Meta-Analysis of Public RNA Sequencing Data Revealed Potential Key Genes Associated with Reproductive Division of Labor in Social Hymenoptera and Termites
Abstract
1. Introduction
2. Results
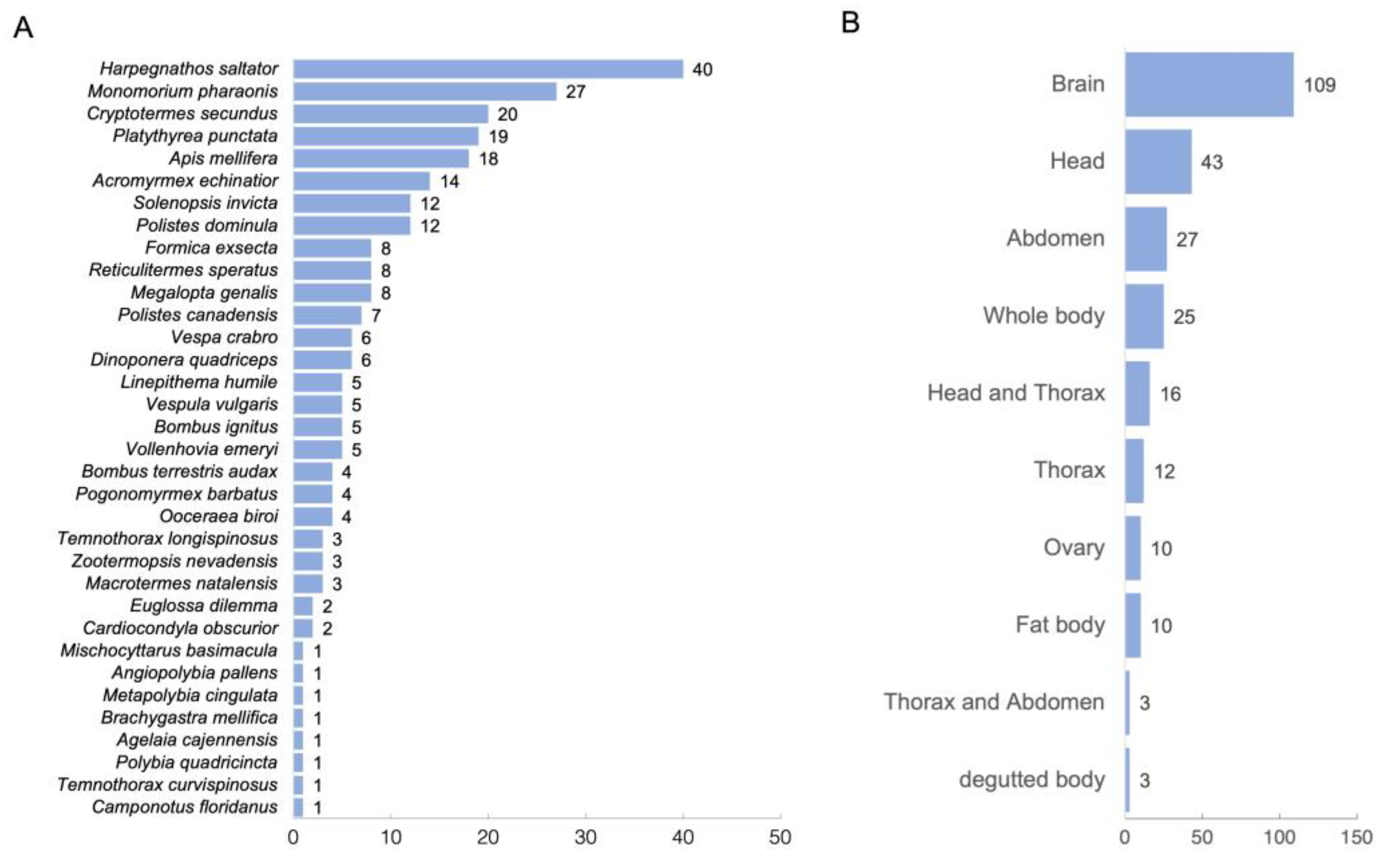
2.1. Data Collection of Transcriptomes Related to Queens and Workers
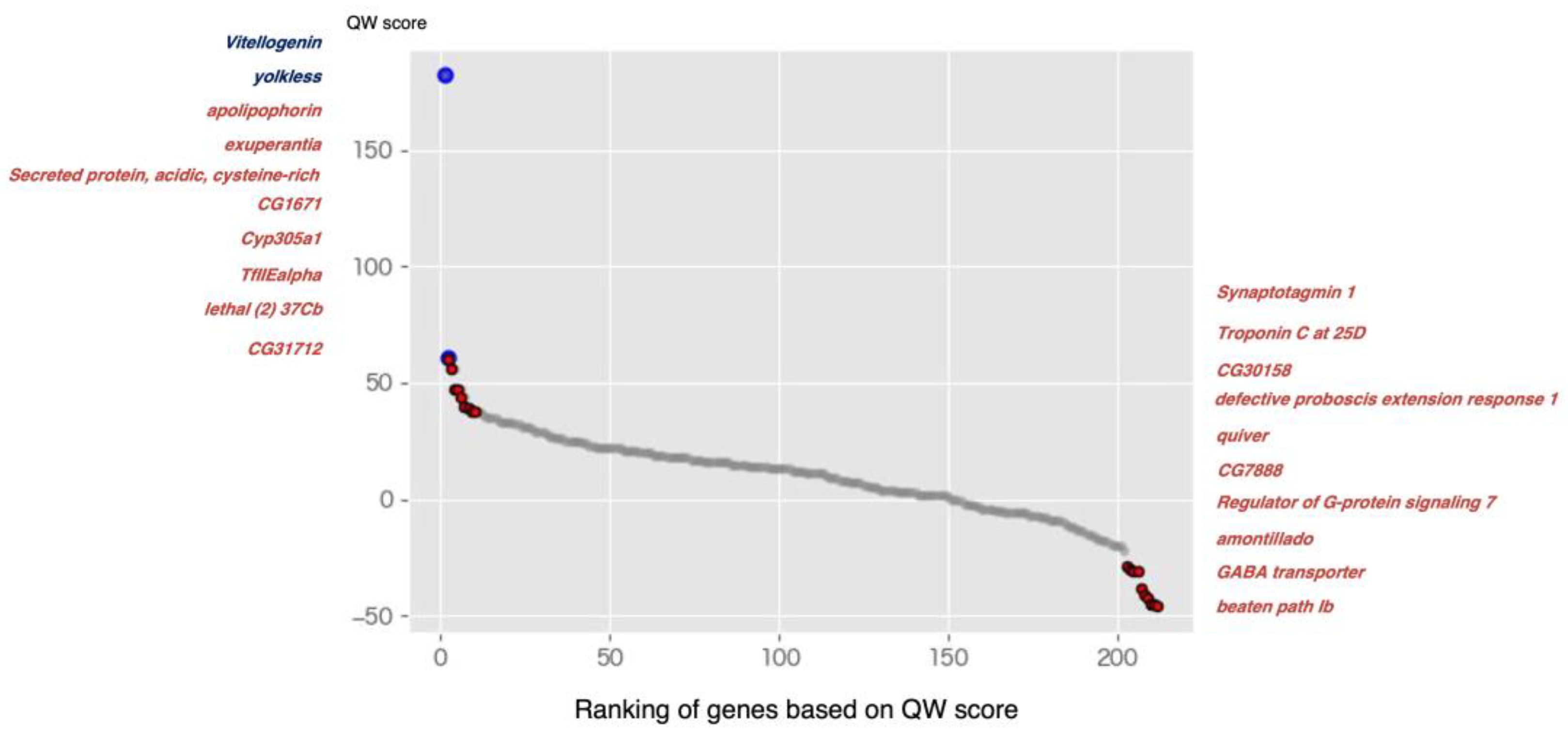
2.2. Meta-Analysis of RNA Sequencing Data in Social Insects
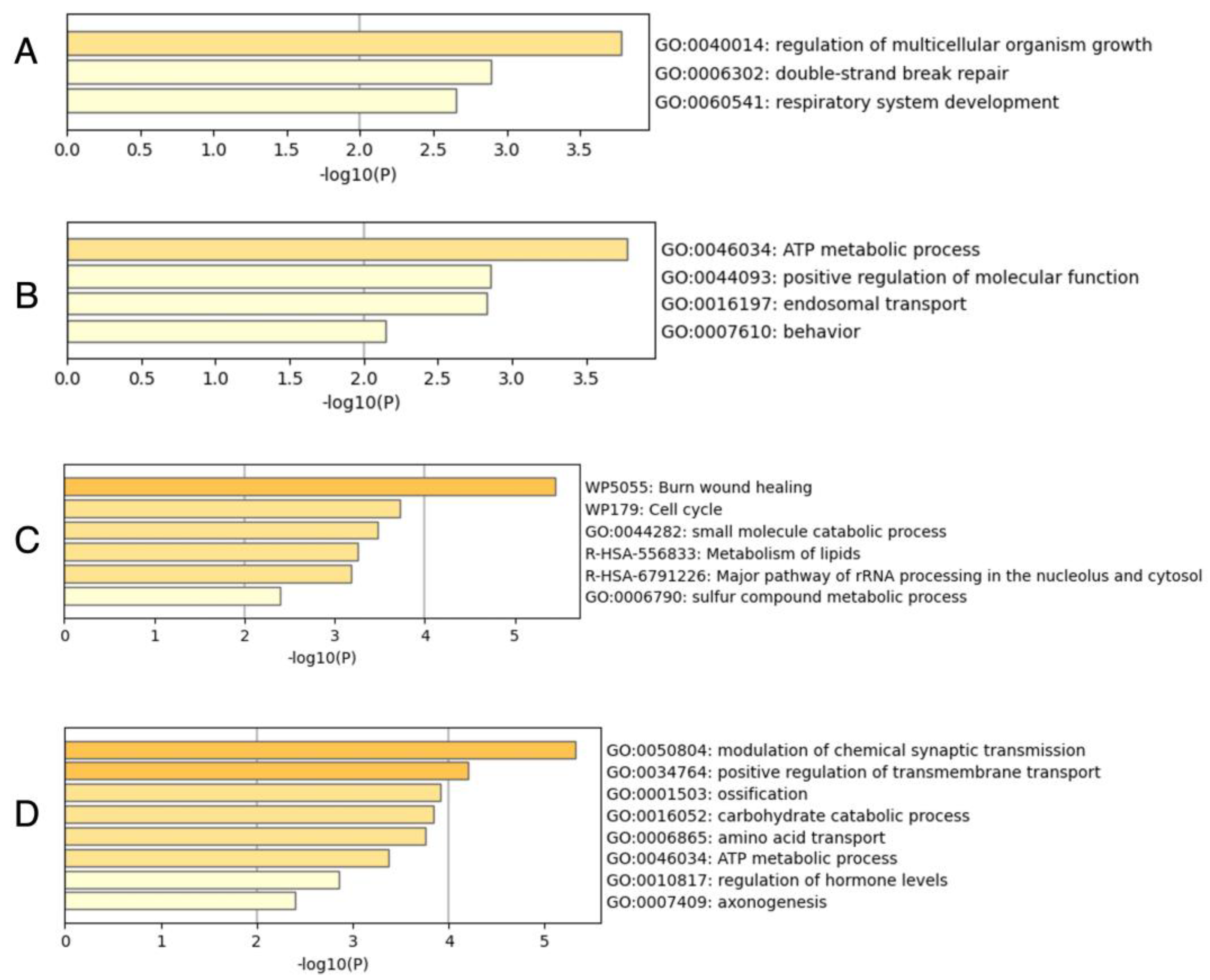
2.3. Enrichment Analysis
3. Discussion
3.1. Upregulated Genes in Queen
3.1.1. Oogenesis, Juvenile Hormone (JH) Binding, and Synthesis (Vitellogenin, yolkless, Cyp305a1, apolipophorin, and exuperantia)
3.1.2. The Regulation of Insulin Secretion (SPARC (Secreted Protein Acidic and Cysteine Rich) and RSG7 (Regulator of G Protein Signaling 7))
3.2. Upregulated Genes in Workers
3.2.1. The Regulation of Secretion of Neuropeptide and Neurotransmitter (amon (Amontillado) and Syt1 (Synaptotagmin 1))
3.2.2. Regulation of Circadian Sleep (Gat (GABA Transporter), quiver (qvr), and CG30158)
4. Materials and Methods
4.1. Curation of Public RNA Sequencing Data
4.2. RNA-Seq Data Retrieval, Processing, and Quantification
4.3. The Detection of Genes Related to the Reproductive Division of Labor
4.4. Annotation for Transcripts and Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Szathmáry, E.; Smith, J.M. The Major Evolutionary Transitions. Nature 1995, 374, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.E.; Grozinger, C.M.; Whitfield, C.W. Sociogenomics: Social Life in Molecular Terms. Nat. Rev. Genet. 2005, 6, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.; Weissing, F.J.; Pen, I.; Keller, L. An Evolutionary Perspective on Self-Organized Division of Labor in Social Insects. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 91–110. [Google Scholar] [CrossRef]
- Anderson, M. The Evolution of Eusociality. Annu. Rev. Ecol. Syst. 1984, 15, 165–189. [Google Scholar] [CrossRef]
- Fischman, B.J.; Woodard, S.H.; Robinson, G.E. Molecular Evolutionary Analyses of Insect Societies. Proc. Natl. Acad. Sci. USA 2011, 108, 10847–10854. [Google Scholar] [CrossRef]
- Tian, L.; Zhou, X. The Soldiers in Societies: Defense, Regulation, and Evolution. Int. J. Biol. Sci. 2014, 10, 296–308. [Google Scholar] [CrossRef]
- Woodard, S.H.; Fischman, B.J.; Venkat, A.; Hudson, M.E.; Varala, K.; Cameron, S.A.; Clark, A.G.; Robinson, G.E. Genes Involved in Convergent Evolution of Eusociality in Bees. Proc. Natl. Acad. Sci. USA 2011, 108, 7472–7477. [Google Scholar] [CrossRef]
- Simola, D.F.; Wissler, L.; Donahue, G.; Waterhouse, R.M.; Helmkampf, M.; Roux, J.; Nygaard, S.; Glastad, K.M.; Hagen, D.E.; Viljakainen, L.; et al. Social Insect Genomes Exhibit Dramatic Evolution in Gene Composition and Regulation While Preserving Regulatory Features Linked to Sociality. Genome Res. 2013, 23, 1235–1247. [Google Scholar] [CrossRef]
- Roux, J.; Privman, E.; Moretti, S.; Daub, J.T.; Robinson-Rechavi, M.; Keller, L. Patterns of Positive Selection in Seven Ant Genomes. Mol. Biol. Evol. 2014, 31, 1661–1685. [Google Scholar] [CrossRef]
- Kapheim, K.M.; Pan, H.; Li, C.; Salzberg, S.L.; Puiu, D.; Magoc, T.; Robertson, H.M.; Hudson, M.E.; Venkat, A.; Fischman, B.J.; et al. Genomic Signatures of Evolutionary Transitions from Solitary to Group Living. Science 2015, 348, 1139–1143. [Google Scholar] [CrossRef]
- Warner, M.R.; Qiu, L.; Holmes, M.J.; Mikheyev, A.S.; Linksvayer, T.A. Convergent Eusocial Evolution Is Based on a Shared Reproductive Groundplan plus Lineage-Specific Plastic Genes. Nat. Commun. 2019, 10, 2651. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, Z.; Lian, J.; Schiøtt, M.; Jin, L.; Zhang, P.; Zhang, Y.; Nygaard, S.; Peng, Z.; Zhou, Y.; et al. Caste-Specific RNA Editomes in the Leaf-Cutting Ant Acromyrmex echinatior. Nat. Commun. 2014, 5, 4943. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, C.; Bentley, M.; Taylor, D.; Favreau, E.; Brock, R.; Taylor, B.; Bell, E. Social Complexity, Life-History and Lineage Innuence the Molecular Basis of Castes in vespid wasps. Nat. Commun. 2023, 14, 1046. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Yokoi, K.; Toga, K. Bumble Bee Queens Activate Dopamine Production and Gene Expression in Nutritional Signaling Pathways in the Brain. Sci. Rep. 2021, 11, 5526. [Google Scholar] [CrossRef] [PubMed]
- Colgan, T.J.; Fletcher, I.K.; Arce, A.N.; Gill, R.J.; Ramos Rodrigues, A.; Stolle, E.; Chittka, L.; Wurm, Y. Caste- and Pesticide-specific Effects of Neonicotinoid Pesticide Exposure on Gene Expression in Bumblebees. Mol. Ecol. 2019, 28, 1964–1974. [Google Scholar] [CrossRef]
- Gospocic, J.; Shields, E.J.; Glastad, K.M.; Lin, Y.; Penick, C.A.; Yan, H.; Mikheyev, A.S.; Linksvayer, T.A.; Garcia, B.A.; Berger, S.L.; et al. The Neuropeptide Corazonin Controls Social Behavior and Caste Identity in Ants. Cell 2017, 170, 748–759.e12. [Google Scholar] [CrossRef] [PubMed]
- Feldmeyer, B.; Gstöttl, C.; Wallner, J.; Jongepier, E.; Séguret, A.; Grasso, D.A.; Bornberg-Bauer, E.; Foitzik, S.; Heinze, J. Evidence for a Conserved Queen-worker Genetic Toolkit across Slave-making Ants and Their Ant Hosts. Mol. Ecol. 2022, 31, 4991–5004. [Google Scholar] [CrossRef] [PubMed]
- Rau, V.; Korb, J. The Effect of Environmental Stress on Ageing in a Termite Species with Low Social Complexity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20190739. [Google Scholar] [CrossRef] [PubMed]
- Monroy Kuhn, J.M.; Meusemann, K.; Korb, J. Long Live the Queen, the King and the Commoner? Transcript Expression Differences between Old and Young in the Termite Cryptotermes secundus. PLoS ONE 2019, 14, e0210371. [Google Scholar] [CrossRef]
- Patalano, S.; Vlasova, A.; Wyatt, C.; Ewels, P.; Camara, F.; Ferreira, P.G.; Asher, C.L.; Jurkowski, T.P.; Segonds-Pichon, A.; Bachman, M.; et al. Molecular Signatures of Plastic Phenotypes in Two Eusocial Insect Species with Simple Societies. Proc. Natl. Acad. Sci. USA 2015, 112, 13970–13975. [Google Scholar] [CrossRef] [PubMed]
- Séguret, A.; Stolle, E.; Fleites-Ayil, F.A.; Quezada-Euán, J.J.G.; Hartfelder, K.; Meusemann, K.; Harrison, M.C.; Soro, A.; Paxton, R.J. Transcriptomic Signatures of Ageing Vary in Solitary and Social Forms of an Orchid Bee. Genome Biol. Evol. 2021, 13, evab075. [Google Scholar] [CrossRef] [PubMed]
- Morandin, C.; Brendel, V.P.; Sundström, L.; Helanterä, H.; Mikheyev, A.S. Changes in Gene DNA Methylation and Expression Networks Accompany Caste Specialization and Age-related Physiological Changes in a Social Insect. Mol. Ecol. 2019, 28, 1975–1993. [Google Scholar] [CrossRef] [PubMed]
- Qiu, B.; Larsen, R.S.; Chang, N.-C.; Wang, J.; Boomsma, J.J.; Zhang, G. Towards Reconstructing the Ancestral Brain Gene-Network Regulating Caste Differentiation in Ants. Nat. Ecol. Evol. 2018, 2, 1782–1791. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.C.; Jongepier, E.; Robertson, H.M.; Arning, N.; Bitard-Feildel, T.; Chao, H.; Childers, C.P.; Dinh, H.; Doddapaneni, H.; Dugan, S.; et al. Hemimetabolous Genomes Reveal Molecular Basis of Termite Eusociality. Nat. Ecol. Evol. 2018, 2, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.M.; Kingwell, C.J.; Wcislo, W.T.; Robinson, G.E. Caste-Biased Gene Expression in a Facultatively Eusocial Bee Suggests a Role for Genetic Accommodation in the Evolution of Eusociality. Proc. R. Soc. B 2017, 284, 20162228. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, Y.; Wen, T.; Liu, W.; Gao, Q.; Zhao, J.; Xiong, Z.; Wang, Z.; Jiang, W.; Yu, Y.; et al. Chromatin Accessibility and Transcriptome Landscapes of Monomorium pharaonis Brain. Sci. Data 2020, 7, 217. [Google Scholar] [CrossRef] [PubMed]
- Korb, J.; Meusemann, K.; Aumer, D.; Bernadou, A.; Elsner, D.; Feldmeyer, B.; Foitzik, S.; Heinze, J.; Libbrecht, R.; Lin, S.; et al. Comparative Transcriptomic Analysis of the Mechanisms Underpinning Ageing and Fecundity in Social Insects. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20190728. [Google Scholar] [CrossRef]
- Smith, C.R.; Helms Cahan, S.; Kemena, C.; Brady, S.G.; Yang, W.; Bornberg-Bauer, E.; Eriksson, T.; Gadau, J.; Helmkampf, M.; Gotzek, D.; et al. How Do Genomes Create Novel Phenotypes? Insights from the Loss of the Worker Caste in Ant Social Parasites. Mol. Biol. Evol. 2015, 32, 2919–2931. [Google Scholar] [CrossRef]
- Saiki, R.; Hayashi, Y.; Toga, K.; Yaguchi, H.; Masuoka, Y.; Suzuki, R.; Fujiwara, K.; Shigenobu, S.; Maekawa, K. Comparison of Gene Expression Profiles among Caste Differentiations in the Termite Reticulitermes speratus. Sci. Rep. 2022, 12, 11947. [Google Scholar] [CrossRef]
- Feldmeyer, B.; Elsner, D.; Foitzik, S. Gene Expression Patterns Associated with Caste and Reproductive Status in Ants: Worker-Specific Genes Are More Derived than Queen-Specific Ones. Mol. Ecol. 2014, 23, 151–161. [Google Scholar] [CrossRef]
- Mitaka, Y.; Kobayashi, K.; Matsuura, K. Caste-, Sex-, and Age-Dependent Expression of Immune-Related Genes in a Japanese Subterranean Termite, Reticulitermes speratus. PLoS ONE 2017, 12, e0175417. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, M.O.; Mikheyev, A.S. QTL Mapping of Sex Determination Loci Supports an Ancient Pathway in Ants and Honey Bees. PLoS Genet. 2015, 11, e1005656. [Google Scholar] [CrossRef] [PubMed]
- Glastad, K.M.; Gokhale, K.; Liebig, J.; Goodisman, M.A.D. The Caste- and Sex-Specific DNA Methylome of the Termite Zootermopsis nevadensis. Sci. Rep. 2016, 6, 37110. [Google Scholar] [CrossRef] [PubMed]
- Hartfelder, K. Insect Juvenile Hormone: From “status quo” to high society. Braz. J. Med. Biol. Res. 2000, 33, 157–177. [Google Scholar] [CrossRef]
- Saiki, R.; Gotoh, H.; Toga, K.; Miura, T.; Maekawa, K. High Juvenile Hormone Titre and Abdominal Activation of JH Signalling May Induce Reproduction of Termite Neotenics. Insect Mol. Biol. 2015, 24, 432–441. [Google Scholar] [CrossRef]
- Brent, C.S.; Vargo, E.L. Changes in Juvenile Hormone Biosynthetic Rate and Whole Body Content in Maturing Virgin Queens of Solenopsis invicta. J. Insect Physiol. 2003, 49, 967–974. [Google Scholar] [CrossRef]
- Cornette, R.; Gotoh, H.; Koshikawa, S.; Miura, T. Juvenile Hormone Titers and Caste Differentiation in the Damp-Wood Termite Hodotermopsis sjostedti (Isoptera, Termopsidae). J. Insect Physiol. 2008, 54, 922–930. [Google Scholar] [CrossRef]
- Bloch, G.; Borst, D.W.; Huang, Z.-Y.; Robinson, G.E.; Cnaani, J.; Hefetz, A. Juvenile Hormone Titers, Juvenile Hormone Biosynthesis, Ovarian Development and Social Environment in Bombus terrestris. J. Insect Physiol. 2000, 46, 47–57. [Google Scholar] [CrossRef]
- Waring, G.L.; Diorio, J.P.; Hennen, S. Isolation of Germ Line-Dependent Female-Sterile Mutation That Affects Yolk Specific Sequestration and Chorion Formation in Drosophila. Dev. Biol. 1983, 100, 452–463. [Google Scholar] [CrossRef]
- Sappington, T.W.; Raikhel, A.S. Molecular Characteristics of Insect Vitellogenins and Vitellogenin Receptors. Insect Biochem. Mol. Biol. 1998, 28, 277–300. [Google Scholar] [CrossRef]
- Soller, M.; Bownes, M.; Kubli, E. Control of Oocyte Maturation in Sexually Mature Drosophila Females. Dev. Biol. 1999, 208, 337–351. [Google Scholar] [CrossRef]
- Saiki, R.; Maekawa, K. Imaginal Organ Development and Vitellogenin Gene Expression Changes during the Dierentiation of Nymphoids of the Termite Reticulitermes speratus. Sociobiology 2011, 58, 499–512. [Google Scholar]
- Zhang, W.; Wang, L.; Zhao, Y.; Wang, Y.; Chen, C.; Hu, Y.; Zhu, Y.; Sun, H.; Cheng, Y.; Sun, Q.; et al. Single-Cell Transcriptomic Analysis of Honeybee Brains Identifies Vitellogenin as Caste Differentiation-Related Factor. iScience 2022, 25, 104643. [Google Scholar] [CrossRef] [PubMed]
- Corona, M.; Libbrecht, R.; Wurm, Y.; Riba-Grognuz, O.; Studer, R.A.; Keller, L. Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant Pogonomyrmex barbatus. PLoS Genet. 2013, 9, e1003730. [Google Scholar] [CrossRef] [PubMed]
- Yaguchi, H.; Suzuki, S.; Kanasaki, N.; Masuoka, Y.; Suzuki, R.; Suzuki, R.H.; Hayashi, Y.; Shigenobu, S.; Maekawa, K. Evolution and Functionalization of Vitellogenin Genes in the Termite Reticulitermes speratus. J. Exp. Zool. B Mol. Dev. Evol. 2022, 34, 68–80. [Google Scholar] [CrossRef]
- Wurm, Y.; Wang, J.; Riba-Grognuz, O.; Corona, M.; Nygaard, S.; Hunt, B.G.; Ingram, K.K.; Falquet, L.; Nipitwattanaphon, M.; Gotzek, D.; et al. The Genome of the Fire Ant Solenopsis invicta. Proc. Natl. Acad. Sci. USA 2011, 108, 5679–5684. [Google Scholar] [CrossRef]
- Miyazaki, S.; Shimoji, H.; Suzuki, R.; Chinushi, I.; Takayanagi, H.; Yaguchi, H.; Miura, T.; Maekawa, K. Expressions of Conventional Vitellogenin and Vitellogenin-like A in Worker Brains Are Associated with a Nursing Task in a Ponerine Ant. Insect Mol. Biol. 2021, 30, 113–121. [Google Scholar] [CrossRef]
- Chandra, V.; Fetter-Pruneda, I.; Oxley, P.R.; Ritger, A.L.; McKenzie, S.K.; Libbrecht, R.; Kronauer, D.J.C. Social Regulation of Insulin Signaling and the Evolution of Eusociality in Ants. Science 2018, 361, 398–402. [Google Scholar] [CrossRef]
- Séité, S.; Harrison, M.C.; Sillam-Dussès, D.; Lupoli, R.; van Dooren, T.J.M.; Robert, A.; Poissonnier, L.-A.; Lemainque, A.; Renault, D.; Acket, S.; et al. Lifespan Prolonging Mechanisms and Insulin Upregulation without Fat Accumulation in Long-Lived Reproductives of a Higher Termite. Commun. Biol. 2022, 5, 44. [Google Scholar] [CrossRef]
- Corona, M.; Velarde, R.A.; Remolina, S.; Moran-Lauter, A.; Wang, Y.; Hughes, K.A.; Robinson, G.E. Vitellogenin, Juvenile Hormone, Insulin Signaling, and Queen Honey Bee Longevity. Proc. Natl. Acad. Sci. USA 2007, 104, 7128–7133. [Google Scholar] [CrossRef]
- Sasaki, K.; Okada, Y.; Shimoji, H.; Aonuma, H.; Miura, T.; Tsuji, K. Social Evolution with Decoupling of Multiple Roles of Biogenic Amines into Different Phenotypes in Hymenoptera. Front. Ecol. Evol. 2021, 9, 659160. [Google Scholar] [CrossRef]
- Meta-Analysis in Basic Biology. Nat. Methods 2016, 13, 959. [CrossRef]
- Bono, H. Meta-Analysis of Oxidative Transcriptomes in Insects. Antioxidants 2021, 10, 345. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.R.; Petralia, M.C.; Ciurleo, R.; Bramanti, A.; Fagone, P.; Shahjaman, M.; Wu, L.; Sun, Y.; Turanli, B.; Arga, K.Y.; et al. Comprehensive Analysis of RNA-Seq Gene Expression Profiling of Brain Transcriptomes Reveals Novel Genes, Regulators, and Pathways in Autism Spectrum Disorder. Brain Sci. 2020, 10, 747. [Google Scholar] [CrossRef] [PubMed]
- Shaar-Moshe, L.; Hübner, S.; Peleg, Z. Identification of Conserved Drought-Adaptive Genes Using a Cross-Species Meta-Analysis Approach. BMC Plant Biol. 2015, 15, 111. [Google Scholar] [CrossRef]
- Ono, Y.; Bono, H. Multi-Omic Meta-Analysis of Transcriptomes and the Bibliome Uncovers Novel Hypoxia-Inducible Genes. Biomedicines 2021, 9, 582. [Google Scholar] [CrossRef]
- Suzuki, T.; Ono, Y.; Bono, H. Comparison of Oxidative and Hypoxic Stress Responsive Genes from Meta-Analysis of Public Transcriptomes. Biomedicines 2021, 9, 1830. [Google Scholar] [CrossRef]
- Tamura, K.; Bono, H. Meta-Analysis of RNA Sequencing Data of Arabidopsis and Rice under Hypoxia. Life 2022, 12, 1079. [Google Scholar] [CrossRef]
- Toga, K.; Yokoi, K.; Bono, H. Meta-Analysis of Transcriptomes in Insects Showing Density-Dependent Polyphenism. Insects 2022, 13, 864. [Google Scholar] [CrossRef]
- Yokoi, K.; Wakamiya, T.; Bono, H. Meta-Analysis of the Public RNA-Seq Data of the Western Honeybee Apis Mellifera to Construct Reference Transcriptome Data. Insects 2022, 13, 931. [Google Scholar] [CrossRef]
- Sevala, V.L.; Bachmann, J.A.S.; Schal, C. Lipophorin: A Hemolymph Juvenile Hormone Binding Protein in the German Cockroach, Blattella germanica. Insect Biochem. Mol. Biol. 1997, 27, 663–670. [Google Scholar] [CrossRef]
- Cardoen, D.; Wenseleers, T.; Ernst, U.R.; Danneels, E.L.; Laget, D.; de Graaf, D.C.; Schoofs, L.; Verleyen, P. Genome-Wide Analysis of Alternative Reproductive Phenotypes in Honeybee Workers. Mol. Ecol. 2011, 20, 4070–4084. [Google Scholar] [CrossRef] [PubMed]
- Ephrussi, A.; Johnston, D.S. Seeing Is Believing. Cell 2004, 116, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; He, F.; Huang, M.; Zhao, Q.; Cheng, L.; Said, N.; Zhou, Z.; Liu, F.; Dai, Y.-S. SPARC Promotes Insulin Secretion through Down-Regulation of RGS4 Protein in Pancreatic β Cells. Sci. Rep. 2020, 10, 17581. [Google Scholar] [CrossRef]
- Kressler, D.; Hurt, E.; Baßler, J. A Puzzle of Life: Crafting Ribosomal Subunits. Trends Biochem. Sci. 2017, 42, 640–654. [Google Scholar] [CrossRef]
- Rahman, M.M.; Franch-Marro, X.; Maestro, J.L.; Martin, D.; Casali, A. Local Juvenile Hormone Activity Regulates Gut Homeostasis and Tumor Growth in Adult Drosophila. Sci. Rep. 2017, 7, 11677. [Google Scholar] [CrossRef]
- Phan, T.; Maity, P.; Ludwig, C.; Streit, L.; Michaelis, J.; Tsesmelis, M.; Scharffetter-Kochanek, K.; Iben, S. Nucleolar TFIIE Plays a Role in Ribosomal Biogenesis and Performance. Nucleic Acids Res. 2021, 49, 11197–11210. [Google Scholar] [CrossRef]
- Yang, H.; Beutler, B.; Zhang, D. Emerging Roles of Spliceosome in Cancer and Immunity. Protein Cell 2022, 13, 559–579. [Google Scholar] [CrossRef]
- Guruharsha, K.G.; Rual, J.-F.; Zhai, B.; Mintseris, J.; Vaidya, P.; Vaidya, N.; Beekman, C.; Wong, C.; Rhee, D.Y.; Cenaj, O.; et al. A Protein Complex Network of Drosophila melanogaster. Cell 2011, 147, 690–703. [Google Scholar] [CrossRef]
- Manfredini, F.; Lucas, C.; Nicolas, M.; Keller, L.; Shoemaker, D.; Grozinger, C.M. Molecular and Social Regulation of Worker Division of Labour in Fire Ants. Mol. Ecol. 2014, 23, 660–672. [Google Scholar] [CrossRef]
- de Wit, H.; Walter, A.M.; Milosevic, I.; Gulyás-Kovács, A.; Riedel, D.; Sørensen, J.B.; Verhage, M. Synaptotagmin-1 Docks Secretory Vesicles to Syntaxin-1/SNAP-25 Acceptor Complexes. Cell 2009, 138, 935–946. [Google Scholar] [CrossRef]
- Cheng, H.-Y.M.; Obrietan, K.; Cain, S.W.; Lee, B.Y.; Agostino, P.V.; Joza, N.A.; Harrington, M.E.; Ralph, M.R.; Penninger, J.M. Dexras1 Potentiates Photic and Suppresses Nonphotic Responses of the Circadian Clock. Neuron 2004, 43, 715–728. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-Y.M.; Obrietan, K. Dexras1: Shaping the Responsiveness of the Circadian Clock. Semin. Cell Dev. Biol. 2006, 17, 345–351. [Google Scholar] [CrossRef]
- Sanes, J.R.; Zipursky, S.L. Synaptic Specificity, Recognition Molecules, and Assembly of Neural Circuits. Cell 2020, 181, 536–556. [Google Scholar] [CrossRef] [PubMed]
- de Souza Araujo, N.; Arias, M.C. Gene Expression and Epigenetics Reveal Species-Specific Mechanisms Acting upon Common Molecular Pathways in the Evolution of Task Division in Bees. Sci. Rep. 2021, 11, 3654. [Google Scholar] [CrossRef]
- Koh, K.; Joiner, W.J.; Wu, M.N.; Yue, Z.; Smith, C.J.; Sehgal, A. Identification of SLEEPLESS, a Sleep-Promoting Factor. Science 2008, 321, 372–376. [Google Scholar] [CrossRef]
- Chaturvedi, R.; Stork, T.; Yuan, C.; Freeman, M.R.; Emery, P. Astrocytic GABA Transporter Controls Sleep by Modulating GABAergic Signaling in Drosophila Circadian Neurons. Curr. Biol. 2022, 32, 1895–1908.e5. [Google Scholar] [CrossRef] [PubMed]
- Wegener, C.; Herbert, H.; Kahnt, J.; Bender, M.; Rhea, J.M. Deficiency of Prohormone Convertase DPC2 (AMONTILLADO) Results in Impaired Production of Bioactive Neuropeptide Hormones in Drosophila. J. Neurochem. 2011, 118, 581–595. [Google Scholar] [CrossRef]
- Pipes, G.C.T.; Lin, Q.; Riley, S.E.; Goodman, C.S. The Beat Generation: A Multigene Family Encoding IgSF Proteins Related to the Beat Axon Guidance Molecule in Drosophila. Development 2001, 128, 4545–4552. [Google Scholar] [CrossRef]
- Robinson, G.E.; Vargo, E.L. Juvenile Hormone in Adult Eusocial Hymenoptera: Gonadotropin and Behavioral Pacemaker. Arch. Insect Biochem. Physiol. 1997, 35, 559–583. [Google Scholar] [CrossRef]
- Penick, C.A.; Liebig, J.; Brent, C.S. Reproduction, Dominance, and Caste: Endocrine Profiles of Queens and Workers of the Ant Harpegnathos saltator. J. Comp. Physiol. A 2011, 197, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Krause, S.A.; Overend, G.; Dow, J.A.T.; Leader, D.P. FlyAtlas 2 in 2022: Enhancements to the Drosophila melanogaster Expression Atlas. Nucleic Acids Res. 2022, 50, D1010–D1015. [Google Scholar] [CrossRef]
- Ferreira, P.G.; Patalano, S.; Chauhan, R.; Ffrench-Constant, R.; Gabaldón, T.; Guigó, R.; Sumner, S. Transcriptome Analyses of Primitively Eusocial Wasps Reveal Novel Insights into the Evolution of Sociality and the Origin of Alternative Phenotypes. Genome Biol. 2013, 14, R20. [Google Scholar] [CrossRef]
- Muscedere, M.L.; Johnson, N.; Gillis, B.C.; Kamhi, J.F.; Traniello, J.F.A. Serotonin Modulates Worker Responsiveness to Trail Pheromone in the Ant Pheidole dentata. J. Comp. Physiol. A 2012, 198, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Klein, B.A.; Klein, A.; Wray, M.K.; Mueller, U.G.; Seeley, T.D. Sleep Deprivation Impairs Precision of Waggle Dance Signaling in Honey Bees. Proc. Natl. Acad. Sci. USA 2010, 107, 22705–22709. [Google Scholar] [CrossRef]
- Fuchikawa, T.; Shimizu, I. Circadian Rhythm of Locomotor Activity in the Japanese Honeybee, Apis Cerana japonica. Physiol. Entomol. 2007, 32, 73–80. [Google Scholar] [CrossRef]
- Kaiser, W.; Steiner-Kaiser, J. Neuronal Correlates of Sleep, Wakefulness and Arousal in a Diurnal Insect. Nature 1983, 301, 707–709. [Google Scholar] [CrossRef]
- Klein, B.A.; Seeley, T.D. Work or Sleep? Honeybee Foragers Opportunistically Nap during the Day When Forage Is Not Available. Anim. Behav. 2011, 82, 77–83. [Google Scholar] [CrossRef]
- Bono, H. All of Gene Expression (AOE): An Integrated Index for Public Gene Expression Databases. PLoS ONE 2020, 15, e0227076. [Google Scholar] [CrossRef]
- The NCBI SRA (Sequence Read Archive); NCBI—National Center for Biotechnology Information/NLM/NIH: Bethesda, MD, USA, 2021.
- Krueger, F. Trim Galore. Available online: https://www.Bioinformatics.Babraham.Ac.Uk/Projects/Trim_galore/ (accessed on 30 April 2021).
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-Level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000research 2020, 9, 304. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
| RANK | QW Score | Gene ID of A. mellifera | Gene Symbol of D. melanogaster | Gene Symbol of H. sapiens | Tissue-Expression Profile in Female Adult D. melanogaster | Caste with High Expression that Was Reported in Previous Study | Biological/ Molecular Function |
|---|---|---|---|---|---|---|---|
| 1 | 182 | NP_001011578.1 (Vitellogenin) | - | - | - | Queen in many species [42,43,44,45,47] | Oogenesis [39,40] |
| 2 | 61 | XP_026295652.1 | yl | LRP2 | ovary | Queen in Diacamma sp.; A. mellifera; M. pharaonis; T. longispinosus [11,30,47] | Oogenesis [39,40] |
| 3 | 60 | XP_026298285.1 | apolpp | GTPBP2 | fat body, spermatheca, head, eye | Queen in R. speratus [31] | JH binding [61] |
| 4 | 56 | XP_623977.2 | exu | - | ovary | Queen in A. mellifera; T. longispinosus [30,62] | Oogenesis [63] |
| 5 | 47 | XP_623079.2 | SPARC | SPARC | heart, eye, head thoracicoabdominal ganglion | - | Insulin secretion [64] |
| 6 | 47 | XP_001120493.3 | CG1671 | TBL3 | relatively high in ovary | - | Ribosome biogenesis [65] |
| 7 | 44 | NP_001314895.1 | Cyp305a1 | CYP2A13 | spermatheca, head | - | JH synthesis [66] |
| 8 | 40 | XP_395253.2 | TfIIEalpha | GTF2E1 | relatively high in ovary | - | Ribosome biogenesis [67] |
| 9 | 39 | XP_026300350.1 | lethal (2) 37Cb | DHX16 | relatively high in heart | - | Splicesome [68] |
| 10 | 38 | XP_395437.4 | CG31712 | GKAP1 | brain, thoracicoabdominal ganglion, head | - | Splicesome [69] |
| 203 | −29 | XP_026295297.1 | Syt1 | SYT1 | thoracicoabdominal ganglion, brain, head | worker (forager) in A. mellifera; P. metricus; S. invicta [70] | Neurotransmitter release at synapses [71] |
| 204 | −30 | XP_026299328.1 | TpnC25D | CALM3 | head, eye, gut | - | A calcium signal transduction pathway (Uniprot) |
| 205 | −31 | XP_026298569.1 | CG30158 | RASD1 | brain, head, thoracicoabdominal ganglion | - | Circadian entrainment [72,73] |
| 206 | −31 | XP_026300722.1 | dpr1 | CNTN4 | brain, thoracicoabdominal ganglion, head | - | Immunoglobulin superfamily [74] |
| 207 | −38 | XP_026302000.1 | qvr | LYPD6 | thoracicoabdominal ganglion, head, eye | worker (forager) in B. terrestris [75] | Sleep [76] |
| 208 | −41 | XP_393138.2 | CG7888 | SLC36A1 | hindgut, thoracicoabdominal ganglion, brain | - | Amino acid transport (Uniprot) |
| 209 | −42 | XP_393403.3 | RSG7 | RGS7 | brain, thoracicoabdominal ganglion, head | - | Insulin secretion [64] |
| 210 | −45 | XP_006571520.1 | Gat | SLC6A1 | thoracicoabdominal ganglion, brain, head | Worker in A. mellifera [62] | regulation of circadian sleep [77] |
| 211 | −45 | XP_392366.2 | amon | PCSK2 | brain, thoracicoabdominal ganglion, head | - | The production of bioactive neuropeptide hormones [78] |
| 212 | −46 | XP_026298737.1 | beat | PSG4 | brain, thoracicoabdominal ganglion, head | - | Immunoglobulin superfamily [74,79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toga, K.; Bono, H. Meta-Analysis of Public RNA Sequencing Data Revealed Potential Key Genes Associated with Reproductive Division of Labor in Social Hymenoptera and Termites. Int. J. Mol. Sci. 2023, 24, 8353. https://doi.org/10.3390/ijms24098353
Toga K, Bono H. Meta-Analysis of Public RNA Sequencing Data Revealed Potential Key Genes Associated with Reproductive Division of Labor in Social Hymenoptera and Termites. International Journal of Molecular Sciences. 2023; 24(9):8353. https://doi.org/10.3390/ijms24098353
Chicago/Turabian StyleToga, Kouhei, and Hidemasa Bono. 2023. "Meta-Analysis of Public RNA Sequencing Data Revealed Potential Key Genes Associated with Reproductive Division of Labor in Social Hymenoptera and Termites" International Journal of Molecular Sciences 24, no. 9: 8353. https://doi.org/10.3390/ijms24098353
APA StyleToga, K., & Bono, H. (2023). Meta-Analysis of Public RNA Sequencing Data Revealed Potential Key Genes Associated with Reproductive Division of Labor in Social Hymenoptera and Termites. International Journal of Molecular Sciences, 24(9), 8353. https://doi.org/10.3390/ijms24098353







