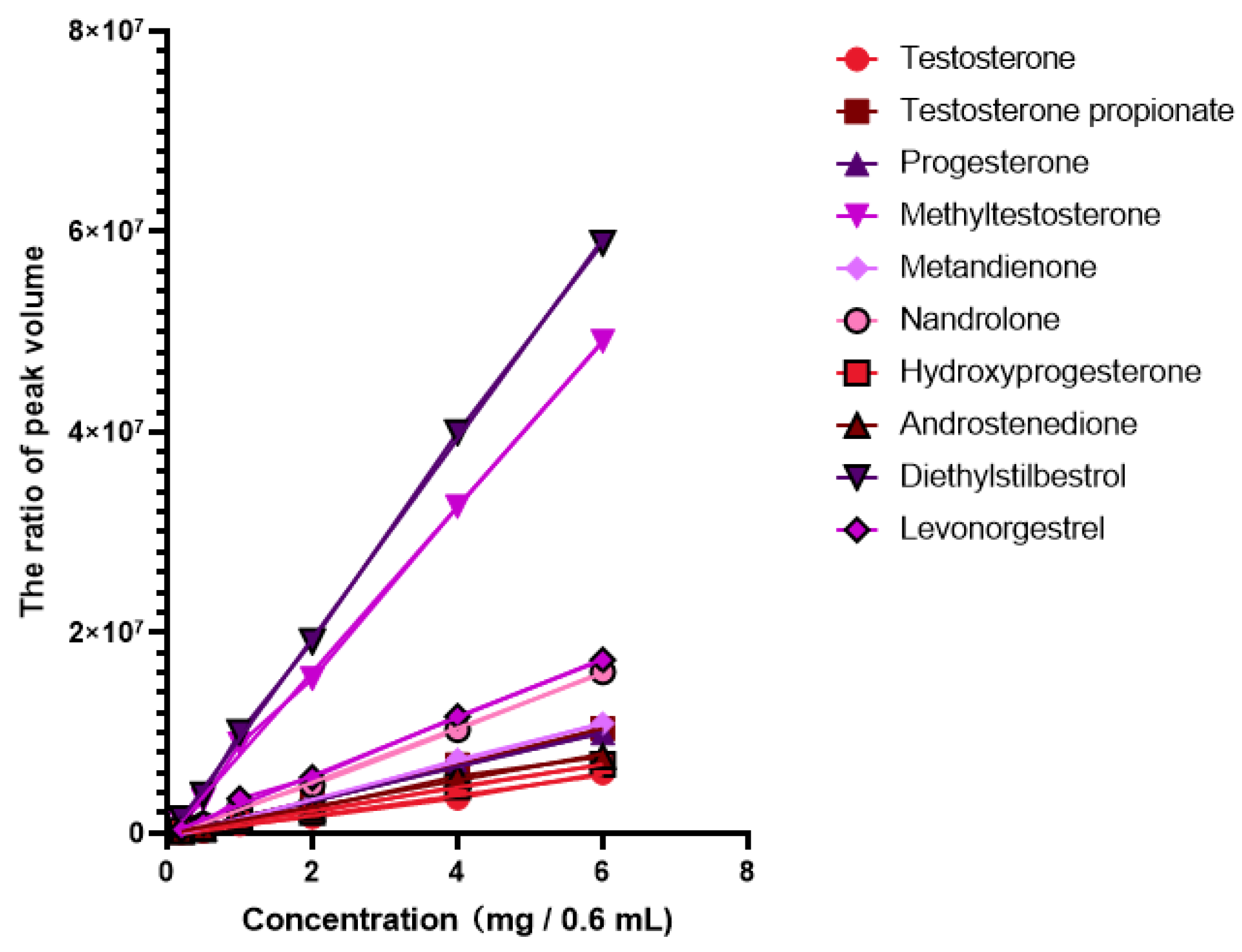
| Analyte | Label | 13C | 1H | Assignment | Linear Range mg/0.6 mL | Equation | R2 |
|---|---|---|---|---|---|---|---|
| Testosterone | Testosterone-1 | 31.3 | 0.91 | C12/H12 in Testosterone | 0.2–6 | Y = 901581 × X − 240413 | 0.9991 |
| Testosterone-2 * | 37.0 | 0.97 | C8/H8 in Testosterone | 0.2–6 | Y = 1012040 × X − 280875 | 0.9932 | |
| Testosterone-3 | 50.4 | 0.89 | C3/H3 in Testosterone | 0.2–6 | Y = 1126412 × X − 96553 | 0.9998 | |
| Testosterone propionate | Testosterone propionate-1 * | 82.2 | 4.54 | C11/H11 in Testosterone propionate | 0.2–6 | Y = 1781890 × X − 285200 | 0.9998 |
| Testosterone propionate-2 | 12.0 | 0.81 | C25/H25 in Testosterone propionate | 0.2–6 | Y = 1425738 × X − 37469 | 0.9955 | |
| Testosterone propionate-3 | 50.0 | 1.07 | C4/H4 in Testosterone propionate | 0.2–6.0 | Y = 1349384 × X − 281588 | 0.9944 | |
| Testosterone propionate-4 | 27.1 | 2.29 | C9/H9 in Testosterone propionate | 0.2–6.0 | Y = 2018563 × X − 151609 | 0.9993 | |
| Progesterone | Progesterone-1 | 13.1 | 0.59 | C15/H15 in Progesterone | 0.2–6.0 | Y = 2316192 × X − 144738 | 0.9979 |
| Progesterone-2 * | 63.1 | 2.56 | C8/H8 in Progesterone | 0.2–6.0 | Y = 1709294 × X − 208664 | 0.9997 | |
| Progesterone-3 | 55.8 | 1.14 | C6/H6 in Progesterone | 0.2–6.0 | Y = 851794 × X − 83156 | 0.9989 | |
| Progesterone-4 | 22.0 | 2.06 | C13/H13 in Progesterone | 0.2–6.0 | Y = 307373 × X + 64381 | 0.984 | |
| Progesterone-5 | 31.7 | 2.07 | C5/H5 in Progesterone | 0.2–6.0 | Y = 2002516 × X − 243307 | 0.9936 | |
| Methyltestosterone | Methyltestosterone-1 * | 34.2 | 2.16 | C16/H16 in Methyltestosterone | 0.2–6.0 | Y = 8248066 × X − 451751 | 0.999 |
| Methyltestosterone-2 | 14.4 | 0.80 | C14/H14 in Methyltestosterone | 0.2–6.0 | Y = 2629071 × X − 243818 | 0.9994 | |
| Metandienone | Metandienone-1 * | 156.3 | 7.19 | C19/H19 in Metandienone | 0.2–6.0 | Y = 1902488 × X − 447472 | 0.9984 |
| Metandienone-2 | 126.8 | 6.11 | C21/H21 in Metandienone | 0.2–6.0 | Y = 2381153 × X − 364067 | 0.999 | |
| Metandienone-3 | 123.2 | 5.97 | C20/H20 in Metandienone | 0.2–6.0 | Y = 2470837 × X − 65284 | 0.9986 | |
| Metandienone-4 | 26.6 | 1.07 | C17/H17 in Metandienone | 0.2–6.0 | Y = 3340572 × X − 159253 | 0.9989 | |
| Nandrolone | Nandrolone-1 | 49.8 | 0.93 | C4/H4 in Nandrolone | 0.2–6.0 | Y = 693588 × X + 136010 | 0.9901 |
| Nandrolone-2 * | 26.4 | 2.20 | C9/H9 in Nandrolone | 0.2–6.0 | Y = 2721231 × X − 391520 | 0.9989 | |
| Nandrolone-3 | 35.0 | 2.27 | C10/H10 in Nandrolone | 0.2–6.0 | Y = 1420652 × X − 24608 | 0.999 | |
| Hydroxyprogesterone | Hydroxyprogesterone-1 | 15.1 | 0.55 | C19/H19 in Hydroxyprogesterone | 0.2–6.0 | Y = 2378870 × X − 195147 | 0.9991 |
| Hydroxyprogesterone-2 | 32.6 | 2.57 | C21/H21 in Hydroxyprogesterone | 0.2–6.0 | Y = 404375 × X + 16640 | 0.9331 | |
| Hydroxyprogesterone-3 | 26.8 | 2.11 | C12/H12 in Hydroxyprogesterone | 0.2–6.0 | Y = 2003262 × X − 39184 | 0.9987 | |
| Hydroxyprogesterone-4 * | 32.6 | 1.38 | C14/H14 in Hydroxyprogesterone | 0.2–6.0 | Y = 1156806 × X − 75744 | 0.9981 | |
| Androstenedione | Androstenedione-1 | 21.7 | 1.88 | C8/H8 in Androstenedione | 0.2–6.0 | Y = 1142997 × X − 24802 | 0.9993 |
| Androstenedione-2 * | 50.4 | 1.27 | C5/H5 in Androstenedione | 0.2–6.0 | Y = 1319332 × X − 9290 | 0.9926 | |
| Androstenedione-3 | 34.8 | 1.74 | C9/H9 in Androstenedione | 0.2–6.0 | Y = 733839 × X − 133902 | 0.9938 | |
| Diethylstilbestrol | Diethylstilbestrol-1 | 115.3 | 6.78 | C15/H15 in Diethylstilbestrol | 0.2–6.0 | Y = 8407064 × X + 1321 | 0.999 |
| Diethylstilbestrol-2 * | 129.4 | 6.98 | C11/H11 in Diethylstilbestrol | 0.2–6.0 | Y = 9963598 × X − 540050 | 0.9994 | |
| Levonorgestrel | Levonorgestrel-1 | 9.7 | 0.93 | C18/H18 in Levonorgestrel | 0.2–6.0 | Y = 1605386 × X + 7647 | 0.9988 |
| Levonorgestrel-2 * | 42.0 | 2.15 | C20/H20 in Levonorgestrel | 0.2–6.0 | Y = 2909640 × X − 87386 | 0.9976 | |
| Levonorgestrel-3 | 40.6 | 1.44 | C5/H5 in Levonorgestrel | 0.2–6.0 | Y = 756719 × X − 151625 | 0.9964 | |
| Levonorgestrel-4 | 28.9 | 1.94 | C17/H17 in Levonorgestrel | 0.2–6.0 | Y = 791756 × X − 210286 | 0.9971 | |
| Levonorgestrel-5 | 18.5 | 1.41 | C14/H14 in Levonorgestrel | 0.2–6.0 | Y = 1894622 × X − 257453 | 0.9984 |
| Compound | Label | Precision RSD (%) (2 mg/0.6 mL, n = 9) | Recovery (2 mg/0.6 mL, n = 5) | |
|---|---|---|---|---|
| Average Recovery (%) | RSD (%) | |||
| Testosterone | Testosterone-2 | 5.78 | 104.2 | 1.09 |
| Testosterone propionate | Testosterone propionate-1 | 3.61 | 103.6 | 1.71 |
| Progesterone | Progesterone-2 | 4.95 | 104.2 | 1.79 |
| Methyltestosterone | Methyltestosterone-1 | 2.53 | 106.6 | 3.15 |
| Metandienone | Metandienone-1 | 5.66 | 101.6 | 1.75 |
| Nandrolone | Nandrolone-2 | 2.92 | 103.4 | 2.41 |
| Hydroxyprogesterone | Hydroxyprogesterone-4 | 5.22 | 101.7 | 1.64 |
| Androstenedione | Androstenedione-2 | 2.22 | 104.0 | 3.72 |
| Diethylstilbestrol | Diethylstilbestrol-2 | 3.32 | 104.3 | 1.04 |
| Levonorgestrel | Levonorgestrel-2 | 1.23 | 105.3 | 3.51 |
| Samples | No. | Concentration (mg/20 g) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Testosterone | Testosterone Propionate | Progesterone | Methyltestosterone | Metandienone | Nandrolone | Hydroxyprogesterone | Androstenedione | Diethylstilbestrol | Levonorgestrel | ||
| Bird’s nest | 1 | - | - | - | - | - | - | - | - | - | - |
| 2 | - | - | - | - | - | - | - | - | - | - | |
| 3 | - | - | - | - | - | - | - | - | - | - | |
| 4 | - | - | - | - | - | - | - | - | - | - | |
| 5 | - | - | - | - | - | - | - | - | - | - | |
| Colla corii asini | 1 | - | - | - | - | - | - | - | - | - | - |
| 2 | - | - | - | - | - | - | - | - | - | - | |
| 3 | - | - | - | - | - | - | - | - | - | - | |
| 4 | - | - | - | - | - | - | - | - | - | - | |
| 5 | - | - | - | - | - | - | - | - | - | - | |
| Chicken gizzard membrane | 1 | 1.2 | - | - | 0.8 | - | - | - | - | - | - |
| 2 | 0.9 | - | - | - | - | - | 0.3 | - | - | - | |
| 3 | - | - | - | 0.5 | - | - | - | - | - | - | |
| 4 | 0.3 | - | - | - | - | - | - | - | - | - | |
| 5 | 1.3 | - | - | - | - | - | - | - | - | - | |
| Propolis | 1 | - | - | - | - | - | - | - | - | - | - |
| 2 | 0.6 | - | - | - | 0.8 | - | - | - | - | - | |
| 3 | - | - | - | - | 1.3 | - | - | - | - | - | |
| 4 | - | - | - | - | - | - | - | - | - | - | |
| 5 | - | - | - | - | - | - | - | - | - | - | |
| Gecko | 1 | - | - | - | - | - | - | - | - | - | - |
| 2 | - | - | - | - | - | - | - | - | - | - | |
| 3 | - | - | - | - | - | - | - | - | - | - | |
| 4 | - | - | - | - | - | - | - | - | - | - | |
| 5 | - | - | - | - | - | - | - | - | - | - | |
| Oyster | 1 | 0.5 | - | - | 0.6 | - | - | - | - | - | - |
| 2 | 1.1 | - | - | 1.3 | - | - | - | - | - | - | |
| 3 | 0.7 | - | - | 0.2 | - | - | - | - | - | - | |
| 4 | - | - | - | - | - | - | - | - | - | - | |
| 5 | - | - | - | - | - | - | - | - | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Liu, Q.-Z.; Yang, J.-Y.; Du, Y.-J.; Liu, N.-S.; Cui, W.-L.; Yuan, M.; Zhang, Y.; Wang, J.-Q.; Wang, D.-L.; et al. The Development of a High-Throughput Homonuclear Decoupling HSQC NMR Platform for the Determination of 10 Sex Hormones in Animal-Source Food and Medicines. Separations 2024, 11, 328. https://doi.org/10.3390/separations11110328
Wang B, Liu Q-Z, Yang J-Y, Du Y-J, Liu N-S, Cui W-L, Yuan M, Zhang Y, Wang J-Q, Wang D-L, et al. The Development of a High-Throughput Homonuclear Decoupling HSQC NMR Platform for the Determination of 10 Sex Hormones in Animal-Source Food and Medicines. Separations. 2024; 11(11):328. https://doi.org/10.3390/separations11110328
Chicago/Turabian StyleWang, Bing, Qing-Zhi Liu, Jing-Ya Yang, Yu-Jie Du, Nai-Shuo Liu, Wei-Liang Cui, Man Yuan, Yong Zhang, Jing-Qi Wang, Dong-Liang Wang, and et al. 2024. "The Development of a High-Throughput Homonuclear Decoupling HSQC NMR Platform for the Determination of 10 Sex Hormones in Animal-Source Food and Medicines" Separations 11, no. 11: 328. https://doi.org/10.3390/separations11110328
APA StyleWang, B., Liu, Q.-Z., Yang, J.-Y., Du, Y.-J., Liu, N.-S., Cui, W.-L., Yuan, M., Zhang, Y., Wang, J.-Q., Wang, D.-L., & Wang, S.-Q. (2024). The Development of a High-Throughput Homonuclear Decoupling HSQC NMR Platform for the Determination of 10 Sex Hormones in Animal-Source Food and Medicines. Separations, 11(11), 328. https://doi.org/10.3390/separations11110328







