Abstract
Advanced biofuels possess superior characteristics to serve for gasoline substitutes. In this study, a whole cell biocatalysis system was employed for production of short-chain alcohols from corresponding fatty acids. To do so, Escherichia coli strain was equipped with a biocatalytic pathway consisting of endogenous atoDA and Clostridium acetobutylicum adhE2. The strain was further reprogrammed to improve its biocatalytic activity by direction the glycolytic flux to acetyl-CoA and recycling acetate. The production of 1-propanol and n-pentanol were exemplified with the engineered strain. By substrate (glucose and propionate) feeding, the strain enabled production of 5.4 g/L 1-propanol with productivity reaching 0.15 g/L/h. In addition, the strain with a heavy inoculum was implemented for the n-pentanol production from n-pentanoic acid. The production titer and productivity finally attained 4.3 g/L and 0.86 g/L/h, respectively. Overall, the result indicates that this developed system is useful and effective for biocatalytic production of short-chain alcohols.
1. Introduction
Our daily life which mainly depends on fossil resources has caused the enormous emission of greenhouse gases, consequently leading to global climate change. It urges us to search for sustainable energy alternatives and environment-friendly chemicals [1]. Biofuels that are produced from renewable feedstock apparently fulfill the need. Ethanol appears to be a representative biofuel. However, it has unfavorable properties including low energy density, high vapor pressure, and hygroscopicity [2]. Advanced biofuels involving longer chain alcohols (C3–C6) are of particular interest because they have superior characteristics applied for gasoline substitutes. For instance, n-butanol (C4) which is blended with gasoline at any concentrations readily serves as the transportation fuel. There is no need to modify the existing pipeline infrastructure for transport of n-butanol [3]. The fuel property of isobutanol is similar to n-butanol whereas it possesses higher octane number. Isopentanol (C5) is a branched-chain alcohol and shares similarity in the physiochemical property with gasoline [4]. In particular, it has a higher volumetric energy density than n-butanol. As a linear-chain alcohol, n-pentanol (C5) is exploited as an additive in diesel [5]. It results in diesel fuel with a better performance in terms of fuel combustion and particulate emission. Likewise, hexanol (C6) in the form of a gasoline blend improves the fuel property [6]. In addition to a gasoline additive, 1-propanol (C3) serves as a precursor for the synthesis of propylene. Propylene is used for the production of plastics, and it is an important platform chemical in industry as well [7]. Bulk chemicals derived from propylene include propylene oxide, acrylonitrile, cumene, butyraldehyde, and acrylic acid.
Naturally-occurring microbes harbor inherent metabolic pathways for the synthesis of advanced alcohols. The best known example is the production of n-butanol in Clostridium species which undergo the mixed acetone-butanol-ethanol (ABE) fermentation [8]. The ABE fermentation proceeds with the acidogenesis phase, followed by the solventogenesis phase [9]. However, the sluggish growth and sporulation of Clostridium usually render the operation difficult and complicated. Propionibacteria natively produce propionic acid through the Wood-Werkman pathway [10]. The production of 1-propanol occurs when the carbon source changes to the more reduced one for fermentation. Nevertheless, propionic acid still remains the major product. In the absence of oxygen, Clostridium propionicum enables utilization of ethanol or lactate via the acrylate pathway [11]. The fermentation product ends up with propionic acid and acetic acid while 1-propanol is marginally produced. In addition, C. carboxidivorans is an acetogen that relies on the Wood-Ljungdahl pathway to grow on syngas (CO or CO2 and H2) [12]. By lowering the metabolic activity, the acetogenic fermentation resulted in the improvement in the production of ethanol, butanol, and hexanol. However, this production process is afflicted by low titer and productivity.
The recent advance in the technology of synthetic biology has successfully transformed various microbes into producers capable of synthesizing advanced biofuels. Recognized as the biotechnology workhorse, Escherichia coli has been subject to rational reprogramming of metabolic pathways, mainly including the 2-ketoacid pathway, the fatty acid synthesis pathway, and the reverse β-oxidation pathway [13,14]. The engineered E. coli generally shows promise for fermentative production of C3–C6 alcohols. Nevertheless, the technical bottleneck still exists and remains to be overcome. To address this issue, we have recently designed a synthetic consortium for production of n-butanol [15]. In this system, an E. coli strain responsible for conversion of butyrate to n-butanol was equipped with a biocatalytic route consisting of endogenous atoDA and C. acetobutylicum adhE2. With supplemented butyrate, this butyrate-conversion strain effectively produced 6.2 g/L n-butanol. In this study, the biocatalysis biocatalysis system based on this strain was exploited for the synthesis of other C3–C6 alcohols from corresponding fatty acids. The central metabolism of the strain was further refined to improve the biocatalytic activity. The result shows that this platform system is useful and effective for production of short-chain fatty alcohols.
2. Materials and Methods
2.1. Strain Construction
The strain development started with BuT-3EA strain. This strain was derived from BuT-3E strain and carried the enhanced level of acs [15]. The strain’s modification was carried out as follows. Following the pervious study [16], the DNA cassette was amplified from pPR-aceE plasmid with RC12060/RC12086 primers. The PCR DNA was then introduced into the strain by electroporation. By the λRed-mediated homologous recombination, the DNA cassette was integrated into the strain’s genome. The inserted LE*-kan-RE* cassette was removed by the act of Cre. This construction resulted in the strain carrying the aceEF operon under the control of the λPL promoter (PλPL). Moreover, pLam-LpdA* plasmid which contained the NADH-insensitive lpdA mutant (lpdA*) fused to PλPL (PλPL-lpdA*) was used for the enhanced expression of lpdA. The DNA containing PλPL-lpdA* was integrated into the λ attachment site of the strain, followed by removal of the inserted marker. The resulting strain was renamed BuT-3EP.
2.2. Fatty Alcohol Production
E. coli strains were grown on Luria-Bertani medium at 37 °C overnight. The bacterial growth was measured with a spectrometer set at 550 nm (OD550). The overnight culture was seeded to Erlenmeyer flasks (125 mL) containing M9Y medium (20 mL) supplemented with glucose (10 g/L) and fatty acids (4 g/L). The medium was composed of Na2HPO4 (6 g/L), KH2PO4 (3 g/L), NaCl (0.5 g/L), NH4Cl (1 g/L), MgSO4 (1 mM), CaCl2 (0.1 mM), and yeast extract (5 g/L). The initial cell density was maintained at OD550 of 0.2. The shake-flask culture was conducted under the oxygen-limited condition according to the previous study [15]. The biocatalytic production of fatty alcohols was terminated at 48 h. The mode of substrate feeding was performed by adding extra glucose (10 g/L) and propionic acid (4 g/L) to the culture medium at 24 h. Incubated with n-pentanoic acid (6 g/L), the strain with the initial cell density at OD550 of 9 was applied for the n-pentanol production.
2.3. Analytic Methods
The analysis of glucose, alcohols, and organic acids essentially followed the reported protocol [15]. Glucose was determined using high-performance liquid chromatography (HPLC) with the ICSep ICE-ION-300 column (Transgenomic, Omaha, NE, USA). HPLC with the reflective index (RID-10A, Shimadzu, Japan) was used for the measurement of organic acids. Alcohols were analyzed by gas chromatograph (Trace 1300, Thermo Scientific, Waltham, MA, USA) which was equipped with flam ionization detector and a DB-WAX capillary column. The oven temperature was initially held at 50 °C for 1 min and raised to 150 °C with a gradient of 15 °C/min, and 150 °C was maintained for 5 min. The injector and detector were maintained at 250 °C
3. Results
3.1. Production of Fatty Alcohols
In this study, a whole cell biocatalysis system was proposed to produce short-chain fatty alcohols. BuT-3E strain enabled production of n-butanol from n-butyric acid and was implemented for the engineering purpose [15]. As indicated in Figure 1, this strain was equipped with a synthetic pathway consisting of endogenous atoDA and Clostridium adhE2. Acetoacetyl-CoA transferase encoded by atoDA has a physiological function for activation of short-chain fatty acids (C4–C6) to corresponding thioesters associated with acetic acid [17]. The subsequent conversion of thioesters to respective alcohols proceeds through the reaction catalyzed by alcohol dehydrogenase (encoded by adhE2). In addition, the undesired pathways were blocked to conserve NADH and to reduce the production of waste products.
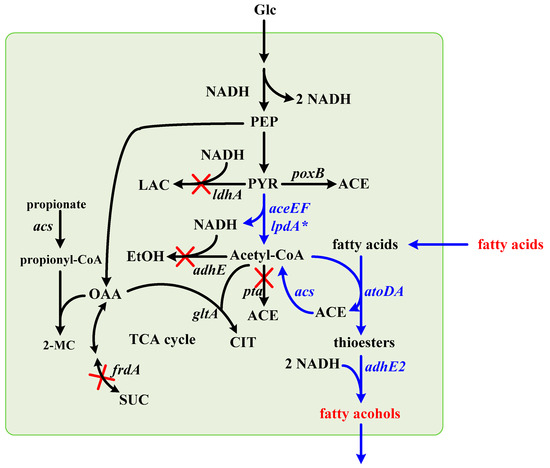
Figure 1.
The biocatalytic pathway leading to fatty alcohols in E. coli. The biocatalytic pathway leading to fatty alcohols from short-chain fatty acids was introduced into E. coli by metabolic engineering. Undesired pathways were blocked by deletion of responsible genes as marked by “X”. The genes involved in the metabolic pathway: aceEF-lpdA*: pyruvate dehydrogenase complex; acs, acetyl-CoA synthetase; adhE, aldehyde-alcohol dehydrogenase; adhE2, butyraldehyde-butanol dehydrogenase; atoDA, acetoacetyl-CoA transferase; ldhA, lactate dehydrogenase; frdA, subunit of fumarate reductase; gltA, citrate synthase; poxB, pyruvate oxidase; pta, phosphate acetyltransferase. Abbreviations: ACE, acetate; CIT, citrate; EtOH, ethanol; Glc, glucose; LAC, lactate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PYR, pyruvate; SUC, succinate; 2-MC, 2-methylcitrate.
It was intriguing to learn the performance of BuT-3E strain for production of alternative fatty alcohols. Therefore, the shake-flask culture was conducted with the medium containing short-chain fatty acids (C3–C7) under the oxygen-limited condition. At the end of the fermentation, the strain produced 1-propanol, isobutanol, n-pentanol, isopentanol, and n-hexanol with a titer reaching 2.3, 1.8, 1.5, 1.2 and 1.3 g/L, respectively (Figure 2a). As shown in Figure 2b, propionic acid (3.4 g/L), isobutanoic acid (2.4 g/L), n-pentanoic acid (2 g/L), isopentanoic acid (1.8 g/L), and n-hexanoic acid (1.7 g/L) were consumed in the strain to serve as the precursor for the synthesis of alcohols. Consequently, the conversion yield of 1-propanol, isobutanol, n-pentanol, isopentanol, and n-hexanol based on corresponding acids accounts for 83.4%, 89.2%, 86.9%, 77.3%, and 87.0% of the theoretical yield (ca., 0.81, 0.84, 0.86, 0.88 g/g), respectively. The result indicates that BuT-3E strain shows promise for production of C3–C6 alcohols. In addition, n-heptanol was not detected. This is attributed to the failure of atoDA in activation of heptanoic acid (Figure 2b).
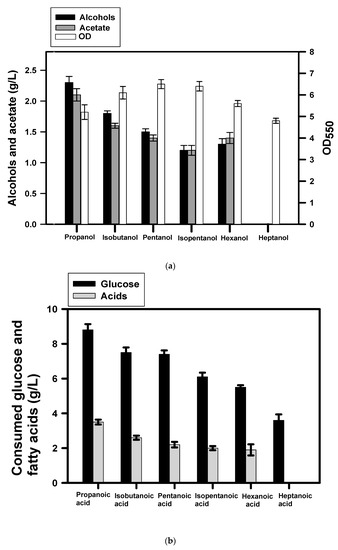
Figure 2.
Production of short-chain alcohols in engineered E. coli. The fermentation was conducted with BuT-3E strain in the presence of glucose (10 g/L) and short-chain fatty acids (4 g/L) as indicated for 24 h. The experiments were conducted in triplicate. The fermentation products and the cell density (OD550) were determined at the end of the fermentation (a). The consumption of glucose and fatty acids were measured for further analyses (b).
3.2. Propanol Production by Substrate Feeding
As illustrated above, BuT-3E strain enabled production of C3-C6 fatty alcohols with a high conversion yield. Nevertheless, the alcohol production decreased with an increase in the chain length. Taking 1-propanol as an example, our next task was undertaken to improve the production yield. It is well recognized that short-chain fatty acids with high solubility are detrimental to microbes because they penetrate into cell membrane at a high concentration, which in turn damages cell membrane [18]. The bacterial growth was greatly inhibited by propionic acid at a level higher than 4 g/L (data not shown). Therefore, the fermentation of 1-propanol was conducted with the substrate feeding. In a similar way, BuT-3E strain was first cultivated with medium containing 10 g/L glucose and 4 g/L propionic acid. At 24 h, 10 g/L glucose and 4 g/L propionic acid were additionally fed to the culture. As a result, the 1-propanol production leveled off at 36 h and reached 4.2 g/L (Figure 3a). The strain consumed around 13 g/L glucose and 6 g/L propionic acid at the end (Figure 3b). It leads to the conversion yield based on propionic acid reaching 86.4% of the theoretical yield. In addition, the cell density was not affected by the extra addition of glucose (Figure 3b). The result indicates that the cell growth is limited and, however, the strain remains metabolically active to provide acetyl-CoA for the synthesis of 1-propanol.
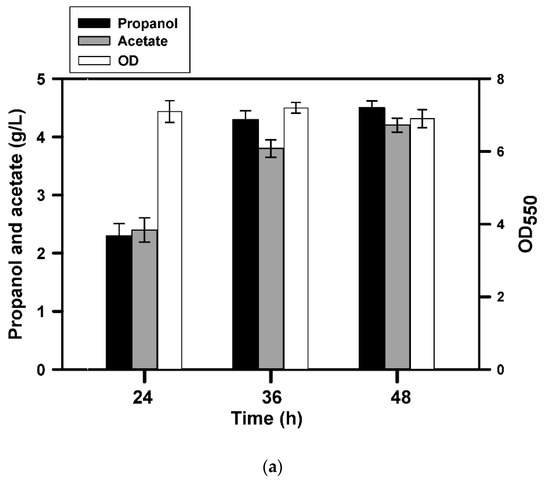
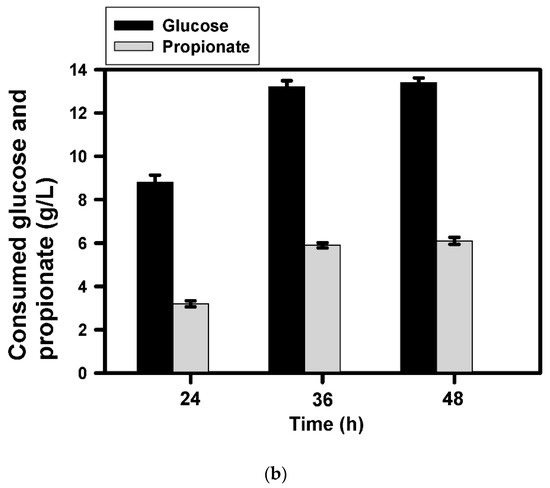
Figure 3.
Production of 1-propanol by the approach of substrate feeding. The fermentation of BuT-3E strain was carried out with glucose (10 g/L) and propionate (4 g/L). At 24 h, the substrate containing glucose (10 g/L) and propionate (4 g/L) was added to the culture medium and the fermentation result was analyzed at a predetermined interval. The experiment was conducted in triplicate. The fermentation products and the cell density (OD550) were determined at the end of the fermentation (a). The consumption of glucose and fatty acids were measured for further analyses (b).
3.3. Improvement of Propanol Production
Acetic acid appears to be an associated product in the synthetic pathway of fatty alcohols in the strain. However, it disables the growth of E. coli at a low level and imparts an inhibitory effect on methionine biosynthesis [19]. Acetyl-CoA synthetase (encoded by acs) activates acetate to acetyl-CoA at the expense of ATP and plays a physiological role in scavenging acetate under the anaerobic condition [20]. It also involves in the 2-methylcitrate (2-MC) cycle for oxidation of propionate and produce propionyl-CoA from propionate [21]. In addition, pyruvate oxidase (encoded by poxB) catalyzes oxidative decarboxylation of pyruvate to acetate. Therefore, the expression of acs and aceEF operon were enhanced to improve the fermentation performance of the strain. The increased activity of pyruvate dehydrogenase (encoded by aceEF) directs pyruvate to acetyl-CoA and away from the poxB pathway. The recruitment of ACS facilitates re-utilization of acetate. The resulting strain was designated as BuT-3EP. The substrate-feeding mode was then performed with BuT-3EP strain. As a consequence, the strain consumed propionic acid of 7.4 g/L and produced 1-propanol of 5.4 g/L at 36 h (Figure 4a,b). The conversion yield based on propionic acid accounts for 90% of the theoretical yield. The production of acetic acid reduced to 2.4 g/L from 4 g/L (i.e., BuT-3E strain in Figure 3a) at the end. Nevertheless, the bacterial growth was arrested and not further improved by the supplement of extra glucose. It is likely that the toxicity of acetic acid in part limits the strain’s growth. Moreover, the combined function of AtoDA and ACS is expected to raise the propionyl-CoA level, which likely perturbs the CoA homeostasis and increases the production of 2-MC via the 2-MC pathway. 2-MC is known to be a potent inhibitor of citrate synthase in the tricarboxylic acid (TCA) cycle [22].
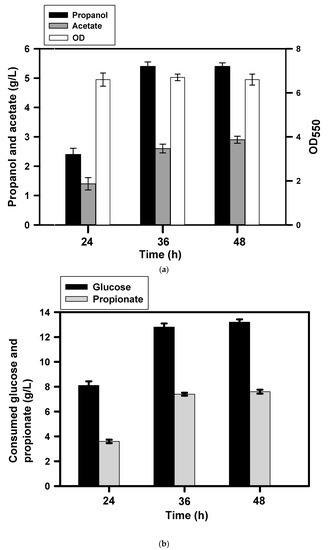
Figure 4.
Production of 1-propanol by the genetically-modified strain. Derived from BuT-3E strain, BuT-3EP strain was employed for the fermentation in the presence of glucose (10 g/L) and propionate (4 g/L). At 24 h, the substrate containing glucose (10 g/L) and propionate (4 g/L) was added to the culture medium and the fermentation result was analyzed at a predetermined interval. The experiment was conducted in triplicate. The fermentation products and the cell density (OD550) were determined at the end of the fermentation (a). The consumption of glucose and fatty acids were measured for further analyses (b).
3.4. n-Pentanol Production by Heavy Inoculum
Our last task was undertaken to improve productivity. This issue was addressed by implementation of the fermentation mode based on the heavy inoculum. In this case, the n-pentanol production was chosen for illustration. The fermentation was carried out using BuT-3EP strain with the cell density reaching 9 at OD550 while 10 g/L glucose and 6 g/L n-pentanoic acid were supplemented in the medium. As shown in Figure 5, the strain produced 4.3 g/L n-pentanol at the expense of 5.5 g/L n-pentanoic acid at 5 h. Consequently, the conversion yield based on n-pentanoic acid accounts for 90.5% of the theoretical yield.
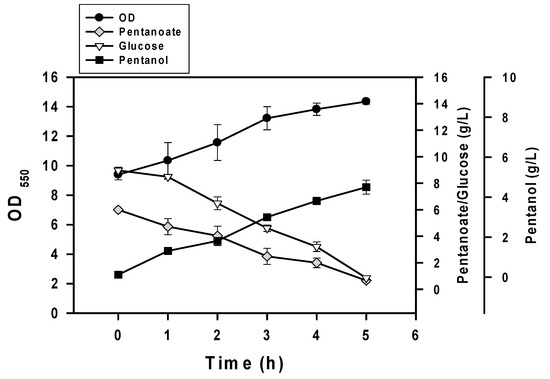
Figure 5.
Improved production of n-pentanol. The fermentation of BuT-3EP strain was carried out with glucose (10 g/L) and n-pentanote (6 g/L). The fermentation was then followed along the time course. The experiment was conducted in triplicate.
4. Discussion
Metabolic engineering is an enabling technology that efficiently reprograms living cells for the desired trait. This technology has been applied for the microbial fermentation production of short-chain alcohols. E. coli still remains the top choice for engineering to develop a production scheme of industrial usefulness. As exemplified by 1-propanol production, two metabolic pathways leading to the formation of 2-ketobutyrate (2-KB) were constructed in E. coli. The synthesis pathway of L-threonine involving thrABC and ilvA was manipulated to direct the glycolytic flux from oxaloacetate (OAA) to 2-KB. The recruitment of cimA and leuBCD constituted the heteroglogous citramalate pathway responsible for the synthesis of 2-KB from pyruvate (PYR). Moreover, the inactivation of four engaged genes conserved the intracellular 2-KB pool which was further converted to 1-propanol through the synthesis pathway consisting of kivd and adhE. As a result, the engineered E. coli utilized glucose to produce 1-propanol of 8 g/L with productivity reaching 0.12 g/L/h [23]. In another study, the ‘sleeping beauty mutase’ operon was activated by overexpression of sbm, ygfD, and ygfG. Equipped with the additional function of sucCD and adh2, E. coli strain was subject to fed-batch fermentation and enabled production of 7 g/L 1-propanol based on glycerol with productivity of 0.04 g/L/h [24]. To synthesize 1-propanol, the methylglyoxal bypass pathway was modified by expression of heterologous budC and ppdABC. This modified pathway leads to the formation of 1,2-propanediol (1,2-PDO) as an intermediate metabolite. The resulting strain produced 0.25 g/L 1-propanol from glucose and the productivity was 0.005 g/L/h [25]. The 1-propanol production was further optimized by using the co-cultivation strategy of two E. coli strains responsible for production of and conversion of 1,2-PDO, respectively. Finally, the result gave 1-propanol of 3 g/L and productivity of 0.014 g/L/h [26]. In contrast to these studies, this work proposed a whole cell biocatalysis system. The core of this system relies on the native atoDAEB operon which is engaged in the degradation of C4-C6 fatty acids [27]. The physiological function of AtoDA activates acetoacetate to acetoacetyl-CoA. Fed with propionic acid, BuT-3E strain bearing functional AtoDA enabled production of 1-propanol (Figure 2a,b). The result indicates that AtoDA functions to activate propionate to propionly-CoA which is subsequently oxidized to 1-propanol by AdhE2. The producer strain was improved by the approach of metabolic engineering, which aims to recycle acetic acid and refine the glycolysis pathway for availability of acetyl-CoA (Figure 1). By the substrate-feeding strategy, the engineered strain (i.e., BuT-3EP) produced 5.4 g/L 1-propanol and attained productivity of 0.15 g/L/h (Figure 4a,b). It is apparent that our proposed system shows high efficiency in production of 1-propanol.
The application of this biocatalysis system was further illustrated with the production of n-pentanol. Initiated with a heavy inoculum, the batch culturing of BuT-3EP strain produced 4.3 g/L n-pentanol with productivity reaching 0.86 g/L/h (Figure 5). Intensive efforts have been devoted to the fermentative production of n-pentanol. In combination with the 2-ketoacid pathway, the reversed β-oxidation pathway in the strain was employed for the synthesis of n-pentanol with a titer and productivity reaching 0.4 g/L and 0.004 g/L/h, respectively [28]. However, the production of n-pentanol was associated with many alcohol byproducts of various chain length. Another study reported the implementation of the iterative elongation cycle involved in the 2-ketoacid pathway. To improve the n-pentanol synthesis, Lactococcus lactis ketoisovalerate decarboxylase (Kivd) was subject to mutation. As a result, the Kivd mutant strain enabled production of pentanol accounting for 90% of the total alcohol content. The application of in situ extraction with oleyl alcohol led to the production titer of 4.3 g/L and productivity of 0.029 g/L/h [29]. Nevertheless, this previous work generally has the disadvantage of low productivity and product promiscuity.
In summary, this study proposed a promising system for biocatalytic production of short-chain fatty alcohols. The developed strain is, however, susceptible to toxicity at a high concentration of fatty acids and fatty alcohols. The method of metabolic evolution is useful for selection of mutant strains that tolerate high levels of fatty acids and alcohols [30]. Meanwhile, the approach by in situ extraction of alcohols facilitates the bacterial survival. The usefulness of this bioconversion platform would be greatly acknowledged by the successful implementation of these combined strategies to reshape the producer strain with the desired trait.
Author Contributions
Writing—original draft preparation, L.-J.L.; methodology and analysis, M.S.; supervision, C.-J.C.; writing—review and editing, Y.-P.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by Ministry of Science and Technology (MOST 109-2320-B-036-MY3 and 108-2221-E-035-052-MY3), Taiwan.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schubert, C. Can biofuels finally take center stage? Nat. Biotechnol. 2006, 24, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Mussatto, S.; Dragone, G.; Guimarães, P.M.; Silva, J.P.; Carneiro, L.M.; Roberto, I.C.; Vicentea, A.; Dominguesa, L.; Teixeira, J.A. Technological trends, global market, and challenges of bio-ethanol production. Biotechnol. Adv. 2010, 28, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Dürre, P. Biobutanol: An attractive biofuel. Biotechnol. J. 2007, 2, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Tsujimura, T.; Pitz, W.J.; Gillespie, F.; Curran, H.J.; Weber, B.W.; Zhang, Y.; Sung, C.J. Development of isopentanol reaction mechanism reproducing autoignition character at high and low temperatures. Energy Fuels 2012, 26, 4871–4886. [Google Scholar] [CrossRef]
- Wei, L.; Cheung, C.S.; Huang, Z. Effect of n-pentanol addition on the combustion, performance and emission characteristics of a direct-injection diesel engine. Energy 2014, 70, 172–180. [Google Scholar] [CrossRef]
- Masum, B.M.; Masjuki, H.H.; Kalam, M.A.; Palash, S.M.; Wakil, M.A.; Imtenan, S. Tailoring the key fuel properties using different alcohols (c2–c6) and their evaluation in gasoline engine. Energy Convers. Manag. 2014, 88, 382–390. [Google Scholar] [CrossRef]
- Walther, T.; François, J.M. Microbial production of propanol. Biotechnol. Adv. 2016, 34, 984–996. [Google Scholar] [CrossRef]
- Jones, D.T.; Woods, D.R. Acetone-butanol fermentation revisited. Microbiol. Rev. 1986, 50, 484–524. [Google Scholar] [CrossRef]
- Lee, S.Y.; Park, J.H.; Jang, S.H.; Nielsen, L.K.; Kim, J.; Jung, K.S. Fermentative butanol production by clostridia. Biotechnol. Bioeng. 2008, 101, 209–228. [Google Scholar] [CrossRef] [PubMed]
- Wood, H.G. Metabolic cycles in the fermentation by propionic acid bacteria. Curr. Top. Cell. Regul. 1981, 18, 255–287. [Google Scholar]
- Tholozan, J.L.; Touzel, J.P.; Samain, E.; Grivet, J.P.; Prensier, G.; Albagnac, G. Clostridium neopropionicum sp. Nov., a strict anaerobic bacterium fermenting ethanol to propionate through acrylate pathway. Arch. Microbiol. 1992, 157, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Ramió-Pujol, S.; Ganigué, R.; Bañeras, L.; Colprim, J. Incubation at 25 °C prevents acid crash and enhances alcohol production in clostridium carboxidivorans p7. Bioresour. Technol. 2015, 192, 296–303. [Google Scholar] [CrossRef]
- Cheon, S.; Kim, H.M.; Gustavsson, M.; Lee, S.Y. Recent trends in metabolic engineering of microorganisms for the production of advanced biofuels. Curr. Opin. Chem. Biol. 2016, 35, 10–21. [Google Scholar] [CrossRef]
- Das, M.; Patra, P.; Ghosh, A. Metabolic engineering for enhancing microbial biosynthesis of advanced biofuels. Renew. Sust. Energy Rev. 2020, 119, 109562. [Google Scholar] [CrossRef]
- Saini, M.; Chen, M.H.; Chiang, C.J.; Chao, Y.P. Potential production platform of n-butanol in Escherichia coli. Metab. Eng. 2015, 27, 76–82. [Google Scholar] [CrossRef]
- Saini, M.; Li, S.Y.; Chiang, C.J.; Chao, Y.P. Systematic engineering of the central metabolism in Escherichia coli for effective production of n-butanol. Biotechnol. Biofuels 2016, 9, 69. [Google Scholar] [CrossRef]
- Sramek, S.J.; Frerman, F.E. Purification and properties of Escherichia coli coenzyme a-transferase. Arch. Biochem. Biophys. 1975, 171, 14–26. [Google Scholar] [CrossRef]
- Bell, G.H. Solubilities of normal aliphatic acids, alcohols and alkanes in water. Chem. Phys. Lipids 1973, 10, 1–10. [Google Scholar] [CrossRef]
- Roe, A.J.; O’Byrne, C.; McLaggan, D.; Booth, I.R. Inhibition of Escherichia coli growth by acetic acid: A problem with methionine biosynthesis and homocysteine toxicity. Microbiology 2002, 148, 2215–2222. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Beatty, C.M.; Browning, D.F.; Busby, S.J.; Simel, E.J.; Hovel-Miner, G.; Wolfe, A.J. Regulation of acetyl coenzyme a synthetase in Escherichia coli. J. Bacteriol. 2000, 182, 4173–4179. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Gu, J.; Wang, X.; Zhang, X.E.; Deng, J. Acs is essential for propionate utilization in Escherichia coli. Biochem. Biophys. Res. Commun. 2014, 449, 272–277. [Google Scholar] [CrossRef]
- Horswill, A.R.; Dudding, A.R.; Escalante-Semerena, J.C. Studies of propionate toxicity in Salmonella enterica identify 2-methylcitrate as a potent inhibitor of cell growth. J. Biol. Chem. 2001, 276, 19094–19101. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.R.; Liao, J.C. Synergy as design principle for metabolic engineering of 1-propanol production in Escherichia coli. Metab. Eng. 2013, 17. [Google Scholar] [CrossRef]
- Srirangan, K.; Liu, X.; Westbrook, A.; Akawi, L.; Pyne, M.E.; Moo-Young, M.; Chou, C.P. Biochemical, genetic, and metabolic engineering strategies to enhance coproduction of 1-propanol and ethanol in engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2014, 98, 9499–9515. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Yan, Y. Dehydratase mediated 1-propanol production in metabolically engineered Escherichia coli. Microb. Cell Factories 2011, 10, 97. [Google Scholar] [CrossRef]
- Jain, R.; Sun, X.; Yuan, Q.; Yan, Y. Systematically engineering Escherichia coli for enhanced production of 1,2-propanediol and 1-propanol. ACS Synth. Biol. 2014, 4, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, L.S.; Nunn, W.D. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: The ato system. J. Bacteriol. 1987, 169, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Machado, H.B.; Dekishima, Y.; Luo, H.; Lan, E.I.; Liao, J.C. A selection platform for carbon chain elongation using the coa-dependent pathway to produce linear higher alcohols. Metab. Eng. 2012, 14, 504–511. [Google Scholar] [CrossRef]
- Chen, G.S.; Siao, S.W.; Shen, C.R. Saturated mutagenesis of ketoisovalerate decarboxylase V461 enabled specific synthesis of 1-pentanol via the ketoacid elongation cycle. Sci. Rep. 2017, 7, 11284. [Google Scholar] [CrossRef]
- Polen, T.; Rittmann, D.; Wendisch, V.F.; Sahm, H. DNA microarray analyses of the long-term adaptive response of Escherichia coli to acetate and propionate. Appl. Environ. Microbiol. 2003, 69, 1759–1774. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).