Abstract
Quorum quenching (QQ) is a novel anti-biofouling strategy for membrane bioreactors (MBRs) used in wastewater treatment. However, actual operation of QQ-MBR systems for wastewater treatment needs to be systematically studied to evaluate the comprehensive effects of QQ on wastewater treatment engineering applications. In this study, a novel QQ strain, Acinetobacter pittii HITSZ001, was encapsulated and applied to a MBR system to evaluate the effects of this organism on real wastewater treatment. To verify the effectiveness of immobilized QQ beads in the MBR system, we examined the MBR effluent quality and sludge characteristics. We also measured the extracellular polymeric substances (EPS) and soluble microbial products (SMP) in the system to determine the effects of the organism on membrane biofouling inhibition. Additionally, changes in microbial communities in the system were analyzed by high-throughput sequencing. The results indicated that Acinetobacter pittii HITSZ001 is a promising strain for biofouling reduction in MBRs treating real wastewater, and that immobilization does not affect the biofouling control potential of QQ bacteria.
1. Introduction
Membrane bioreactors (MBRs) are widely used in wastewater treatment systems as effective wastewater treatment strategies because of their low sludge production, small area, and high treatment efficiency [1]. Membrane bioreactors have been used for more than three decades and their practical application has grown rapidly worldwide [2,3,4,5,6]. As the number of large (≥10,000 m3/d) and extra-large (≥100,000 m3/d) wastewater treatment plants in operation has increased, MBR systems have comprised an increasing number of wastewater treatment systems. The main advantages of MBRs are their high effluent quality, small footprint, and ability to separate hydraulic retention time (HRT) and solids retention time (SRT), while the main disadvantage is their high energy consumption [7,8]. Many factors affect MBR operating costs, such as power requirements as well as membrane cleaning and replacement costs [9,10], among which the costs associated with membrane biofouling are a major issue preventing the widespread applications of MBR [11]. Therefore, the main challenge currently facing widespread application of MBR systems is identification of better approaches to solve membrane biofouling.
Currently, physical and chemical methods such as backwashing and adding chemical reagents are still the most commonly used ways of controlling membrane biofouling [12]. Although these methods are somewhat effective at controlling membrane biofouling, they have many shortcomings. For example, physical methods usually only remove reversible fouling, and intense mechanical cleaning can cause membrane damage. Moreover, frequent chemical cleaning can reduce the membrane lifetime and membrane permeability [13]. In addition, residual NaClO in the MBR can cause severe bio-thermolysis and subsequent formation of toxic halogenated aromatic by-products, which pose a great threat to aquatic environments [14]. Furthermore, long-term application of chemical biocides for removal of membrane biofouling can cause bacteria to gradually develop resistance, reducing the effectiveness of biocides [15]. However, compared with physical and chemical methods, biological methods can effectively remove biofouling from membranes with less impact on ecological environments affected by wastewater treatment systems. Therefore, many biological membrane biofouling control strategies have been rapidly developed in recent years [16].
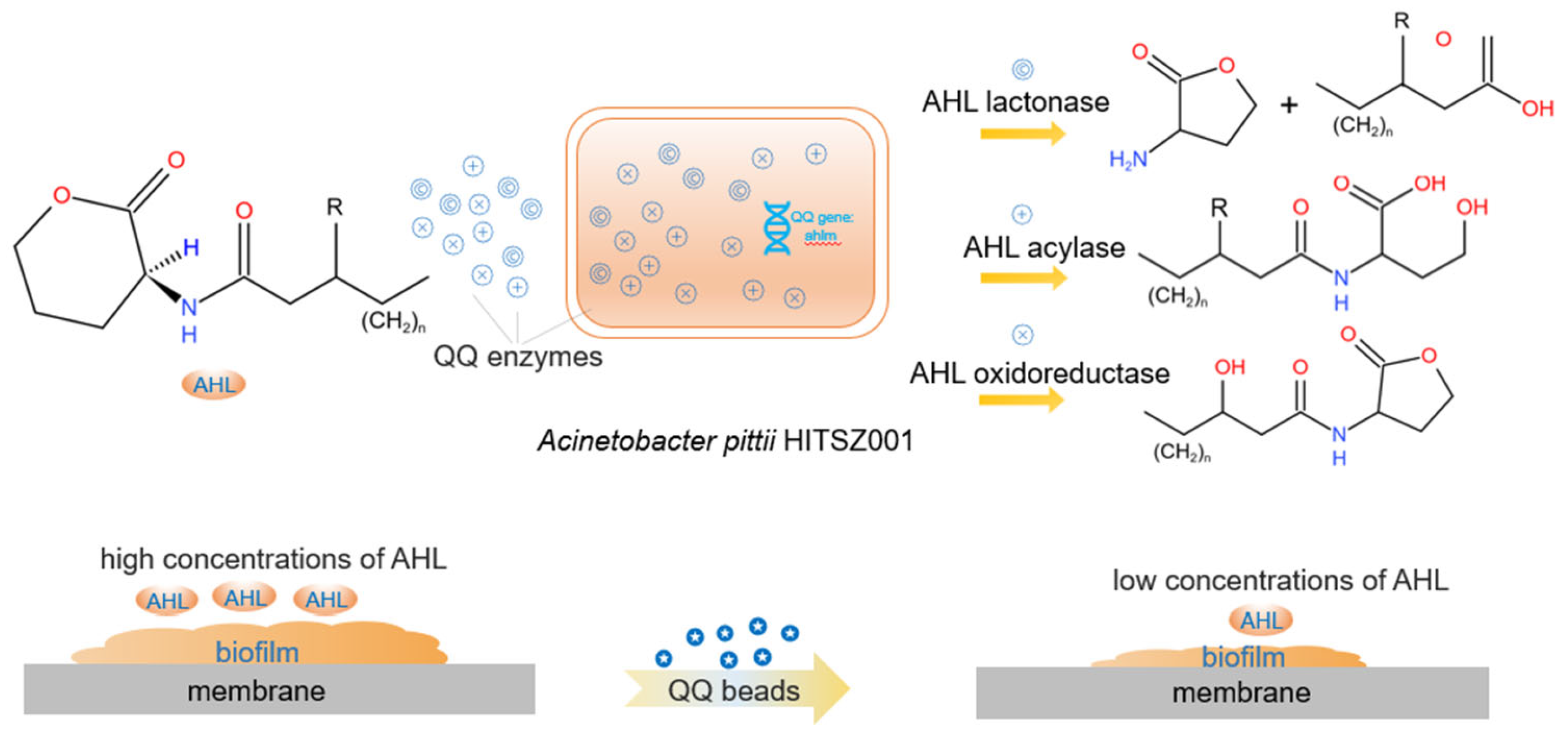
Quorum sensing (QS) is a mechanism of chemical communication among bacteria that relies on the synthesis, detection, and response to extracellular signal molecules known as autoinducers [17]. As shown in Figure 1, when the amount of bacteria increases, resulting in a threshold concentration of signal molecules, these molecules can enter cells and activate the expression of target genes to regulate various physiological functions, such as biofilm formation and expression of virulence factors [18,19]. All membrane biofouling processes, such as microbial growth within membrane pores and bacterial aggregation on membrane surfaces, are associated with QS [20]. Quorum quenching (QQ) is the inhibition of quorum sensing processes by QQ enzymes and QQ bacteria [21]. Quorum quenching enzymes can inactivate or degrade signal molecules, and the bacteria that can secrete QQ enzymes are called QQ bacteria [22]. QQ enzymes bring a new paradigm for MBR membrane biofouling, however, the practical application is limited due to the high cost and low stability [23,24]. As an alternative, QQ bacteria, which produce QQ enzymes, are a better choice. Although the activity is not as high as the QQ enzyme, QQ bacteria application is preferred for the following reasons: (1) it is more readily available and less costly; (2) stable nature, as the intracellular QQ enzyme is not affected by the external environment; (3) bacterial cells are easier to handle than enzymes. In addition, QQ bacteria can survive without an external nutrient supply because the wastewater contains nutrients and dissolved oxygen, etc., which are necessary for bacterial growth. Therefore, an in-depth study of the effects of QQ bacteria on the inhibition of MBR membrane biofouling is greatly needed.
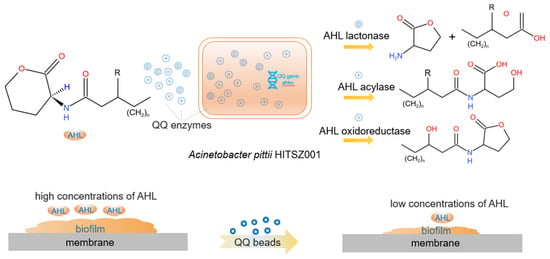
Figure 1.
Quorum quenching process in MBRs. When QQ beads encapsulated with QQ bacteria were added, the QQ bacteria secrete QQ enzymes that degrade signal molecules, prevent bacterial aggregation and biofilm formation, and inhibit membrane biofouling [33].
Kim et al., immobilized QQ enzyme (acylase) into nanofiltration membranes, which slowed down the formation of mushroom-shaped mature biofilms by reduction of EPS secretion [25]. Hyun-Suk Oh et al., encapsulated the recombinant Escherichia coli secreting N-acyl-homoserine lactase or quorum quenching Rhodococcus sp. isolated from a real MBR plant to the lumen of microporous hollow fiber membranes, respectively [26]. The microbial vessels with effective bacteria were placed in the MBR to mitigate biofouling. The effectiveness of biofouling suppression by microbial vessels was tested over 80 days of MBR operation. Jahangir et al. used continuously regenerated QQ bacteria within microbial vessels, which steadily maintained their QQ activity for more than 100 d [27]. This strategy effectively interrupted cell-to-cell communication, interfered with the QS system among bacteria, and resulted in a reduction in MBR aeration, thus offering energy-saving potential. Yeon et al. conducted a laboratory-scale MBR and demonstrated that direct addition of amidotransferase (10 mg/L) derived from the porcine kidney to the MBR could effectively inhibit the trans-membrane pressure (TMP) [28]. Lee et al. investigated the effectiveness of QQ bacteria in a wastewater treatment plant, where they put prepared QQ beads into the MBR to test its effectiveness on membrane biofilm control [29]. The study showed that the increases in the rates of TMP were significantly lower in QQ-MBR and the concentration of EPS in the mixture was also significantly lower compared with conventional MBR. In real wastewater treatment system, QQ bacteria had no negative effect on the effluent quality of QQ-MBR, and the QQ activity and system stability of QQ-MBR were significantly advanced and maintained for at least 4 months. Yu et al., designed a novel bilayer biocarrier with QQ bacteria as the shell and bio-stimulator as the core, which can effectively control membrane biofouling [30]. The structure can inhibit biofilm formation and showed the best membrane biofilm mitigation performance in MBR assays, as well as advanced signal molecule degradation and reduced EPS secretion. Kampouris et al., isolated a novel QQ strain (Lactobacillus sp. SBR04MA) from municipal sewage and examined its performance in reducing membrane fouling [31]. Results showed that the QQ bacteria significantly reduced the production of SMP and EPS and had no effect on the COD removal efficiency. Ham et al. isolated an Enterococcus sp. HEMM-1 from MBR and found that the novel QQ bacteria could effectively degrade AHLs and reduce biofilm on membrane surfaces [32]. These studies proved that QQ bacteria have better economic and technology feasibility in MBR systems. Thus, it is necessary to screen more efficient QQ bacteria and clarify their inhibitory effect. Furthermore, although some studies on quorum quenching against biofouling have appeared, most of them have focused on pure strain experiments, and so we have applied it to a long-time real operating MBR, which is more valuable for practical applications in the future.
The primary aim of this study was to investigate the effects of QQ bacteria on the operation of actual MBR systems and membrane biofouling control. To verify the operational effectiveness of immobilized QQ strains in MBR systems, we examined many water quality indicators and sludge characteristics to evaluate their effects on membrane fouling inhibition. We also examined the EPS and SMP contents in the studied systems. Finally, we examined changes in microbial communities in the studied systems by high-throughput sequencing.
2. Materials and Methods
2.1. Isolation and Identification of QQ Bacteria
To isolate potential QQ bacteria, activated sludge was sampled from the Xili Water Recycling Plant (Shenzhen, China). The sludge sample was centrifuged at 17,500× g for 10 min. The pellet was washed with minimal medium (per liter: NaCl, 1 g; KCl, 0.5 g; Na2SO4, 0.15 g; MgCl2, 0.4 g; CaCl2, 0.1 g; KH2PO4, 2 g; Na2HPO4, 2.25 g). It was acidified to pH 5.5 using 1 M of HCl and trace elements were added to final concentrations of 1 mg FeCl3, 0.1 g MnCl2 and 46 mg ZnCl2 per liter. They were then resuspended in 2 mL of minimal medium. The suspension was inoculated into modified sterile minimal medium containing 3 mM N-hexanoyl-L-homoserine lactone (C6-HSL) (Sigma–Aldrich, St Louis, MO, USA) as the sole carbon source. After 2 d, 10% (v/v) of the culture was transferred to fresh C6-HSL-containing minimal medium to enrich for C6-HSL-metabolizing bacteria. Similar transfers to fresh enrichment medium were performed after 2 d of incubation. In the fourth enrichment cycle, diluted suspension was plated onto LB agar. Single colonies were picked and repeatedly streaked on LB agar medium to obtain pure colonies.
The identification of bacterial species was determined by the 16S rRNA gene sequence. The 16S rRNA gene was PCR-amplified from bacterial DNA using the universal primers 27F (AGTTTGATCMTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT). The PCR conditions were as follows: 94 °C for 4 min; 30 cycles of 94 °C for 45 s, 55 °C for 45 s, and 72 °C for 1 min; and a final step at 72 °C for 10 min. Amplified PCR products were analyzed on a 2% agarose gel and then sequenced by Sangon Biotech Co., Ltd. (Shanghai, China). The primer sequence is shown in the Supplementary Materials. The BLAST algorithm was used to sequence and analyze the 16S rRNA genes. Multiple sequence alignments were generated using ClustalX. Then, the phylogenetic tree was built using the neighbor-joining method implemented in MEGA7 software. Based on 16S rRNA gene sequence analysis, the strain was identified as Acinetobacter pittii and it was named as Acinetobacter pittii HITSZ001 [33].
2.2. Immobilization of Quorum Quenching Strain
Strain Acinetobacter pittii HITSZ001 was incubated for 12 h while being shaken at 220 rpm and 37 °C. The supernatant was then discarded after centrifuging the cells at 4500 g for 10 min to pellet them. Bacterial suspensions were made after the pellet underwent two sterile PBS (phosphate-buffered saline) washes. The bacterial suspension was then combined in a 1:9 (v/v) ratio with a solution containing 4% sodium alginate. Using a syringe and the mixture, calcium chloride (CaCl2) solution at a concentration of 4% (w/v) was added to create the QQ beads. The Ca2+ ions cause the alginate droplets to cross-link, resulting in gelled beads with a typical bead diameter of 3–4 mm. The beads were cross-linked for 12 h at 4 °C, then cleaned three times in sterile water and kept at that temperature. PBS was used in place of the QQ bacterial suspension, and empty beads were created as a blank control in the manner previously described. [34].
2.3. MBR System
A total of three lab-scale MBRs with a working volume of 4 L were constructed in this study. Polyvinylidene fluoride (PVDF) hollow fiber membranes with an effective area of 0.06 m2 and 0.22 μm nominal pore size were used for the membranes. The influent was synthetic municipal wastewater that was pumped into the system using a metering pump. Influent flow into the system was controlled by a liquid level relay installed in the reaction tank. Continuous air sparging was performed at an airflow rate of 1.5 L/min using an air pump and air diffuser. The permeate flux was kept at 8.33 Liter/m2/h (LMH). The MBR hydraulic retention time (HRT) and sludge retention time were both 8 h. The MLSS concentration was 4500 ± 500 mg/L. The QQ bacteria were immobilized onto sodium alginate beads, then added to MBR 3. MBR 1 and MBR 2 were control groups containing no added beads and vacant beads, respectively. The volume of beads added to the MBR was 2% of the working volume.
2.4. Biomass Assay
After the experimental period, 1 cm hollow fiber membranes were cut from three membrane modules. Crystal violet staining was used to measure the biomass of the contaminated membranes [35]. Briefly, the membranes were washed three times with PBS and then placed into an oven (60 °C) for 30 min to remove suspended bacteria and fix the biofilm. Next, they were put into 24-well plates, after which 0.2 mL of crystal violet (0.1%) was added to each well. Samples were then incubated for 30 min in the dark. After staining was complete, the excess dye was washed off with PBS and 1 mL of alcohol (95%) was added to each well to extract the crystal violet. The 24-well plate was then placed on a shaker for 2 h at 100 rpm. A total of fifty microliters of the resulting solution was subsequently transferred to 96-well plates and diluted to 200 μL. Finally, the OD values were read at 585 nm with a microplate reader and the biofilm biomass in the wells was determined from the OD585 values.
2.5. Extraction of EPS and SMP
A total of 50 mL of mixed liquor taken from the MBR was centrifuged at 8000 rpm for 10 min, after which the supernatant was filtered through a 0.45 μm filter. The resulting filtrate, referred to as the SMP fraction, was then analyzed for its protein and polysaccharide content. Next, 0.05% NaCl was added to the remaining sludge, vortexed and shaken for 1 min, then heated in a 60 °C water bath for 30 min. The samples were subsequently centrifuged again at 8000 rpm for 10 min, after which the supernatant was collected as loosely bound EPS (LB-EPS). Next, the remaining sludge was resuspended in 0.05% NaCl, vortexed for 20 s, and then placed in 60 °C water bath for 30 min. Finally, samples were centrifuged at 8000 rpm for 20 min and the supernatant was collected as tightly bound EPS (TB-EPS) [36,37,38].
2.6. Analytical Methods
Proteins and polysaccharides were analyzed using the Lowry method and phenol-sulfuric acid method, respectively [39,40]. The mixed liquor suspended solids (MLSS), settling tests (SV30), sludge volume index (SVI), ammonia nitrogen (AN), total nitrogen (TN), total phosphorus (TP), and chemical oxygen demand (COD) were measured according to the standard methods [41].
2.7. High-Throughput Sequencing
DNA samples were extracted and the purity and concentration were confirmed using a NanoDrop One (Thermo Scientific, Waltham, MA, USA). The V4 region of bacterial 16S rRNA gene sequences was PCR amplified using the 515F and 806R primers and a TaKaRa Premix Taq amplification kit. The backbone was purified by gel extraction using an E.Z.N.A. gel extraction kit (Omega, Biel/Bienne, Switzerland). Samples were sequenced using the NovaSeq 6000 (Illumina, San Diego, CA, USA) platform and data were processed using fastp to obtain paired-end clean reads. Operational taxonomic units (OTUs, Chicago, IL, USA) were clustered at the 0.8 similarity level using the UPARSE method and noise removal was performed using QIIME2. For each OTU, the most abundant sequence was chosen as a representative sequence, and taxonomy assignment was conducted with RDP classifier using the SILVA database.
2.8. Statistical Analyses
The results shown in the figures represent the averages of three independent replicate treatments. The data obtained in this study are presented as the means ± standard deviations, which were determined using the SPSS software (version 24, IBM, Chicago, IL, USA).
3. Results and Discussion
3.1. Biofouling Inhibition by Quorum Quenching in MBR System
3.1.1. Variations of Trans-Membrane Pressure (TMP)
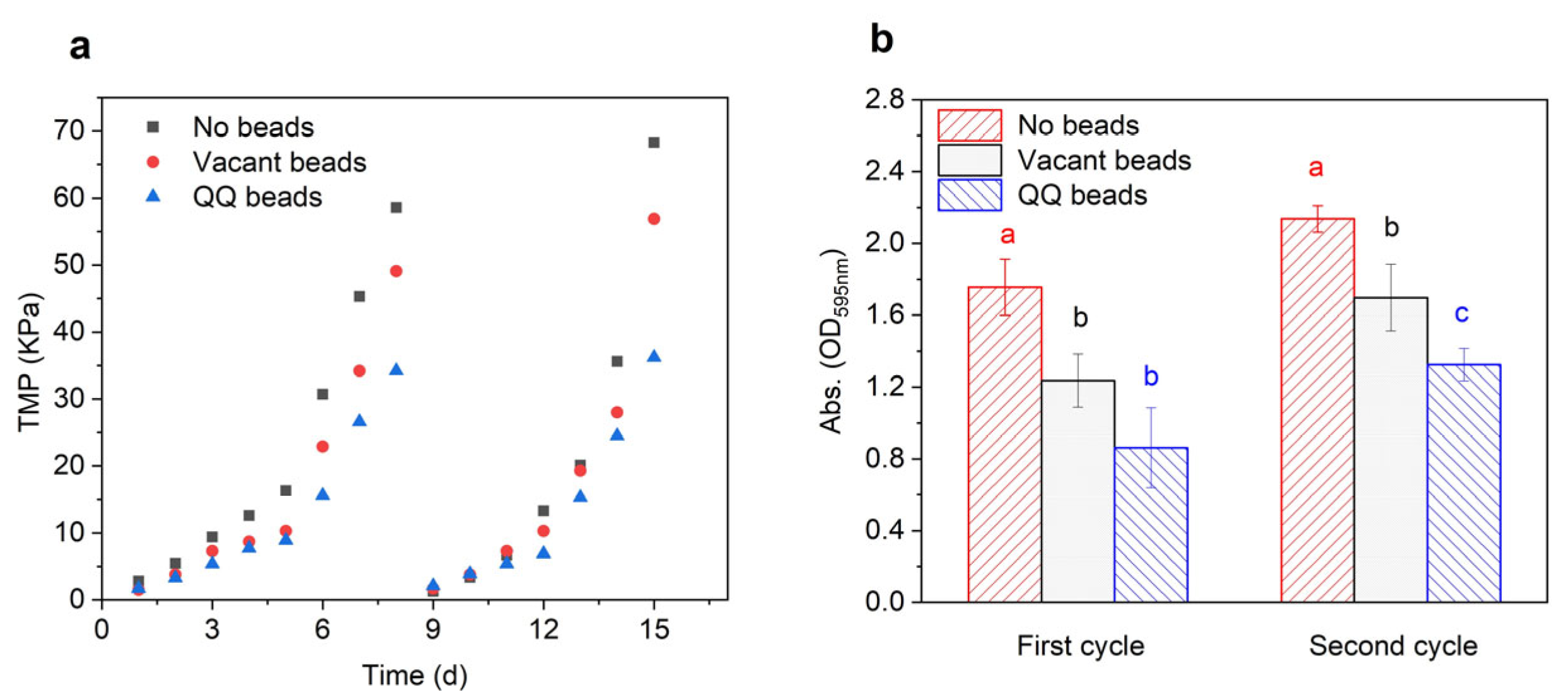
The aim of this study was to investigate the effects of QQ beads on membrane biofouling. TMP was used as an indicator for the assessment of biofouling control. When MBR biofouling increases, the TMP will also increase obviously. In this study, MBRs were operated continuously for two cycles with a critical flux of 8.33 LMH, during which time TMP was monitored in real time using pressure gauges which were mounted on the top of the MBRs. The membranes were considered to be completely biofouled when the TMP increased to 35 Kpa. When the TMP reaches the maximum, the filtration cycle ends. As shown in Figure 2a, the increase in TMP occurred in three phases. During the first phase, which was the initial growth period of TMP, the TMP of the three groups increased rather slowly. The flux membranes in the three bioreactors were maintained at a high level during the first 3 days, and there were no obvious differences among groups. In the second phase, the TMP growth rate of TMP increased greatly from day 4 to day 6, indicating that different levels of membrane biofouling might have occurred in the three bioreactors. Specifically, the TMP of the no beads group increased the fastest and generated the highest pressure, followed by that of the vacant beads group, and then the QQ beads group. The third phase, which occurred during the last 1–2 days, was characterized by a sharp increase in TMP. In this phase, a large amount of pollutant accumulated on the membrane, the flux decreased rapidly, and membrane pores became severely clogged. Concentration polarization, cake layer formation, organic adsorption, and inorganic precipitation, which are all interlinked and influence each other, contributed to membrane biofouling and eventually led to a sharp rise in TMP. Overall, the results showed that there were significant differences among the three groups in this phase. Notably, the value of the no beads group was about twice that of the QQ beads group during this phase. These results demonstrated that the addition of QQ beads played a significant role in alleviating the rise in TMP and had a great inhibitory effect on membrane biofouling. The weak inhibition effect of the vacant beads group may be attributed to the physical collision of sodium alginate beads in the bioreactor. Because the aeration device is located on the bottom of the bioreactor, the beads will roll or rock back and forth inside the reactor during aeration. While moving, they may impact the pollutants or flocs attached to the membrane surface when they touch the membrane, which controls membrane biofouling to some extent.
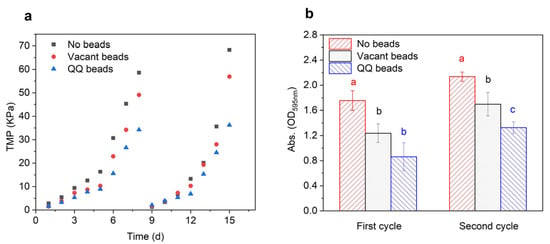
Figure 2.
TMP of MBR (a) and biomass on the membrane surface after each filtration cycle (b). Statistical analysis was carried out using analysis of variance. The significant difference of treatment was shown in different letters.
3.1.2. Variations in Biomass
After each filtration cycle, the membranes were removed, and 1 cm of the membrane filaments were cut off and analyzed to evaluate the biomass on the membrane surface. As shown in Figure 2b, the biomass of the no beads group was twice that of the QQ group at the end of the first filtration cycle, indicating that the no beads group had many more biological pollutants accumulated on the membrane than the QQ group. After the second filtration cycle, the accumulation on the membrane in the QQ group was still much lower than that on membranes of the other groups, demonstrating that QQ beads effectively mitigated membrane biofouling.
3.2. EPS and SMP Concentration in the Bioreactor Mixed Liquor
EPS is a polymeric substance secreted by microorganisms, mainly composed of polysaccharides, proteins, small amounts of humic acids and nucleic acids. It is considered to be a key factor in membrane biofouling and can be divided into the loosely bound EPS (LB-EPS) and tightly bound EPS (TB-EPS). The main roles of EPS are removal of pollutants by adsorption of inorganic ions; adsorption of external organic matter to aggregate nutrients in the water environment; digestion of external macromolecules to meet nutrient requirements; and formation of a protective layer to impede harmful external environments. In the case of relatively low organic load, EPS can act as a carbon and energy source, and can aggregate flocs and biofilms of bacterial cells. The main components of EPS are polysaccharides and proteins, which account for about 70% to 80% of the total EPS, while humic substances, nucleic acids, glyoxylates, lipids and amino acids are less abundant. SMP is the product of microbial metabolic substrate or biomass decay, which is biotoxic, reducing microbial activity and inhibiting nitrification, with polysaccharides, proteins and humic substances as the main components [41].
3.2.1. EPS
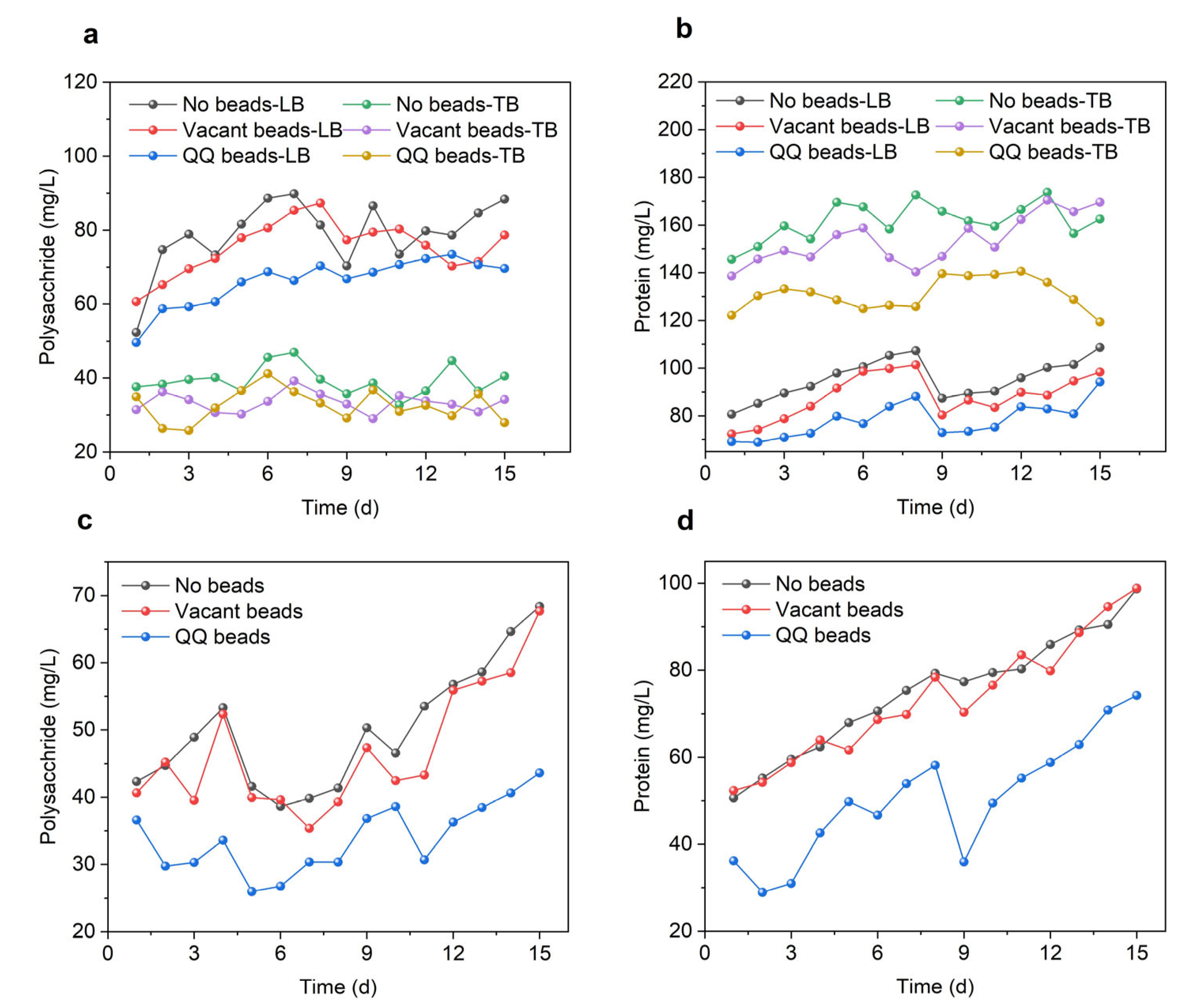
As shown in Figure 3a, the concentrations of polysaccharides were significantly higher in the no beads group, and the vacant beads group, than in the QQ group during the filtration period, which demonstrates that the addition of QQ beads inhibited microbial polysaccharide secretion. This trend was more obvious in LB-EPS than in TB-EPS, indicating that the QQ strain was effective at inhibiting the loose extracellular polymer flowing in the system, but had limited ability to inhibit substances tightly adhered to the cell surface with poor mobility. As shown in Figure 3a, the polysaccharide content in LB-EPS was significantly higher than that in TB-EPS; therefore, the QQ beads could repress most of the polysaccharides. However, proteins were more abundant in TB-EPS than in LB-EPS. In LB-EPS, the MBR protein content maintained a similar trend in the three groups, while the protein content differed in TB-EPS. These differences may have originated from changes in the microbial community in the MBR system. The presence of QQ bacteria could lead to transformations in the microbial community structure in the system, resulting in significant differences in protein secretion from the other two groups. As shown in Figure 3b, protein in TB-EPS of the QQ group decreased by 29.63% and 26.56%, respectively, compared with the other two groups. In sum, QQ beads resulted in decreased EPS secretion in the MBR system, and its main components, polysaccharides and proteins, were significantly reduced, which effectively mitigated membrane biofouling.
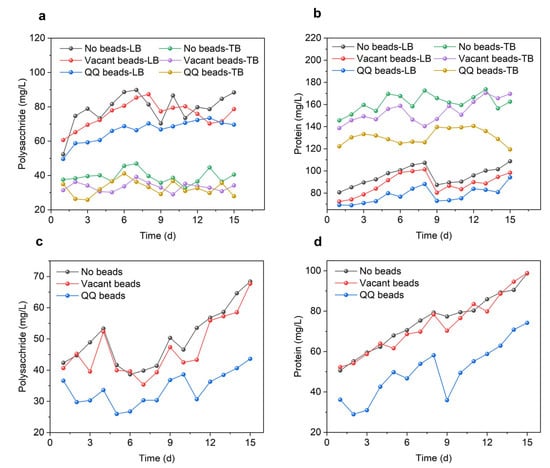
Figure 3.
EPS and SMP variations during continuous MBR operation in two cycles. (a) Polysaccharides of LB-EPS and TB-EPS; (b) proteins of LB-EPS and TB-EPS; (c) polysaccharides of SMP; (d) proteins of SMP.
3.2.2. SMP
As shown in Figure 3c,d, the SMP polysaccharides and proteins in all three groups increased during the filtration cycle. In this cycle, there was little difference between the no beads and vacant beads groups, but the levels of polysaccharides and proteins in the QQ beads group was significantly lower than in the other two groups. At the end of the first cycle, the polysaccharides in the QQ beads group had decreased by 26.67% and 22.81%, while the proteins had decreased by 26.68% and 25.81% compared with the no beads and vacant beads groups, respectively. At the end of the second cycle, the polysaccharides in the QQ bead group had decreased by 36.21% and 35.56% compared with the no beads and vacant beads groups, respectively, while the proteins had decreased by 24.80% and 24.94%, respectively. The reduction of polysaccharides and proteins indicated that QQ enhanced the MBR performance by affecting the production of biopolymers of sludge microflora.
3.3. Effect of Quorum Quenching on Sludge Characteristics
Because of the complex microbial environments in the MBR systems, it is necessary to determine whether the addition of QQ bacteria would affect biochemical reactions and growth of activated sludge. Therefore, MLSS, SVI and SV 30 were measured to determine the effects of QQ on activated sludge.
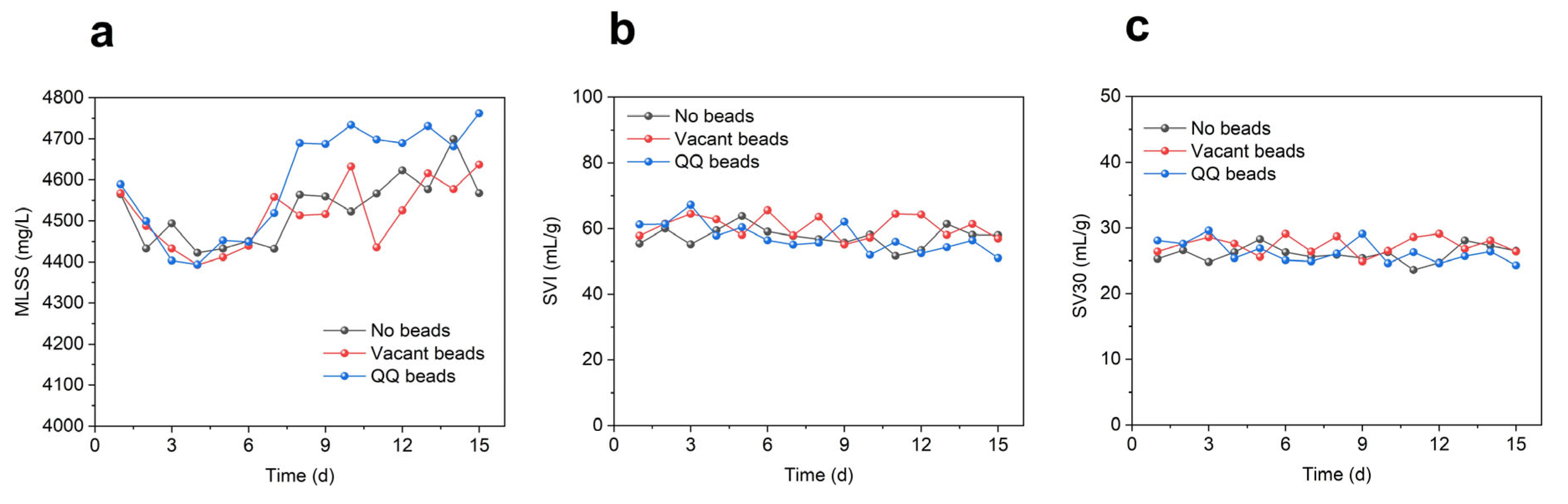
As shown in Figure 4, the MLSS in all three groups of MBRs was between 4400 and 4800 mg/L, the SVI was basically maintained between 50% and 70%, and the SV 30 was always below 30%, indicating good sludge growth and performance. The MLSS of the QQ beads group was slightly higher than that of the other two groups during anaphase, indicating that the presence of QQ bacteria might promote the growth of microorganisms to some extent. The SVI and SV 30 were similar among the three groups, indicating that the settling properties and loosening degree of sludge were basically unchanged.
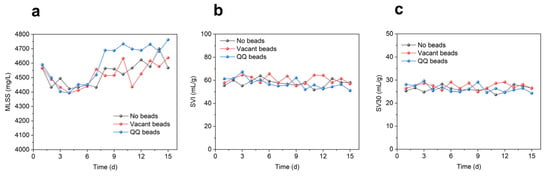
Figure 4.
Sludge variations during continuous MBR operation in two cycles; (a) MLSS; (b) SVI; (c) SV30.
3.4. Effect of Quorum Quenching on MBR Treatment Efficiencies
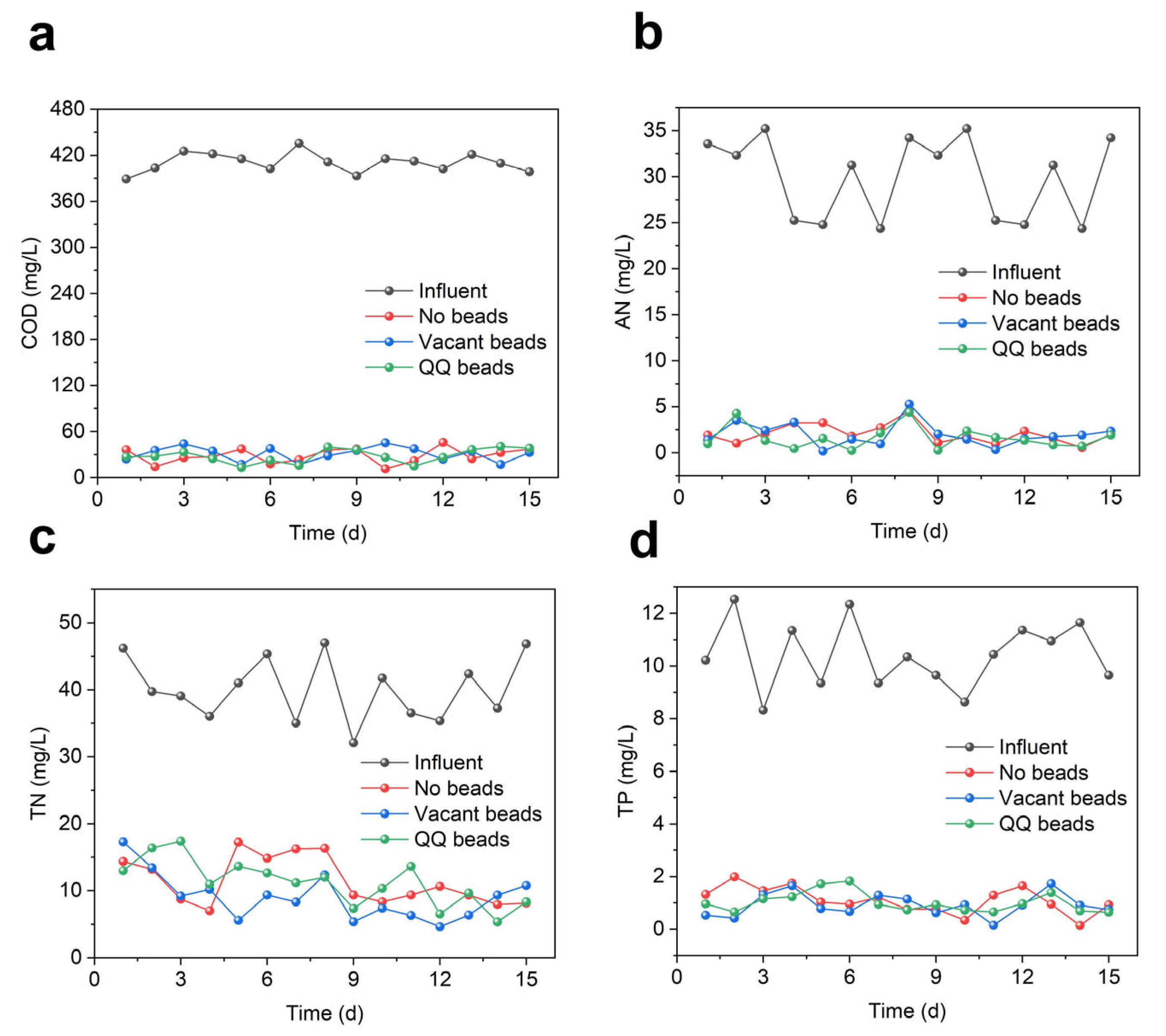
To verify whether the addition of QQ bacteria affects the removal of pollutants in the MBR system, the COD, ammonia nitrogen, total nitrogen, and total phosphorus of the effluent were measured. As shown in Figure 5, there was no significant difference in the removal of COD, nitrogen and phosphorus, demonstrating that the addition of QQ beads had no negative impact on the removal of pollutants by the MBR system.
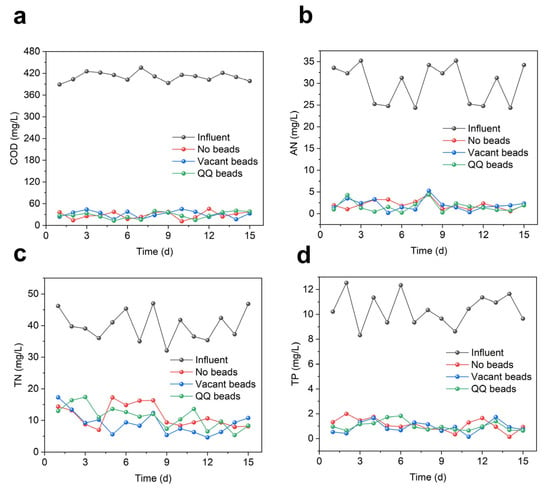
Figure 5.
Effluent quality during continuous MBR operation in two cycles; (a) COD; (b) AN; (c) TN; (d) TP.
3.5. Microbial Community Structure Analysis
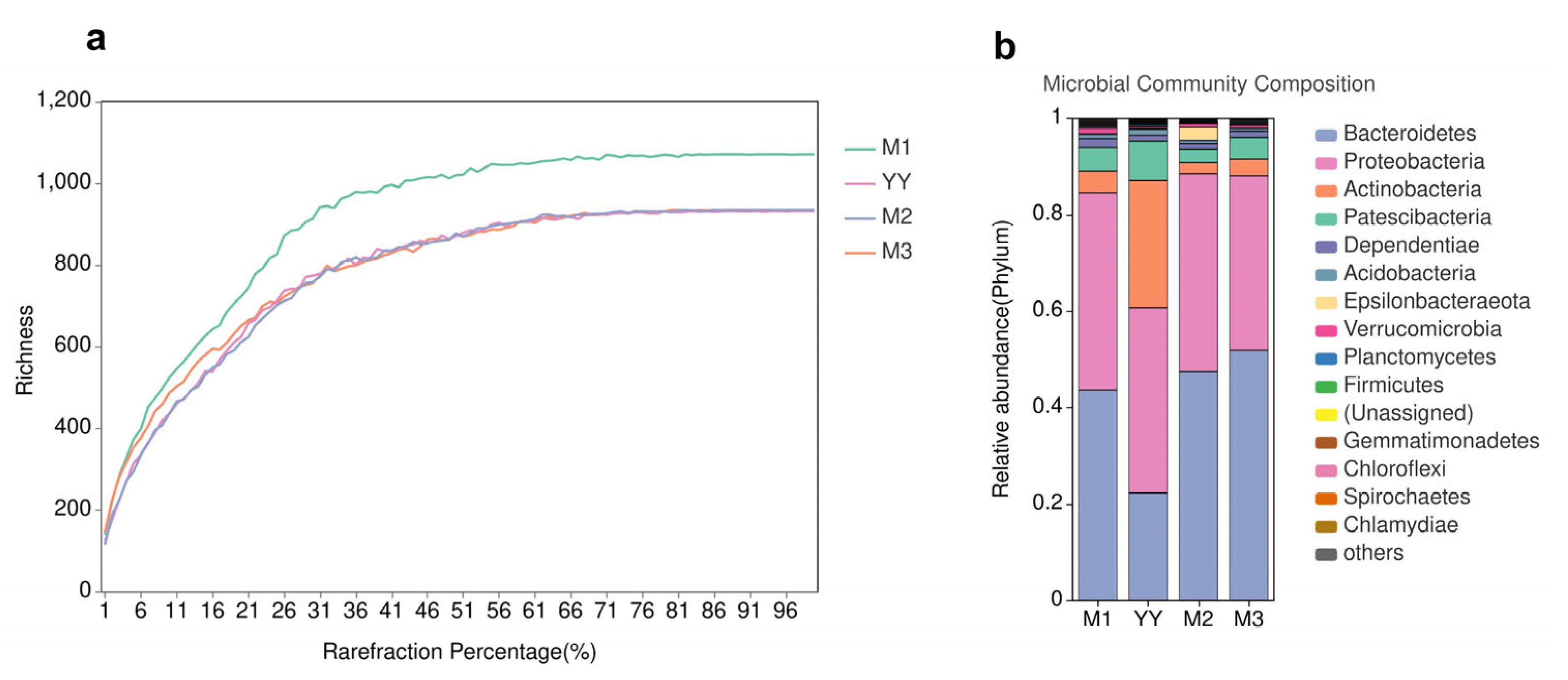
The richness of the microbial communities was estimated using a dilution curve (Figure 6a), which indicates when sequencing has reached the ideal depth and the sample is sufficient and reasonable. Continuing to increase the sequencing depth beyond this point will not identify additional operational taxonomic units (OTUs). As shown in Table 1, the valid sequences of the raw sludge (untreated by the MBR system) and sludge samples from the three MBRs were 60,159, 61,418, 59,082 and 71,215, respectively, and these sequences contained 1029, 1204, 1097 and 1084 OTUs. The Chao1 index in the raw sludge was 1029.4, while the Chao1 indexes for the three MBR samples were 1204.2, 1097.4 and 1084.6, respectively, indicating a decreasing trend. The Chao1 index of the QQ beads group was lowest, indicating that its species richness was reduced and microbial abundance was enriched. In addition, the Shannon index decreased and the Simpson index increased in the QQ beads group, which also indicated a decrease in community diversity within the system.
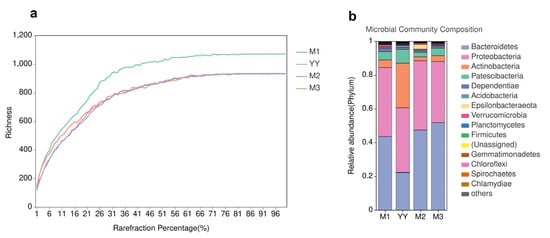
Figure 6.
The dilution curves (a); Microbial community composition (b). Variations in bacterial community structure in the MBR system. YY is raw sludge, M1 is the no beads group, M2 is the vacant beads group, M3 is the QQ beads group.

Table 1.
Analysis of alpha-diversity index. YY is the raw sludge sample, M1 is the no beads sample, M2 is the vacant beads sample and M3 is the QQ beads sample.
The relationship between all samples and phyla in the MBRs is shown in Figure 6b. Community composition analysis indicated that the dominant phyla present in the MBR sludge were Bacteroidetes and Proteobacteria. In addition, there were small amounts of Actinobacteria, Dependentiae and Planctomycetes in the MBR, as well as other phyla that were present in trace amounts. The presence of a large number of bacteria with QS genes belonging to Bacteroidetes and Proteobacteria suggests that a more general QS phenomenon may be occurring in the MBR system. As MBR operation continued, the number of Proteobacteria increased significantly. This indicated that the population induction phenomenon had intensified, which might reflect the gradual formation of biofilm. Additionally, the number of Anaplasma remained basically stable, while there were some fluctuations in the levels of other bacteria present in small amounts. The coexistence of multiple complex microbial communities maintained the stable growth of sludge and the removal of pollutants in the MBR.
4. Conclusions
Membrane biofouling is one of the greatest hindrances limiting the widespread and large-scale application of MBR. To address this issue, a novel QQ strain, A. pittii HITSZ001 was immobilized in sodium alginate beads and applied to the MBR systems. The results indicated that, immobilized QQ bacteria could inhibit the secretion of EPS and SMP without affecting the MBR effluent quality and activated sludge biochemical reactions, thus acting as an anti-membrane biofouling agent. Additionally, the microbial community diversity in the MBR system was reduced to some extent after the addition of QQ beads, which might have been because of the occurrence of directional enrichment in the system.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/separations10020127/s1, Figure S1: Membrane in no beads group; Figure S2. Membrane in vacant beads group; Figure S3. Membrane in QQ beads group.
Author Contributions
Conceptualization, X.F. and Y.W.; methodology, Y.W., W.W. and C.J.; software, Y.W.; validation, Z.X. and X.Z.; data curation, Y.X.; writing, original draft preparation, Y.W. and X.F.; writing, review and editing, X.F. and H.S.; supervision, X.F.; project administration, N.R.; funding acquisition, X.F. All authors have read and agreed to the published version of the manuscript.
Funding
This investigation was funded by the National Natural Science Foundation of China (No. 51908161), Guangdong Basic and Applied Basic Research Foundation (No. 2019A1515010807), State Key Laboratory of Urban Water Resource and Environment (Harbin Institute of Technology) (No. 2021TS30) and Shenzhen Science and Technology Program (Grant No. KQTD20190929172630447, KCXFZ20211020163404007 and GXWD20201230155427003-20200824100026001).
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Bijtenhoorn, P.; Mayerhofer, H.; Müller-Dieckmann, J.; Utpatel, C.; Schipper, C.; Hornung, C.; Szesny, M.; Grond, S.; Thürmer, A.; Brzuszkiewicz, E.; et al. A Novel Metagenomic Short-Chain Dehydrogenase/Reductase Attenuates Pseudomonas Aeruginosa Biofilm Formation and Virulence on Caenorhabditis Elegans. PLoS ONE 2011, 6, e26278. [Google Scholar] [CrossRef]
- Yamamoto, K.; Hiasa, M.; Mahmood, T.; Matsuo, T. Direct solid-liquid separation using hollow fiber membrane in an activated sludge aeration tank. Water Pollut. Res. Control. Brighton 1988, 21, 43–54. [Google Scholar] [CrossRef]
- Huang, L.; Lee, D.J. Membrane Bioreactor: A Mini Review on Recent R&D Works. Bioresour. Technol. 2015, 194, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Xiao, K.; Shen, Y. Recent Advances in Membrane Bioreactor Technology for Wastewater Treatment in China. Front. Environ. Sci. Eng. China 2010, 4, 245–271. [Google Scholar] [CrossRef]
- Judd, S.J. The Status of Industrial and Municipal Effluent Treatment with Membrane Bioreactor Technology. Chem. Eng. J. 2016, 305, 37–45. [Google Scholar] [CrossRef]
- Krzeminski, P.; Leverette, L.; Malamis, S.; Katsou, E. Membrane Bioreactors—A Review on Recent Developments in Energy Reduction, Fouling Control, Novel Configurations, LCA and Market Prospects. J. Memb. Sci. 2017, 527, 207–227. [Google Scholar] [CrossRef]
- Meng, F.; Zhang, S.; Oh, Y.; Zhou, Z.; Shin, H.S.; Chae, S.R. Fouling in Membrane Bioreactors: An Updated Review. Water Res. 2017, 114, 151–180. [Google Scholar] [CrossRef] [PubMed]
- Drews, A. Membrane Fouling in Membrane Bioreactors—Characterisation, Contradictions, Cause and Cures. J. Memb. Sci. 2010, 363, 1–28. [Google Scholar] [CrossRef]
- Young, T.; Muftugil, M.; Smoot, S.; Peeters, J. MBR vs. CAS: Capital and Operating Cost Evaluation. Water Pract. Technol. 2012, 7, wpt2012075. [Google Scholar] [CrossRef]
- Gao, T.; Xiao, K.; Zhang, J.; Zhang, X.; Wang, X.; Liang, S.; Sun, J.; Meng, F.; Huang, X. Cost-Benefit Analysis and Technical Efficiency Evaluation of Full-Scale Membrane Bioreactors for Wastewater Treatment Using Economic Approaches. J. Clean. Prod. 2021, 301, 126984. [Google Scholar] [CrossRef]
- Tang, K.; Xie, J.; Pan, Y.; Zou, X.; Sun, F.; Yu, Y.; Xu, R.; Jiang, W.; Chen, C. The Optimization and Regulation of Energy Consumption for MBR Process: A Critical Review. J. Environ. Chem. Eng. 2022, 10, 108406. [Google Scholar] [CrossRef]
- Gkotsis, P.K.; Batsari, E.L.; Peleka, E.N.; Tolkou, A.K.; Zouboulis, A.I. Fouling Control in a Lab-Scale MBR System: Comparison of Several Commercially Applied Coagulants. J. Environ. Manag. 2017, 203, 838–846. [Google Scholar] [CrossRef]
- Cui, Z.; Wang, J.; Zhang, H.; Ngo, H.H.; Jia, H.; Guo, W.; Gao, F.; Yang, G.; Kang, D. Investigation of Backwashing Effectiveness in Membrane Bioreactor (MBR) Based on Different Membrane Fouling Stages. Bioresour. Technol. 2018, 269, 355–362. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, F.; He, X.; Zhou, Z.; Huang, L.N.; Liang, S. Optimisation and Performance of NaClO-Assisted Maintenance Cleaning for Fouling Control in Membrane Bioreactors. Water Res. 2014, 53, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.K.; Kashif, A.; Fuwad, A.; Choi, Y. Current Advances in Treatment Technologies for Removal of Emerging Contaminants from Water–A Critical Review. Coord. Chem. Rev. 2021, 442, 213993. [Google Scholar] [CrossRef]
- Cui, Y.; Gao, H.; Yu, R.; Gao, L.; Zhan, M. Biological-Based Control Strategies for MBR Membrane Biofouling: A Review. Water Sci. Technol. 2021, 83, 2597–2614. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.G.; Yin, W.F.; Sam, C.K.; Koh, C.L. A Novel Medium for the Isolation of N-Acylhomoserine Lactone-Degrading Bacteria. J. Ind. Microbiol. Biotechnol. 2009, 36, 247. [Google Scholar] [CrossRef] [PubMed]
- Chong, T.M.; Koh, C.L.; Sam, C.K.; Choo, Y.M.; Yin, W.F.; Chan, K.G. Characterization of Quorum Sensing and Quorum Quenching Soil Bacteria Isolated from Malaysian Tropical Montane Forest. Sensors 2012, 12, 4846. [Google Scholar] [CrossRef]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef]
- Fast, W.; Tipton, P.A. The Enzymes of Bacterial Census and Censorship. Trends Biochem. Sci. 2012, 37, 7–14. [Google Scholar] [CrossRef]
- Uroz, S.; Dessaux, Y.; Oger, P. Quorum Sensing and Quorum Quenching: The Yin and Yang of Bacterial Communication. ChemBioChem 2009, 10, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Bzdrenga, J.; Daudé, D.; Rémy, B.; Jacquet, P.; Plener, L.; Elias, M.; Chabrière, E. Biotechnological Applications of Quorum Quenching Enzymes. Chem. Biol. Interact. 2017, 267, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum Quenching: Role in Nature and Applied Developments. FEMS Microbiol. Rev. 2016, 40, 86–116. [Google Scholar] [CrossRef] [PubMed]
- Maddela, N.R.; Sheng, B.; Yuan, S.; Zhou, Z.; Villamar-Torres, R.; Meng, F. Roles of Quorum Sensing in Biological Wastewater Treatment: A Critical Review. Chemosphere 2019, 221, 616–629. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, D.C.; Yeon, K.M.; Kim, S.R.; Lee, C.H. Enzyme-Immobilized Nanofiltration Membrane to Mitigate Biofouling Based on Quorum Quenching. Environ. Sci. Technol. 2011, 45, 1601–1607. [Google Scholar] [CrossRef]
- Oh, H.-S.; Yeon, K.-M.; Yang, C.-S.; Kim, S.-R.; Lee, C.-H.; Park, S.Y.; Han, J.Y.; Lee, J.-K. Control of Membrane Biofouling in MBR for Wastewater Treatment by Quorum Quenching Bacteria Encapsulated in Microporous Membrane. Environ. Sci. Technol. 2012, 46, 4877–4884. [Google Scholar] [CrossRef]
- Jahangir, D.; Oh, H.S.; Kim, S.R.; Park, P.K.; Lee, C.H.; Lee, J.K. Specific Location of Encapsulated Quorum Quenching Bacteria for Biofouling Control in an External Submerged Membrane Bioreactor. J. Memb. Sci. 2012, 411–412, 130–136. [Google Scholar] [CrossRef]
- Yeon, K.M.; Lee, C.H.; Kim, J. Magnetic Enzyme Carrier for Effective Biofouling Control in the Membrane Bioreactor Based on Enzymatic Quorum Quenching. Environ. Sci. Technol. 2009, 43, 7403–7409. [Google Scholar] [CrossRef]
- Lee, S.; Park, S.K.; Kwon, H.; Lee, S.H.; Lee, K.; Nahm, C.H.; Jo, S.J.; Oh, H.S.; Park, P.K.; Choo, K.H.; et al. Crossing the Border between Laboratory and Field: Bacterial Quorum Quenching for Anti-Biofouling Strategy in an MBR. Environ. Sci. Technol. 2016, 50, 1788–1795. [Google Scholar] [CrossRef]
- Yu, H.; Lee, K.; Zhang, X.; Choo, K.H. In Situ versus Pre-Quorum Quenching of Microbial Signaling for Enhanced Biofouling Control in Membrane Bioreactors. J. Memb. Sci. 2019, 592, 117387. [Google Scholar] [CrossRef]
- Kampouris, I.D.; Karayannakidis, P.D.; Banti, D.C.; Sakoula, D.; Konstantinidis, D.; Yiangou, M.; Samaras, P.E. Evaluation of a Novel Quorum Quenching Strain for MBR Biofouling Mitigation. Water Res. 2018, 143, 56–65. [Google Scholar] [CrossRef]
- Ham, S.Y.; Kim, H.S.; Cha, E.; Park, J.H.; Park, H.D. Mitigation of Membrane Biofouling by a Quorum Quenching Bacterium for Membrane Bioreactors. Bioresour. Technol. 2018, 258, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Feng, X.; Shi, H.; Wang, W.; Jiang, C.; Xiao, Z.; Xu, Y.; Ren, N. New Insights into Biofilm Control and Inhibitory Mechanism Analysis Based on the Novel Quorum Quenching Bacterium Acinetobacter Pittii HITSZ001. J. Memb. Sci. 2022, 663, 121012. [Google Scholar] [CrossRef]
- Kim, S.R.; Oh, H.S.; Jo, S.J.; Yeon, K.M.; Lee, C.H.; Lim, D.J.; Lee, C.H.; Lee, J.K. Biofouling Control with Bead-Entrapped Quorum Quenching Bacteria in Membrane Bioreactors: Physical and Biological Effects. Environ. Sci. Technol. 2013, 47, 836–842. [Google Scholar] [CrossRef]
- Feng, X.C.; Guo, W.Q.; Zheng, H.S.; Wu, Q.L.; Luo, H.C.; Ren, N.Q. Effect of Metabolic Uncoupler, 3,3′,4′,5-Tetrachlorosalicylanilide (TCS) on Bacillus Subtilis: Biofilm Formation, Flocculability and Surface Characteristics. RSC Adv. 2018, 8, 16178–16186. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhang, C.; Lian, J.; Lu, C.; Chen, Z.; Song, Y.; Guo, Y.; Xing, Y. Effect of Thiosulfate on Rapid Start-up of Sulfur-Based Reduction of High Concentrated Perchlorate: A Study of Kinetics, Extracellular Polymeric Substances (EPS) and Bacterial Community Structure. Bioresour. Technol. 2017, 243, 932–940. [Google Scholar] [CrossRef]
- Somani, B.L.; Khanade, J.; Sinha, R. A Modified Anthrone-Sulfuric Acid Method for the Determination of Fructose in the Presence of Certain Proteins. Anal. Biochem. 1987, 167, 327–330. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, Z.; Tang, S. Extracellular Polymeric Substances (EPS) Properties and Their Effects on Membrane Fouling in a Submerged Membrane Bioreactor. Water Res. 2009, 43, 2504–2512. [Google Scholar] [CrossRef]
- Nwachukwu, I.D.; Girgih, A.T.; Malomo, S.A.; Onuh, J.O.; Aluko, R.E. Thermoase-Derived Flaxseed Protein Hydrolysates and Membrane Ultrafiltration Peptide Fractions Have Systolic Blood Pressure-Lowering Effects in Spontaneously Hypertensive Rats. Int. J. Mol. Sci. 2014, 15, 18131–18147. [Google Scholar] [CrossRef] [PubMed]
- Leng, F.; Sun, S.; Jing, Y.; Wang, F.; Wei, Q.; Wang, X.; Zhu, X. A Rapid and Sensitive Method for Determination of Trace Amounts of Glucose by Anthrone-Sulfuric Acid Method. Bulg. Chem. Commun. 2016, 48, 109–113. [Google Scholar]
- Rice, E.W.; Baird, R.B.; Eaton, A.D. Standard Methods for the Examination of Water and Wastewater, 23rd ed.; American Public Health Association: Washington, DC, USA, 2012. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).