Abstract
Introduction: Healthcare-associated infections are a major concern for healthcare systems around the world. Microorganisms developing resistance to potent antibiotics are an urgent threat to public health. Methods: The present study is a retrospective, single-center study performed at the Mina Minovici National Institute of Legal Medicine, Bucharest, Romania, over a period of ten years (2011–2020). Autopsies for deaths occurring in the hospital setting for which postmortem bacteriological examination was solicited were screened and the recovered data consisted of demographics, hospital stay duration, autopsy data, and postmortem microbiology. Results: In the 516 autopsies recovered we found that carbapenemase-producing Enterobacteriaceae (CPE) isolates from postmortem bacteriology results increased in 2019. Positive postmortem microbiology results were associated with histological infection in over 80% of cases. Positive results for healthcare-related pathogens were associated with prolonged hospital stay. In our data vancomycin-resistant enterococci were isolated from 2015. Conclusions: Postmortem bacteriology results from medico-legal autopsies mirror antimicrobial resistance trends from hospital settings with several limitations due to the scarcity of solicitations.
Introduction
Healthcare-associated infections are a major concern for healthcare systems around the world. The European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group of Forensic and Postmortem Microbiology (ESGFOR) evaluated postmortem microbiology sampling following in-hospital deaths. The consensus statement of the study group identified three primary reasons to procure postmortem microbiology from biological samples: identification of the etiological agent of a previously undiagnosed infection, confirmation of an ante-mortem presumed diagnosis and determination of the effectiveness of antimicrobial therapy [1]. Regarding multidrug-resistant microorganisms specific to the hospital settings, the urgent threats indicated by the Centers for Disease Control and Prevention and World Health Organization are carbapenem-resistant Enterobacteriaceae, multidrug-resistant Acinetobacter, and multidrug-resistant Pseudomonas aeruginosa [2] occurring in healthcare-associated infections (HCAIs); the most challenging microorganisms developing resistance are summarized by the ESKAPE acronym: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. [3]. The National Institute of Public Health, Bucharest, Romania participates in antimicrobial resistance (AMR) surveillance in Europe with an estimated national population coverage of 21% in 2020, the latest report showing an increase of carbapenem-resistant strains of Klebsiella pneumoniae and Acinetobacter baumannii between 2016 and 2020 [4].
The present study aimed to characterize over a period of ten years the trends of antimicrobial resistance encountered in postmortem microbiological samples taken during medico-legal autopsies performed at the Mina Minovici National Institute of Legal Medicine in Bucharest, Romania, respectively violent or suspect deaths occurring in all hospitals of the capital.
Methods
We performed a retrospective, single-center study at the Mina Minovici National Institute of Legal Medicine, Bucharest, Romania. Autopsy cases from 2011 to 2020 were screened and we selected the cases that met the following inclusion criteria: death during the hospitalization, collection of biological samples for postmortem microbiology examinations, and availability of a full autopsy report. We collected demographic data (age, gender), date of death, date of the autopsy, medical unit, date of hospital admission, date of intensive care unit (ICU) admission, results of hospital microbiology, diagnosis of infection, manner of death, medical cause of death, pathology result, biological sample type and results of microbiological examinations. From the retrieved data we were able to calculate the hospital stay (days), ICU stay (days) and postmortem interval (days, from date of death to date of autopsy/ date of sampling). For data analysis, we used IBM SPSS Statistics for Windows version 24.0 (IBM Corp, USA). As the population did not have a normal distribution, continuous variables were described by median and IQR (25th, 75th percentile) while categorical data was defined as frequencies. Descriptive data were compared using the Chi-Square test, while comparison of differences between groups was performed by using ANOVA. A p-value below 0.05 was considered statistically significant.
The cases included in the study came from 29 medical units in Bucharest which were anonymized using Hn (H1-H29) and divided into 6 categories: 1-Emergency Hospital, 2-Chronic Disease Hospital, 3-Children’s Hospital, 4-Infectious Diseases Hospital, 5-Private Hospital, 6-Maternity.
In the study center, biological samples are collected by a trained assistant and within the first 24 h after sampling they are transported to an external collaborator of the Institute, the laboratory at the “Victor Babeș” Private Medical Clinic. Over the ten years period all postmortem bacteriology results specified the microorganisms and the type of antimicrobial resistance without listing the specific antibiotics tested, respectively extended-spectrum-β-lactamase (ESBL) producers, carbapenem-resistant (CR) isolates either carbapenemase producers (CP) or non-carbapenemase producers (NCP), multidrug-resistant (MDR) variants, vancomycin-resistant (VR) isolates.
Ethics statement
The study was approved by the Institutional Research Ethics Committee of Mina Minovici National Institute of Legal Medicine, Bucharest, Romania. All medico-legal autopsies in Romania are mandated by authorities. Institutional access to the archives was obtained. All data retrieved for the study was anonymized before analysis. Waiver of consent was granted, as the informed consent of relatives was not deemed feasible considering the time frame from which the data was retrieved.
Results
Microbiological sample analysis was requested in few of the autopsies for in-hospital deaths performed in our center (1.6-5.4% cases/year) and included in the study 1.6%-5.0% autopsies/year. We identified 516 medico-legal autopsies that met the inclusion criteria.
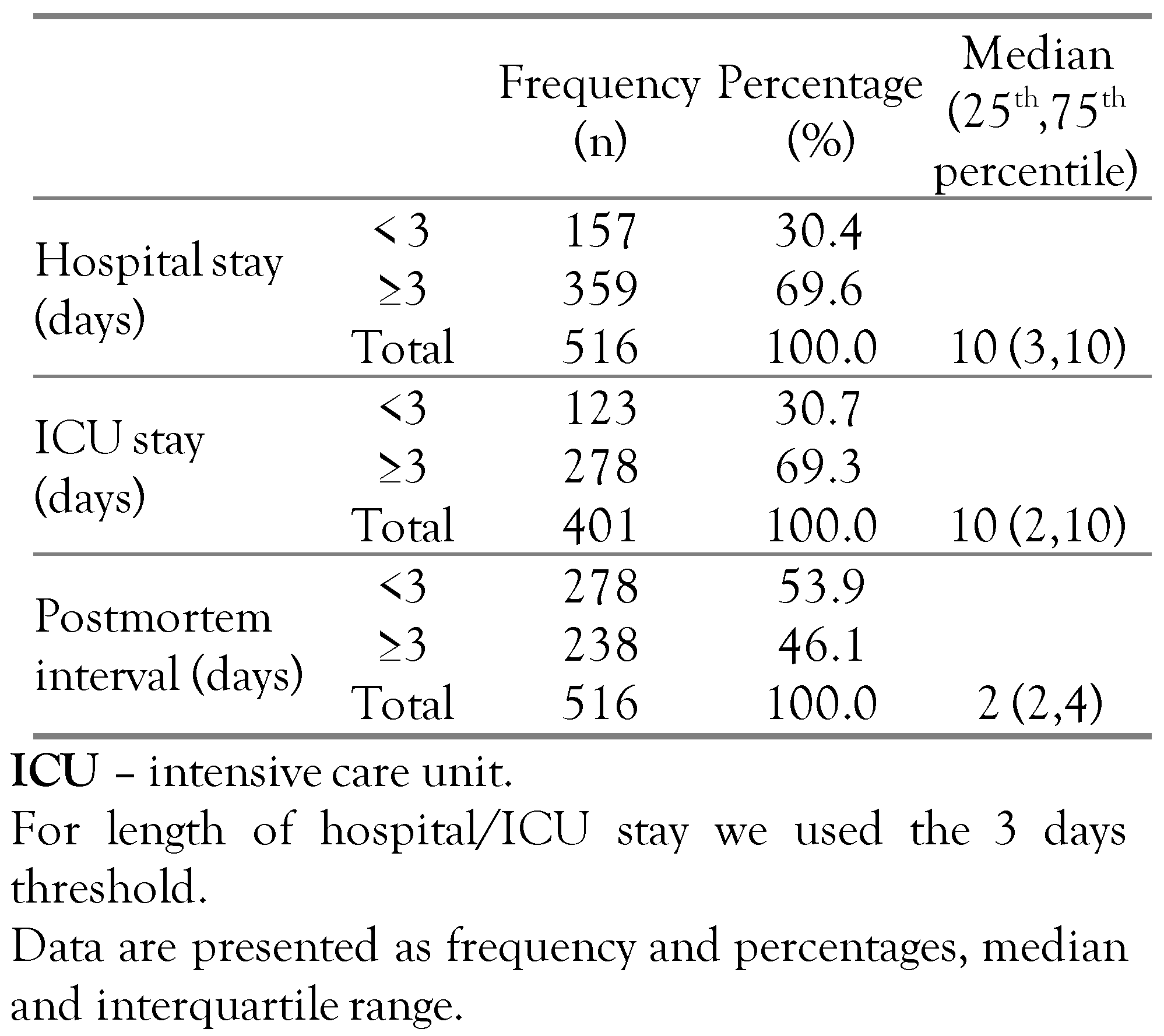
The demographic characteristics of the study population were as follows: 310 males (60.1%) and 206 female (39.9%), age range between 0 and 94 years, with a median and interquartile range age of 57 years (35, 70). The length of hospitalization was between one day (1) and 232 days, with a median of 10 days (3, 10); 401 patients (77.7%) were also admitted to intensive care units (ICU) and had the longest hospitalization periods. The postmortem interval before the forensic autopsy and the collection of biological samples respectively were between one day (1) and 8 days postmortem with a median of 2 days (Table 1).

Table 1.
Descriptive statistics of study population showing hospital and ICU stay and postmortem interval as binary variables.
Cases included in the study came mostly from emergency hospitals (69.8%) followed by chronic disease hospitals (12%). Antimicrobial-resistant strains (ESBL, CP, MDR–predominantly A. baumannii) were mostly identified in emergency hospitals, over 75% of AMR strains, and vancomycin-resistant enterococci were found solely in this type of unit.
According to the medical records provided for forensic autopsies, the diagnosis of infection was recorded in 346 cases (67.1%/study cases, admission and discharge diagnoses) independent of the number of days of hospitalization or the number of days of ICU (p>0.05). Data on microbiological tests performed during the hospital stay was found for 233 (45.2%) cases of which 21 (4.1%) had negative results. Out of all cases, infections were included in the mechanism of death in 437 cases (84.7%). The manner of death was assessed as violent in 185 cases (35.9%) and nonviolent in 315 cases (61.0%). Exceptionally in 16 cases (3.1%) the manner of death was not recorded, as these were postoperative deaths in which medical care errors were identified.
The biological products taken for laboratory examinations varied with a predominance of samples for postmortem blood cultures. Ranking second were lung tissue samples and tracheal secretion swabs, followed by skin/wound tampons and skin fragments. We found 14 variables corresponding to the types of biological samples collected for postmortem bacteriology thus we assessed the combinations in order to evaluate the sampling protocol in the study center. The summary of the missing values revealed that only 12.9% of the studied variables had complete data. We found that 96 sets (combinations) of biological products were used for the 516 cases included in the study.
Except for biological examinations of skin samples and wound swabs which were all positive, microbiological examinations of other biological products sampled postmortem resulted in negative cultures in a relatively small percentage (from 8.6% in blood cultures to 57.1% for pericardial fluid). Positive postmortem cultures identified: Enterobacteriaceae in over 50% of each type of sample, Enterococcus in 21.5% to 40.3%, and staphylococci in 11.4% to 27.7%. Other microorganisms of interest for healthcare-associated infections, Acinetobacter baumannii and Pseudomonas aeruginosa, were found in a high percentage: up to 46.2% of wound and skin cultures.
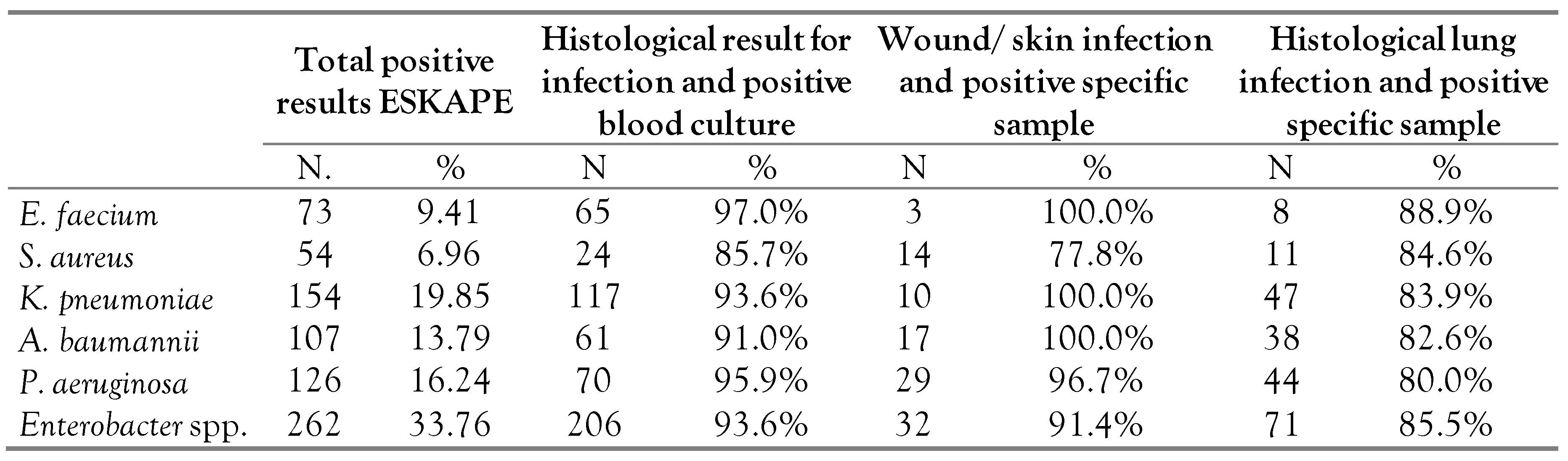
Among the ESKAPE acronym microorganisms, Enterobacter was predominant—identified in 33.7% (262 cases) while S. aureus was the least common—6.9% (54 cases)—Table 2.

Table 2.
Pathogens known under the acronym ESKAPE: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp.
ESKAPE microorganisms identified in wound and skin cultures had a histological result for infection in more than 90% of each microorganism except for S. aureus which appeared in 22.2% of cases without infection (4 cases, 3 methicillin-susceptible Staphylococcus aureus, and 1 methicillin-resistant Staphylococcus aureus)—Table 2.
Lung and tracheal swab cultures were associated with a histologically confirmed lung infection in over 80% of cases, regardless of the ESKAPE microorganism involved (Table 2).
Blood cultures were associated with a histologically confirmed infection in over 85% of cases, regardless of the ESKAPE microorganism involved (Table 2).
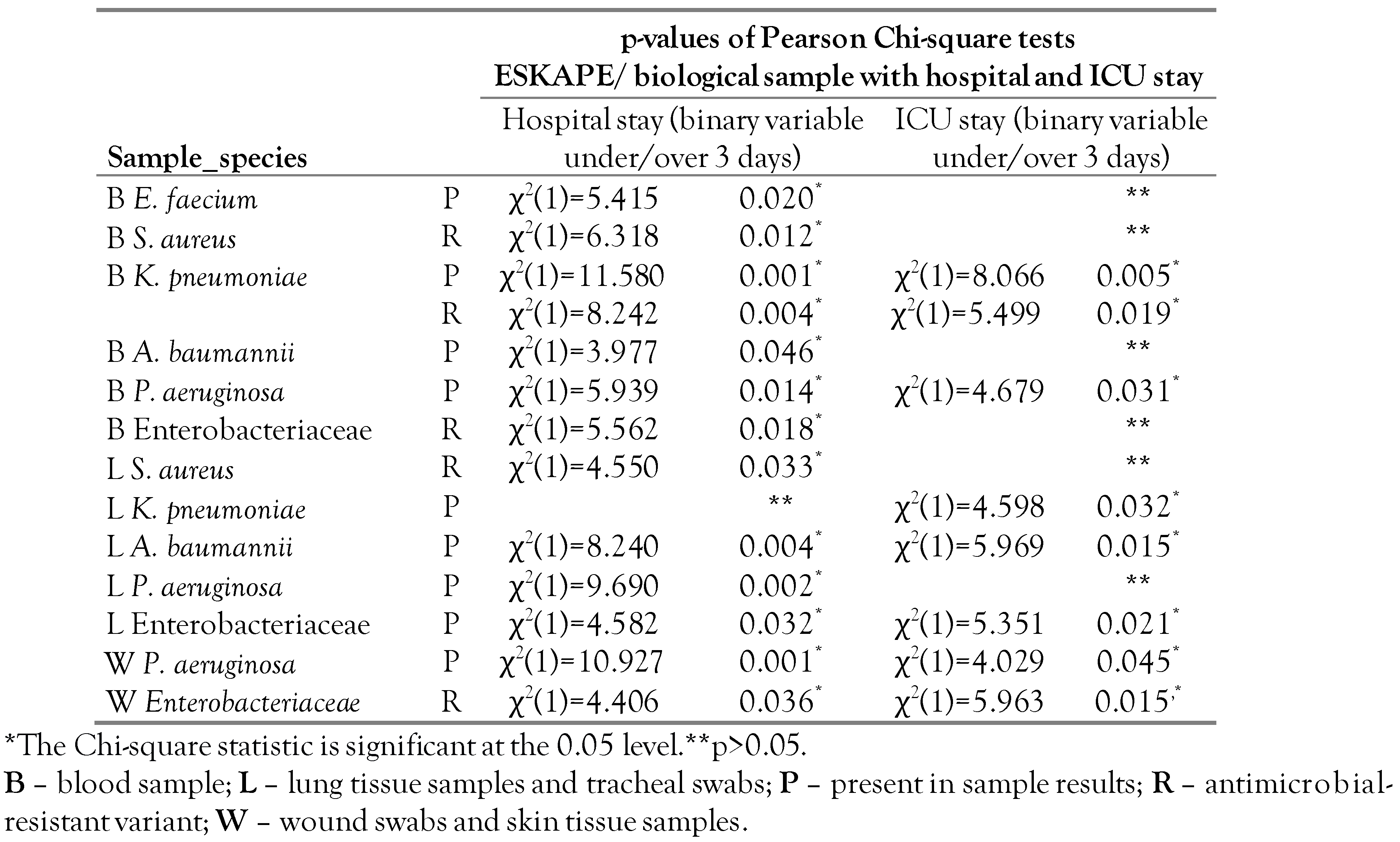
In blood culture, the main species of Enterobacteriaceae identified were Klebsiella pneumoniae (24.2%), Proteus mirabilis (11.8%), and Escherichia coli (9.5%). Antimicrobial resistance (AMR) of Enterobacteriaceae (ESBL and CPE) is associated with prolonged hospitalization (i.e., over 3 days of hospital stay, χ2(1)=5.562, p=0.018) but not with the number of days spent in the ICU (χ2(1)=2.452,p=0.117) or postmortem interval (F(6,213)=1.60, p=0.148). Klebsiella pneumoniae positive blood cultures showed a significant increase of resistant variants with the increase of the extent (number of days) of hospital stay: 57.1% under 3 days and 84.6% over 3 days (χ2(1)=8.240, p=0.004). The number of AMR Enterobacteriaceae species identified in lung fragments and tracheal swabs did not increase with the length of hospitalization (χ2(1)=0.003, p=0.975) or days of ICU (χ2(1)=2.461, p=0.117). Wound swabs and skin tissue samples results showed a statistically significant association of antibiotic resistance with both hospital stay without ICU (χ2(1)=4.406, p=0.036) and admission to the ICU (χ2(1)=5.963, p=0.015) regardless of the postmortem interval. Most cultures of pleural and peritoneal fluid were negative for Enterobacteriaceae, and when enterobacteria were identified, they were antibiotic-resistant variants.
The blood cultures were positive for enterococci in 40.3% (154) of cases, of which the majority were positive for a single microorganism (38.2% of the blood cultures). Blood cultures positive for Enterococcus faecalis and Enterococcus faecium identified strains susceptible to antibiotics in 94.8% and 82.1% respectively. Between 2017 and 2019 there was an increase in vancomycin resistance despite the number of cases for which postmortem blood cultures were requested being low. Vancomycin resistance appeared in lung cultures in 2018 and 2020. Other biological products had positive results only for susceptible variants of enterococci.
Blood cultures were positive for staphylococci in 15.7% (60) of cases. The species identified were Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus lugdunensis, Staphylococcus haemolyticus, coagulase-negative staphylococci and some positive results containing one or more species were reported as Staphylococcus spp. (this type of result known as postmortem contamination).
Methicillin-resistant Staphylococcus aureus (MRSA) was identified in more cases compared to the susceptible strains: 5.2% and 1.6%, respectively; there was an increase in the number of MRSA positive cases between 2015-2017 and a decrease between 2018-2020. In other biological products, staphylococci were rarely identified: most lung cultures were negative for MRSA, although it was identified sporadically in other biological products. The hospital stay was longer for patients with MRSA positive blood cultures (χ2(1)=6.318, p=0.012) or lung cultures (χ2(1)=4.550, p=0.033), but the relationship disappeared when considering the length of ICU stay (χ2(1)=0.048, p=0.827 and χ2(1)=2.500, p=0.114, respectively).
Pseudomonas aeruginosa and Acinetobacter baumannii were identified in blood cultures in less than 20% of cases included in the study. Multidrug-resistant Pseudomonas aeruginosa strains and carbapenemase-resistant variants were found in 5.5% of study cases whereas strains with antibiotic susceptibility were identified in 13.6% of all cases. P. aeruginosa was associated with prolonged hospitalization and ICU stay (p values in Table 3). Resistant strains of P.aeruginosa seemed to be more common with prolonged hospitalization (hospitalization <3 days: 8.3% vs. hospitalization >3 days: 32.8%) without reaching statistical significance (χ2(1)=2.926, p=0.087).

Table 3.
Statistically significant associations of sample-specific results for ESKAPE pathogens with length of hospital stay and ICU stay as binary variables (under 3 days/over 3 days).
Acinetobacter baumannii positive blood cultures were associated with prolonged hospitalization (χ2(1)=3.977, p=0.046,) while A. baumannii positive lung cultures were associated with prolonged ICU stay (χ2(1)=5.964, p=0.015). Multidrug-resistant (MDR) strains were identified in 5.2% of cases as opposed to the sensitive strains, which were identified in 12.3% of blood cultures, regardless of hospital (χ2(1)=0.085, p=0.771) or ICU length of stay (χ2(1)=0.001, p=0.978).
Postmortem microbiology shows a retrospective image of AMR in hospitals in Bucharest, Romania between 2011 and 2020: ESBL variants are predominant with a significant peak in 2016, CR variants (carbapenemase-producing and non-carbapenemase-producing), were scarce (<10 cases/year) until 2019 when they surpassed ESBL variants. Other MDR and VR isolates have a low but constant identification rate of less than 10 cases/year (Figure 1).
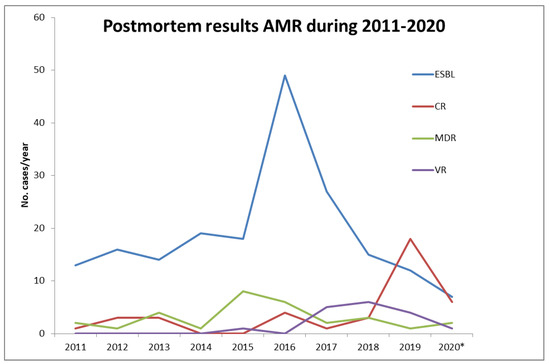
Figure 1.
Annual variation in the distribution of antimicrobial-resistant strains (AMR) between 2011-2020 as recovered from postmortem microbiology reports. *Incomplete data due to unavailable autopsy records. CR—carbapenemase-resistant (carbapenemase producers and non-carbapenemase producers); ESBL—extended-spectrum-β-lactamase; MDR—multidrug-resistant; VR– vancomycin resistant.
In our postmortem microbiology results, ESBL variants of Enterobacteriaceae such as E. coli, P. mirabilis, C. freundii, E. cloacae, K. pneumoniae, and K. oxytoca were identified in samples taken at the beginning of the study in 2011, also CP E.coli and K. pneumoniae. Moreover, in 2011 other MDR variants of A. baumannii, K. pneumoniae, and P. aeruginosa were isolated from postmortem samples. MDR variants of C. freundii and ESBL S. marcescens were recovered in 2012, and MDR variants of E. cloacae in 2015. CP variants of E. cloacae and P. mirabilis were identified between 2018 and 2019. CP variant of P. aeruginosa was recovered first in 2016 and NCP-CR K. pneumoniae in 2019. VR variants were first recovered in 2015 for E. faecium and in 2016 for E. faecalis (Figure 2).

Figure 2.
Representation of antimicrobial resistance (AMR) for ESKAPE pathogens, a retrospective image of AMR type by species (first recovered in postmortem microbiological reports in our center). CP—carbapenemase-producing; ESBL—extended-spectrum-β-lactamase; MDR—multidrug-resistant; NCP-CR—non-carbapenemase producing carbapenemase resistant; VR—vancomycin resistant.
Discussion
Antimicrobial-resistant pathogens in hospitalized patients are of great concern to physicians worldwide. Postmortem microbiology has had a questionable relevance but has gained value in sampling following in-hospital deaths through systematic research. This is the first systematic single-center study describing the identification of AMR variants of microorganisms specific to hospital settings recovered from postmortem sampling during medico-legal autopsies in Romania.
Nevertheless, this retrospective analysis has several limitations that contribute to an underestimation at a clinical level. First, the number of medico-legal autopsies in which postmortem microbiology was requested is small since this type of investigation is not a standard in Romanian autopsy protocols and is requested on a case-by-case-basis by the forensic pathologist. Second, the heterogeneous sampling patterns identified in our study consist of 14 sample types in 96 combinations that interfere with the association of microbiological results and histological confirmed infection/ infection site. This variation determines the need for a standardized sampling protocol. Third, under Romanian law not all in-hospital deaths are subjected to a medico-legal autopsy. Hence, these postmortem results are applicable to a small portion of cases: violent or suspect deaths as mandated by the authorities, which determines an underestimation of AMR in our study; epidemiological indicators such as incidence and prevalence are deemed irrelevant to our study. Other limitations include the unavailability of data regarding medical history and previous hospitalizations, which would provide a more complex epidemiological image.
We did not intend to profile immune-compromised patients as this has been a subject of extensive research in clinical fields and resulted in severity and prognostic scales. The data recovered for this study did not include clinical data or laboratory results (other than bacteriology) for the hospital stay, thus no attempt to apply the specific HCAI case definitions was made. However, 77.7% of our study population consisted of critically ill or severely injured patients—with patient deaths occurring in the ICU after a median stay of 10 days. For analysis regarding the association of antimicrobial resistance and length of hospitalization/ ICU stay, and possible HCAIs, we divided our study population into groups by length of hospital and ICU stay, according to the time frame from European definitions [5,6] of previously mentioned types of infections. Using the onset of 3rd day threshold, we identified similar percentages of cases in groups, independent of the criterion used.
Over half the autopsies were performed in the first two days postmortem and the median for the entire study population was also 2 days; this is possibly due to the protocols regarding medico-legal autopsies for in-hospital deaths. The necessary paperwork (medical records, identification papers, case justification) is submitted to the authorities during the work week; on the date of death or the following day, the corpse is retrieved from the medical unit only after an authority mandate is obtained and autopsy is performed usually within the first two days. If a patient dies during the weekend or holidays, there is a delay in submitting the paperwork and ultimately the autopsy.
Regarding the types of medical units included in our analysis, most cases came from emergency hospitals—these admit a large number of patients with life-threatening conditions (data regarding the number of in-hospital patients and ICU admissions was unavailable). Not all healthcare facilities were included (no data to assess coverage was available) thus adding another limitation to the image of antimicrobial resistance trends and possible HCAIs in Bucharest.
Our statistical analysis was centered on the most challenging resistance-developing microorganisms (ESKAPE) and association to hospital stay over 3 days, possible HCAI sites while focusing on the sample types most frequently retrieved during autopsy (blood, lung, and tracheal swabs, skin tissue, and wound swabs). Urine was scarcely sampled during autopsy and histologic kidney infection was not a common entity in our study population, thus surrogates for possible urinary catheter-related infections (HCAI) were not assessed due to the insufficient number of samples/cases.
Some species of the Enterobacteriaceae family are known contaminants in postmortem microbiology (Escherichia, Enterobacter, and Klebsiella) and/or determine bacterial translocation from the human gastrointestinal tract (Enterococcus groups or Lactobacilli) [7]. As such, our analysis aimed to exclude these contaminants by focusing on ESKAPE pathogens through selecting sample type results associated with histological infection (over 80% of cases). Results showed statistically significant associations for ESKAPE with prolonged hospital and ICU stay independent of postmortem interval, thus showing the positive predicting value of the results. Moreover, AMR isolates of the previously mentioned species were associated with prolonged hospital and/or ICU stay.
Carbapenemase-producing Enterobacteriaceae isolates from our postmortem microbiology results increased in frequency in 2019 following reports from Romanian Public health Authorities [4]. Carbapenem-resistant Enterobacteriaceae occur through a variety of resistance mechanisms and can be divided into carbapenemase producers and non-carbapenemase producers, the latter apparently less virulent and with a lower mortality rate [8,9]. Carbapenemases as a mechanism of resistance have gained importance as the carbapenems in combination therapy have become widely used and effective in improving patient outcomes [10]. Other carbapenem-resistant pathogens of healthcare concern are Acinetobacter baumannii (CRAB) and Pseudomonas aeruginosa (CRPA) [11,12].
Solely ESBL variants were reported less and less, as CP increased possibly due to an increase in usage of carbapenem therapy. CP variants of Enterobacteriaceae species were identified heterogeneously over 10 years with P. mirabilis being the most recently identified (2019).
AMR variants of great concern in present days such as vancomycin-resistant S. aureus or CP A. baumannii were not identified in our samples. Vancomycin-resistant enterococci were not isolated in the first half of the study period (2011-2014). Identified over 30 years ago, vancomycin-resistant enterococci have been spreading rapidly and are becoming a major healthcare problem [13].
Conclusions
Carbapenemase-producing isolates of species from the Enterobacteriaceae family were identified from as early as 2011 and stayed relatively constant up to 2019 when a significant increase caused concern to healthcare facilities.
Vancomycin resistance was identified solely in Enterococcus species (E. faecium and E. faecalis) from 2015.
Postmortem microbiology from the analyzed samples showed a fair association between the tissue sample type and macroscopic or histologically confirmed infection site.
The presence of ESKAPE pathogens and antimicrobial resistant variants were associated with prolonged hospital and intensive care unit stay (over 3 days).
Author Contributions
Conceptualization: ID; methodology: ID and AA; software: ID and AAK; validation: ID and MH; formal analysis: ID and MH; investigation: ID and CD; resources: ID; data curation: ID and CD; writing—original draft preparation: ID; writing—review and editing: ID, AAK and MH; visualization: ID; supervision: CD and AAK; project administration: ID. All authors have read and approved the final version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Burton, J.L.; Saegeman, V.; Arribi, A.; et al. Postmortem microbiology sampling following death in hospital: An ESGFOR task force consensus statement. J Clin Pathol. 2019, 72, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Sievert, D.M.; Ricks, P.; Edwards, J.R.; et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol. 2013, 34, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Regional Office for Europe/European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe 2022–2020. 2022.
- 6.7.2018 L 170/1-74; Commission Implementing Decision (EU) 2018/945 of 22 June 2018 on the communicable diseases and related special health issues to be covered by epidemiological surveillance as well as relevant case definitions. Official Journal of the European Union.
- 27.9.2012 L 262/1-57; Commission Implementing Decision of 8 August 2012 amending Decision 2002/253/EC laying down case definitions for reporting communicable diseases to the Community network under Decision No 2119/98/EC of the European Parliament and of the Council. Official Journal of the European Union.
- Palmiere, C.; Egger, C.; Prod’Hom, G.; Greub, G. Bacterial translocation and sample contamination in postmortem microbiological analyses. J Forensic Sci. 2016, 61, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Goodman, K.E.; Harris, A.D. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin Infect Dis. 2017, 64, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Black, C.A.; So, W.; Dallas, S.S.; et al. Predominance of non-carbapenemase producing carbapenem-resistant Enterobacterales in South Texas. Front Microbiol. 2021, 11, 623574. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, R.; Nusrin, K.S.; Georgy, S.A.; Sreelakshmi, K.S. Role of beta lactamases in antibiotic resistance: A review. Int Res J Pharm. 2014, 5, 37–40. [Google Scholar] [CrossRef]
- Hamidian, M.; Nigro, S.J. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom. 2019, 5, e000306. [Google Scholar] [CrossRef] [PubMed]
- Walters, M.S.; Grass, J.E.; Bulens, S.N.; et al. Carbapenem-resistant Pseudomonas aeruginosa at US Emerging Infections Program Sites, 2015. Emerg Infect Dis. 2019, 25, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Cetinkaya, Y.; Falk, P.; Mayhall, C.G. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000, 13, 686–707. [Google Scholar] [CrossRef] [PubMed]
© GERMS 2022.