1. Introduction
Laboratory automation has received increasing attention from the biotechnology industry over the last two decades. While, at the beginning, biotech automation was primarily employed in analytical laboratories, it has gained in prevalence across the entire sector with the advance of digitalization and Industry 4.0. Indeed, it is well understood that many aspects of the medicine of the future will only become feasible through the aid of automation—ranging from automated analysis of complex data for precision medicine, and healthcare robotics in surgery and care, to automated production and intelligent supply chains for advanced therapies [
1,
2].
This is particularly true for cell-based personalized therapies that require automation to make products affordable and accessible [
3,
4]. Whether the application is in personalized drug development or for autologous cell therapies, the expansion of patient-derived cells is labor-intensive and characterized by a high batch-to-batch variability. Furthermore, the individual batch sizes are small and thus the overall throughput has to be high in order to render personalized medicine economically feasible.
A broad range of personalized treatment modalities are based on patient-derived stem cells, which are amplified in vitro. For example, human mesenchymal stem cells (hMSC) are appealing candidates for research and regenerative therapies [
5,
6,
7,
8,
9]. They cannot only differentiate into adipose, cartilage or bone cells but can also induce regeneration in the surrounding tissue and modulate the immune response [
5]. Like most stem cell types, hMSCs require a surface for adherence and proliferation. Thereby, planar cultivation methods are most suitable if only small quantities are required, for example, for screening.
In this context, automation can provide support in terms of standardization, parallelization and the handling of labor-intensive tasks. For this reason, the industry has been developing automated solutions that are currently transitioning into the second generation [
10]. For example, the production of cell-based therapy products requires the manufacture of high cell numbers. Automated production of stem cells therefore often utilizes bioreactors to generate the necessary volumes. Examples include stirred-tank, fixed-bed and wave reactors [
11,
12,
13,
14], which are typically only partially automated, covering the cultivation step itself. One of the first robot-assisted approaches for scaled production was the CompacT SelecT (Sartorius Stedim Biotech, Royston, UK), designed for production of stem cells in T-flasks [
15,
16]. More recently, a robot-assisted facility for production of mesenchymal stem cells in bioreactors has been presented by the Autostem project [
17].
However, not all applications require large quantities of cells. When it comes to research, disease modeling or drug or toxicity screening, the focus shifts from large volumes to a smaller scale that requires a high throughput. There are a number of commercially available, automated systems that typically consist of a liquid handler, into which additional devices are integrated. Examples include the Freedom EVO from Tecan, the Cellhost from Hamilton Robotics or the Biomek Cell Workstation from Beckman Coulter [
18]. While these systems provide a high flexibility, they are limited in terms of space and thus number of devices to be integrated. Additionally, the handling of material is typically mediated by the liquid handler itself and thus restricted to simple transportation steps. Furthermore, programming of protocols requires expert knowledge for most of these systems.
In this paper, we present the StemCellDiscovery as an addition to the automation landscape that focuses on stem cell production in small scale and high throughput. In contrast to other solutions, the StemCellDiscovery integrates an industrial six-axis robotic arm mounted on a linear axis with a custom gripper for manipulation of disposables and complex handling jobs. This way, many different devices can be integrated simultaneously, thus representing a fully automated laboratory environment. The custom, flexible control software provides an easy-to-use graphical user interface for operation of the system and protocol generation with a drag-and-drop editor. Due to the service-oriented architecture, new devices and modules can be integrated in a “plug-and-produce” manner. Furthermore, the system is provided with a non-invasive in-line technology to monitor cell growth, which will be presented in this paper.
While the system is capable of handling any type of adherent cell, in this publication, we will focus on human adipose-derived mesenchymal stem cells in particular. The StemCellDiscovery contains all equipment required for adherent cell cultivation. It can seed cells on six-well and other multi-well culture plates, change medium and harvest the cells. For splitting of the cell cultures, the cells can also be directly reseeded after the harvest into multiple new culture plates. The cell cultures and disposables are transported and manipulated by an industrial six-axis robotic arm. This way, any direct interaction between user and product becomes obsolete. A central control software monitors the system and cell cultures and processes and logs each step to enable full track and trace.
An integral part of cell cultivation is regular culture assessment. In manual culture, the standard procedure relies on visual inspection through microscopy. In particular, a standard method is phase contrast microscopy, which enhances the image of transparent, unstained cells. The main metric of non-invasive cell culture assessment is estimation of the confluence. When the cells become confluent, i.e., they grow in a dense monolayer, cell growth stagnates, and the culture needs to be passaged.
As the decision to harvest the cells, for example, depends on the cell density, the integration of a microscopic quality control into the automated platform is crucial. Therefore, the StemCellDiscovery robotic laboratory integrates a fully automated high-speed phase contrast microscope that allows scanning of entire culture plates in less than 1 min per well [
19]. In order to evaluate the acquired images, an image processing algorithm is required to assess, for example, the cell density. Thereby, the image processing algorithm should (1) provide meaningful information, (2) be accurate and (3) be fast in order to allow downstream, simultaneous operation to the microscopy itself. A suitable quality control algorithm should provide information on the culture confluence within the timeframe of one minute and should be robust to minor changes in illumination and sharpness.
Deep learning has proven itself in recognition of other cell types such as induced pluripotent stem cells (iPSCs) [
20]. However, the growth behavior of hMSCs differs significantly from iPSCs as they do not form colonies but disperse across the culture surface in a fibroblast-like morphology. For this reason, we developed a deep learning-based approach to assess the confluence by recognizing hMSCs, detached cells and background for in-line quality control in fully automated cell cultivation.
In this publication, we present the StemCellDiscovery as a robotic laboratory for plate-based hMSC production in parallel. Furthermore, we employ automated high-speed microscopy in combination with a deep learning-based image processing algorithm. We demonstrate the suitability of the setup for daily culture assessment and quality control. Finally, we examine the system capacity in the context of a high-throughput operation.
2. Materials and Methods
2.1. Robot-Assisted Automated Cell Culture Laboratory
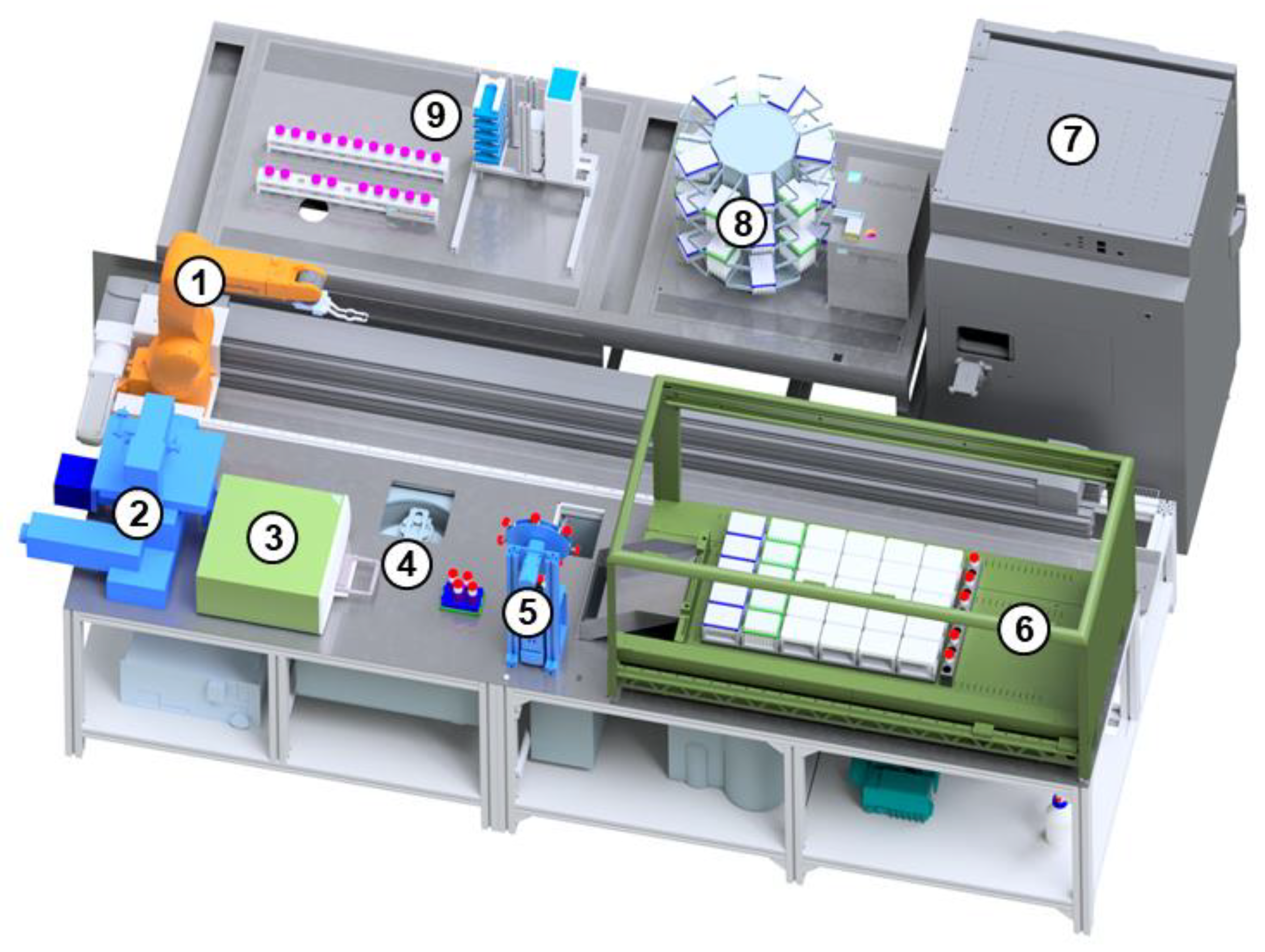
The StemCellDiscovery (
Figure 1) is a robotic platform for fully automated cell cultivation. It consists of several devices for treatment of cells that are enclosed in a cleanroom cabinet with HEPA filters (High Efficiency Particulate Airfilter) (
Figure 2).
In a central position is a robot (VS-087, Denso, Kariya, Japan) that can rotate around six axes and is mounted on a linear axis (Toothed belt axis EGC-185, Festo, Esslingen am Neckar, Germany) to provide full access of the devices. The StemCellDiscovery contains an incubator (Cytomat 24 C-IG, Thermofisher, Waltham, MA, USA) for storage of 360 cell cultures simultaneously at controlled CO2 levels. In addition, the StemCellDiscovery integrates a liquid handling unit (Microlab STAR, Hamilton Company, Reno, NV, USA) to handle medium and reagents, a centrifuge (4–16 KRL, Sigma Laborzentrifugen, Osterode am Harz, Germany) to separate cells and liquid, a decapper (custom build) to open and close tubes, a plate reader (Infinite M200 Pro, Tecan, Männedorf, Switzerland) and a microscope (Eclipse Ti2, Nikon, Minato, Japan) for visual assessment of the cells. The material required for processing is provided by storage hotels (custom build) that have the storage capacity for seven empty cell culture plates, 21 tubes for media and liquids with a capacity of 50 mL each and ten pipette tip racks. It is thereby possible to store media and reagents at ambient and cooled temperatures. After usage, the materials are disposed into a waste container underneath the system. The devices are positioned alongside the linear robot axis.
2.2. Production Monitoring and Control Software
The whole platform is operated through a control software that provides the interface for the user and manages all communication with the devices (
Figure 3). The software does not communicate with each device directly but via separate programs, called software agents. Each agent acts as a translator between control software and the individual devices. It communicates with the dedicated device via the individual software interface and with the control software via a standardized interface. When a device is added or changed, only the software agent needs to be modified while the control software remains unchanged. By adapting the corresponding agent, almost every communication interface such as USB, OPC (Open Platform Communications) and Ethernet can be interacted with [
21]. Via the interface of the main control software, the user can define protocols for entire processes by definition of command sequences in the drag-and-drop protocol editor.
The protocol definition also allows integration of decision nodes that branch the process into different sequences depending on the fulfillment of defined criteria. For example, a decision node could branch off either into a harvesting process if the confluence is above or into a medium change if the confluence is below a threshold value.
The processes can be started at predefined times and can run simultaneously. The software logs the position and status of every cell product, material and disposable in the platform. [
22]
2.3. High-Speed Microscopy
The imaging system integrated into the StemCellDiscovery is based on a phase contrast high-speed microscopy design (
Figure 4A.1) [
19]. The loading and unloading processes of the cell culture multi-well plates as well as image acquisition are fully automated. The system uses a continuous scanning process with flash illumination.
Six-well culture plates are digitized in motion and the focus is continuously adjusted by hardware auto-focus systems. At a magnification of 4×, the resulting image of a well in a six-well plate consists of [12 × 12] stitched single 5 MPx shots, resulting in a total of [25,047 × 24,363] px with a resolution of 1.69 µm per px (compare
Figure 4A.2).
Due to surface tension, the surface of the liquid medium in the wells curves towards the walls of the culture plate. The curving of the liquid acts like a lens. As a result, phase contrast conditions are only present in the flat part of the liquid lens and bright field conditions occur at the border of the well. Therefore, the image is cropped to the phase contrast region with a size of [8208 × 8208] px.
2.4. Training and Validation of the Deep Learning Network for Confluence Detection
Image evaluation is a downstream application for image acquisition with the high-speed microscope. In the StemCellDiscovery, this was implemented as a separate module within the microscope software in order to (1) allow simultaneous processing and (2) make optimal use of the microscope’s computing power.
In order to detect and distinguish cells on the culture well surface with the deep learning-based algorithm, four different classes were defined:
hMSCs: cells that have a fibroblast-like behavior (
Figure 4B.2);
Detached cells: cells in suspension that are visible as white spots (
Figure 4B.3);
Background: the cell-free area, also containing debris of cells and non-classified particles (
Figure 4B.1);
Unknown: pixels that are not defined as a specific class, or are in between two classes, introducing ambiguity (
Figure 4(B.4)). This class is not trained and only implemented in order to prevent mislabeling at class borders. The unknown class is only present in training and test datasets.
The dataset for deep learning was built from 19 example images covering low and high cell densities. The phase contrast region of each image was split in 4 × 4 parts. These parts were divided in training (80%) and test (20%) datasets. In order to segment the training test set, the open source toolbox Phantast was used, which provides tools for cell and background segmentation of phase contrast microscopy images [
23]. The generation of the ground truth followed a step-based algorithm (
Figure 4C):
Furthermore, data augmentation techniques were used to cover grayscale variation, affine transformation and blurring. The image parts were enlarged from [2052 × 2052] px by mirroring the borders to [2236 × 2236] px to preserve the size of the output image. The image part was then split into overlapping tiles sized [412 × 412] px with stride 228 px. In total, 56,700 tiles were used for the training and test datasets. Furthermore, nine full-well images were prepared for an evaluation dataset without augmentation, resulting in 2187 tiles. The training set is the basis for training the algorithm, while the test dataset monitors the training. The evaluation dataset reflects normal use of the training. Images of the microscope are analyzed directly by the algorithm, but the unknown class remains uncalculated for comparison with the Phantast algorithm.
The deep learning network is based on a U-Net architecture [
24] with an additional batch normalization layer before each ReLu layer. For any given [412 × 412] px input image, the network generates a corresponding [228 × 228] px centered segmentation map. By using an overlapping tiling-based approach and mirroring at the image borders, as proposed by Ronneberger et al. [
24], segmentation maps for large images can be produced. Furthermore, convolution layers, as well as transposed convolution layers, were initialized using Xavier initialization [
25]. Softmax loss is used as a loss function. The algorithm is implemented in the Caffe deep learning library [
26]. The training is based on statistic gradient descent, with a base learning rate of 0.0005, batch size of 3, gamma of 0.1, momentum of 0.9 and a weight decay of 0.001 over 12 epochs.
The algorithm allows pixel classification on the whole image. For automation, the confluence Ci is calculated for each class as the ratio of pixels classified as class i in contrast to the number of all pixels. Furthermore, the segmentation masks are provided. On this basis, the individual calculation of the distribution of hMSCs or counting of detached cells is possible.
The segmentation quality is evaluated using the confusion matrix. Here, the agreement of the respective segmentation mask is given as a true positive prediction
tp or a class-wise recall, or a false positive prediction
fp; the background mask is predicted as detached cells or hMSCs, and a false negative prediction
fn; hMSCs and detached cells from the ground truth are predicted as background. Thus, the detection accuracy of individual classes can be investigated. The
F1 score is defined as
and is calculated for each class.
2.5. Automated Cell Culture
In order to demonstrate the feasibility of automated production including the deep learning-based confluence algorithm, human adipose-derived mesenchymal stem cells from the abdominal fat tissue were cultivated on the StemCellDiscovery. The cells were obtained from two female donors (age 34 and 60) from the Domhofklinik, Aachen, with written consent. The cells were seeded in the third passage onto uncoated six-well plates (VWR International GmbH, Radnor, PA, USA) with 2.5 × 103 cells per cm2 (24.000 cells per well) by the liquid handling unit (LHU) in the StemCellDiscovery. To each well, 3 mL DMEM (Dulbecco’s Modified Eagle’s Medium) medium (ThermoFisher Scientific Inc., Waltham, MA, USA) containing 1 g/L glucose and L-glutamine was added. The medium was supplemented with 10% fetal calf serum (Sigma-Aldrich Inc., St. Louis, MO, USA) and 1% penicillin/streptomycin (ThermoFisher Scientific Inc., Waltham, MA, USA). The cells were cultivated on the automated system for six days with daily evaluation by automated microscopy and medium change every other day. In between processing, the cells were stored in the incubator at 37 °C and 5% CO2.
The cells were harvested when at least three of the six wells reached 70% confluence. For this, the medium was aspired and cells were washed with 1.5 mL PBS (ThermoFisher Scientific Inc., Waltham, MA, USA) per well on the LHU. To each well, 600 µL 0.05% Trypsin/EDTA (ThermoFisher Scientific Inc., Waltham, MA, USA) was added and the plate was shaken for 60 s at 1200 rpm on the shaker module of the LHU. Then, the culture plate was transported to the incubator. After incubation for 5 min at 37 °C and 5% CO2, the culture was transported back to the LHU and shaken for 60 s at 1200 rpm. Subsequently, 2.4 mL of medium per well was added to each well of the plate and cells were resuspended by pipetting the liquid up and down repeatedly. The cell suspension from all wells was aspired and pooled in a 50 mL tube. The robot transferred the tube to the centrifuge, where cells and medium were separated through centrifugation at 1000 rpm (approximately 150 rcf) for 5 min at ambient temperature. The supernatant was aspired from the tube and the pellet was resuspended in 5 mL medium. The cell count was determined outside of the StemCellDiscovery on a cell counter (NucleoCounter® NC-3000™, ChemoMetec A/S, Allerod, Denmark) using a Via1-Cassette™ (ChemoMetec A/S, Allerod, Denmark).
2.6. Modeling of the Production Environment
In order to estimate the production capacity of the StemCellDiscovery, the process and production environment were modeled in JaamSim (
http://jaamsim.com (accessed on 24 March 2021)), a discrete event-based simulation software for emulation of production process networks. Simulation entities were used to represent the cell cultures. The simulation model featured different entity processers to represent the plate reader, microscope and LHU. Each entity processor contains a queue, which handles simulation entities by the first-in-first-out principle. The plate reader station excludes plates from the process with a probability of 1 to 1000 to emulate contamination of plates. The microscope station triggers an external Python script that calculates the confluence value of the cell cultures using the cell growth model described below. Processes on the LHU require resources such as pipette tips and liquids, which are seized permanently from the stock in the required amount. The consumed amounts were modeled according to the protocol. Empty resource stocks are automatically refilled by triggering a process, which occupies the LHU for about one to two minutes. The resource quantities utilized by the LHU and refilled by the restocking processes are given in
Table 1. The simulation models five processes: The quality control process uses the plate reader, which measures turbidity in order to detect clouding of the cultures due to contamination. In the same process, it also measures absorption of light at 430 nm and 560 nm to detect when the pH indicator phenol red in the medium changes from red to yellow, as it typically happens when the medium is acidified due to bacterial contamination. In the second process, the growth of the cells is assessed on the microscope. The remaining processes are medium change, and the harvesting and seeding processes, which use the LHU.
Although the real processes require additional devices, such as the robot or the centrifuge, these devices are not explicitly modeled as they can be implemented as a part of other devices. For example, the centrifuge is only in operation during the harvesting process and its operation is therefore included in the utilization of the LHU. The other devices are therefore implicitly modeled by calculating the processing durations as the sum of individual steps while considering the standard deviations of the durations. The durations of the individual process steps were estimated based on system log files from previous automated cultivation runs and were modeled as standard distributions to reflect the variability of the production environment (see
Table 2).
2.7. Cell Growth Modeling
Since the cells in culture are in the exponential growth phase most of the time and are harvested before they enter the stationary phase, the growth of hMSCs in culture can be described using a simple exponential growth function. The confluence
C(
t) of a culture was mathematically modeled as
with growth rate µ (1/d) and initial confluence
C0 for
t = 0. The culture age
t (in d) is determined by the difference between time of seeding and current time. While, in reality, each well of a six-well plate can be considered as an individual culture with its own growth curve, for simplification, only one confluence is calculated per simulated culture plate.
2.8. Simulation of the Cell Culture Process
The operation of the StemCellDiscovery was simulated for 21 days, assuming different numbers of cell culture plates seeded per day (10, 12, 13 and 14). Each cell culture was represented by a simulation entity that is assigned a value for µ and C0 upon generation (i.e., seeding of a culture plate) by drawing a random number from a defined distribution.
On the first day of the simulation period, the job list on the simulated StemCellDiscovery was empty except for the cell cultures to be seeded. The simulated cultures were put into the job list for every subsequent day to be processed according to the culture protocol (daily microscopy, daily quality control on the plate reader, medium change every other day). When the simulated cell culture was on by the entity processor representing the microscope, the confluence was calculated for each simulated cell culture from equation (1) based on the culture age, assigned µ and C0. If the calculated confluence reached 70%, the culture was harvested and then not further included in the job lists of the subsequent days. For each day of the simulation period, the number of processed cell cultures was counted, and the utilization times of the plate reader and microscope were calculated as well as the overall daily processing time of the system (in which at least one device was busy).
3. Results and Discussion
3.1. Deep Learning-Based Confluence Estimation
After training, the deep learning algorithm computes images directly taken from the microscope. An example of different image tiles with confluences between 15% and 95% obtained from selected times and wells in the cultivation of the donor hMSCs is shown in
Figure 5. To quantify the accuracy of the detection, the
score and confusion matrix were calculated. The values are calculated on the whole evaluation set that has not been part of the network training. The confusion matrix (
Figure 6A) gives the ratio of the ground truth to the prediction by the network for each class. The
score of the background is 0.859, while the hMSC score is 0.833. These
scores indicate that the two main classes are detected with a sufficient accuracy. The class of the detached cells has the lowest value of 0.6811. In detail, it is noticeable that a high proportion of false negative predictions occur, especially in the class hMSCs with 23.7 %. This means that areas segmented as hMSCs in the ground truth are identified as background by the prediction of the network. It is important to note that the
score and confusion matrix are dependent on both the prediction of the network and the correct segmentation of the ground truth. In order to assess the phenomenon, we can examine the images in more detail.
Figure 6B,C show two example images generated from the evaluation set. The red overlay shows the false negative regions, the green overlay shows the class-wise recall for hMSCs and the blue overlay shows the false positive prediction.
Figure 6B shows a representative example of the outcome. It shows that the hMSCs have been identified well and the class-wise recall is high. In accordance with the confusion matrix, the number of pixels that have been detected as false positives is marginal. However, there is a discernible number of false negative pixels in the image. This effect is even more significant in
Figure 6C, which is a part image exhibiting high false negative classification. It is noticeable that particularly the regions without cells are marked red, meaning that they were declared as hMSCs in the ground truth but not recognized as cells by the deep learning algorithm. This is also in line with the high values of false negatives in the confusion matrix, where image sections labeled as “hMSCs” by the Phantast algorithm were detected to be background by the network. An examination of the images, however, shows that the areas labeled as false negative are indeed not covered by cells but belong to the actual background. Other images show similar behavior. This indicates that the automatic generation of the ground truth using the Phantast algorithm [
23] is less accurate than the prediction of the deep learning network. These inaccuracies are also present within the training and test sets. Some of these are covert by using the class unknown, especially at the cell borders. However, more importantly, the inaccuracies are statistically distributed and cover a small area in comparison to the original class the pixels belong to. As deep learning provides an estimation of the class membership, the most probable value is trained. Thus, the output can be more accurate than the input.
Pixel classification with deep learning also has other advantages over conventional algorithms: (1) It is robust against slight variations in initial conditions such as differences in brightness or slight blurring during acquisition. (2) The algorithm allows determination of cell distribution across the culture plate and identification of particularly dense regions. (3) In addition to classical algorithms such as Phantast [
23] and adaptive thresholding [
27], the additional cell class “detached cells” adds new information to the status of the cell culture.
The algorithm achieved a runtime of 29 s per [8208 × 8208] image on an NVidia GeForce 1080Ti graphics card. This is within the period of image capture on the high-speed microscope. Therefore, the confluence determination generally can run in parallel to the image acquisition. All results considered, we can conclude that the confluence detection algorithm is a suitable in-line quality control technology for fully automated, robot-assisted cell culture.
3.2. Fully Automated Cell Cultivation Including In-Line Quality Control
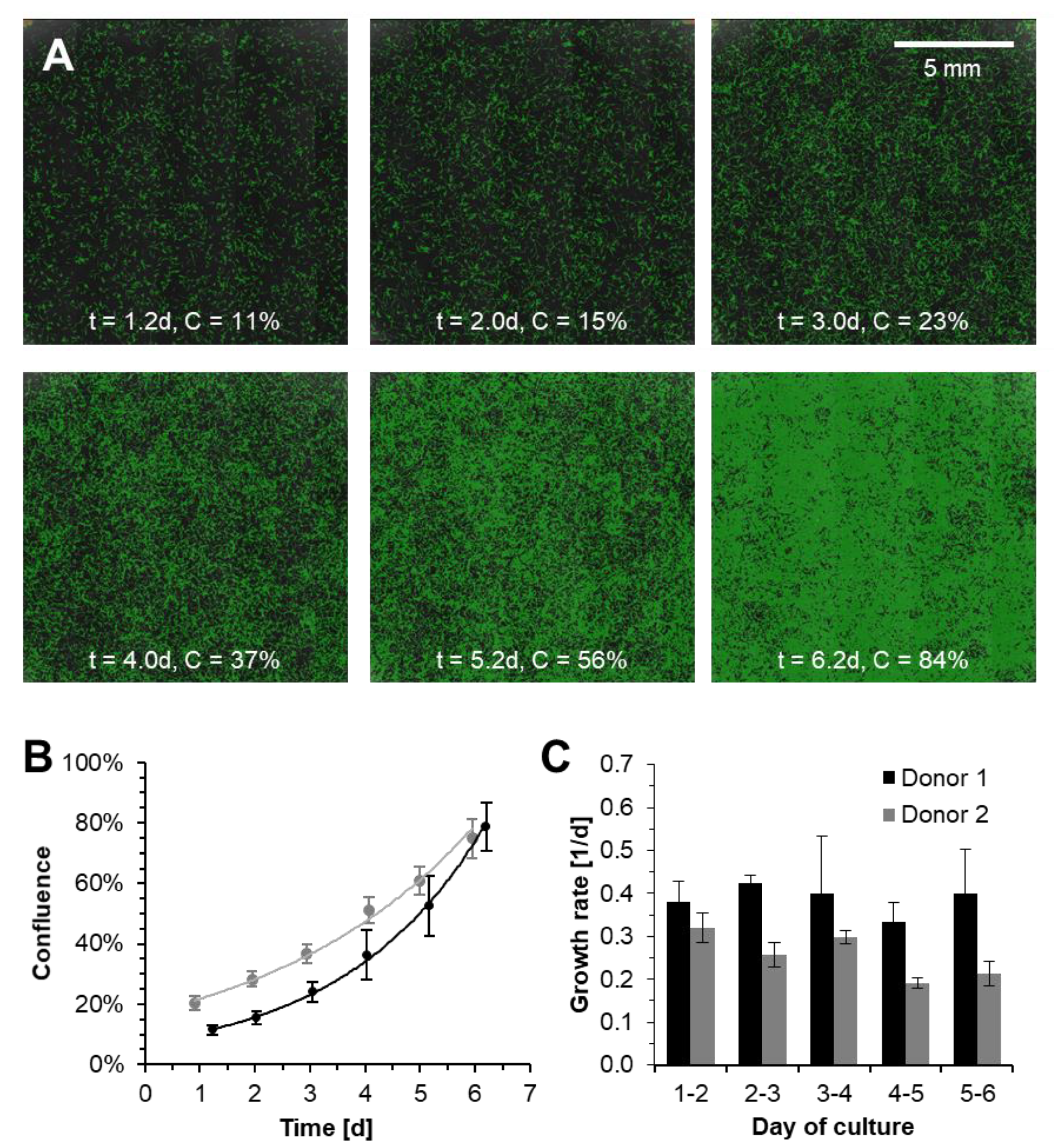
The cultivation process was demonstrated on the StemCellDiscovery using adipose-derived hMSCs from two female donors that were seeded into two six-well plates. Each well was assessed through a microscope daily for six to seven days and processed with the deep learning-based image processing algorithm, giving a total of six confluence curves for each of the six-well culture plates.
Figure 7A shows images of the cells from donor 2 taken from well A2 of the cell culture from days 1–6. The cells are spread evenly across the cell surface and confluences increase exponentially over time, as the graph in
Figure 7B shows clearly.
The figure shows the average confluence across all six wells of a culture plate with standard deviation. Both cell lines exhibit exponential growth in all wells, but confluences are underlying a biological variation across the wells. The confluence values differ with a standard deviation of 1–10 percentage points. The microscope images, such as those shown in
Figure 5, illustrate that the labeling of the deep learning-based algorithm is generally in good agreement with the cells that can be identified with the human eye. Additionally, the images in
Figure 7B show an even distribution of the cells across the cell culture surface. Therefore, it is unlikely that the deviation originates from errors in the confluence detection algorithm but is more likely inherent to the process and the biological variability.
Exponential regression of the growth curves yields an overall average growth rate of 0.39 1/d for donor 1 (t
D = 42.9 h) and 0.26 1/d for donor 2 (t
D = 64.5 h). Based on the confluences, the growth rates per well and per day were calculated, as shown in
Figure 7C. For donor 1, average growth varied between 0.33 and 0.43 1/d, while donor 2 proliferated more slowly with µ = 0.19–0.39 1/d. After harvest, the cultures, however, yielded cell numbers in the same range with 76.5 (donor 1) and 65.0 thousand cells (donor 2).
The significant difference in the growth between the two cultures can most likely be attributed to the biological variability. Indeed, the donors differed significantly from each other in terms of age (34 and 60). It is well understood that increasing age can have a negative impact on cell proliferation [
28,
29,
30]. The doubling times observed are comparable with findings in the literature. Zhu et al. [
28] reported doubling times for adipose-derived stem cells that increased with age between 35.8 and 72.6 h, with an average doubling time of 55.2 h. Yang et al. [
29] confirmed these findings and reported doubling times within the same range.
In addition, we could observe a variation in the confluences between the different wells of the same cultures. Multiple factors can lead to such a variation. The examination of the individual images indicates that the image processing algorithm can detect the cells very well, rendering it unlikely for measurement errors to be the main cause. It is more plausible that the fluctuations in confluence are due to the inherent biological variability of the donor-derived stem cells. Therefore, we can conclude that the system is generally capable of cultivating donor-derived hMSCs.
3.3. Modeling of the System Capacity under High-Troughput Conditions
For economic operation of the system, it is key that the StemCellDiscovery can be operated at high throughput. In order to estimate the capacity of such an automated laboratory, we modeled the process and production environment in JaamSim. The durations of the individual process steps were estimated based on system log files from previous automated cultivation runs. The cell growth was modeled with exponential growth. In order to reproduce the inherent biological variability, values for actual seeding confluence C0 and growth rate µ were drawn randomly as described below.
In order to determine the distribution for C0, the exponential growth curve was fitted to the confluence measurements of each of the twelve wells from the two culture donors described above. The resulting initial confluence of C0 = 12 ± 5% was used to draw a value for C0 by the simulation, assuming a normal distribution. The upper and lower boundaries for C0 were set to 2% and 22%, as these were the minimum and maximum initial confluences that occurred in the cultivation.
For donor-derived hMSCs, it is well known that the growth rate µ can vary significantly from donor to donor [
28,
29,
30]. Therefore, we can neglect technical variances and assume that the main factor determining the growth rate is the biological variability. Thereby, the variance of the doubling times observed in the cultivations on the StemCellDiscovery (40.0–68.5 h) is comparable to that of the doubling times reported in the literature (35.8–72.6 h, [
28]). For the simulation, therefore, we assume an even distribution of doubling times between 40 and 70 h (mean 55 h), corresponding to growth rates of 0.24–0.42 d.
3.4. Estimation of System Capacity through Simulation
The simulation over 21 days assumed different numbers of culture plates to be seeded per day to determine the resulting system utilization rates. For each seeding rate, the simulation was repeated ten times in order to account for stochastic effects of the biological variability.
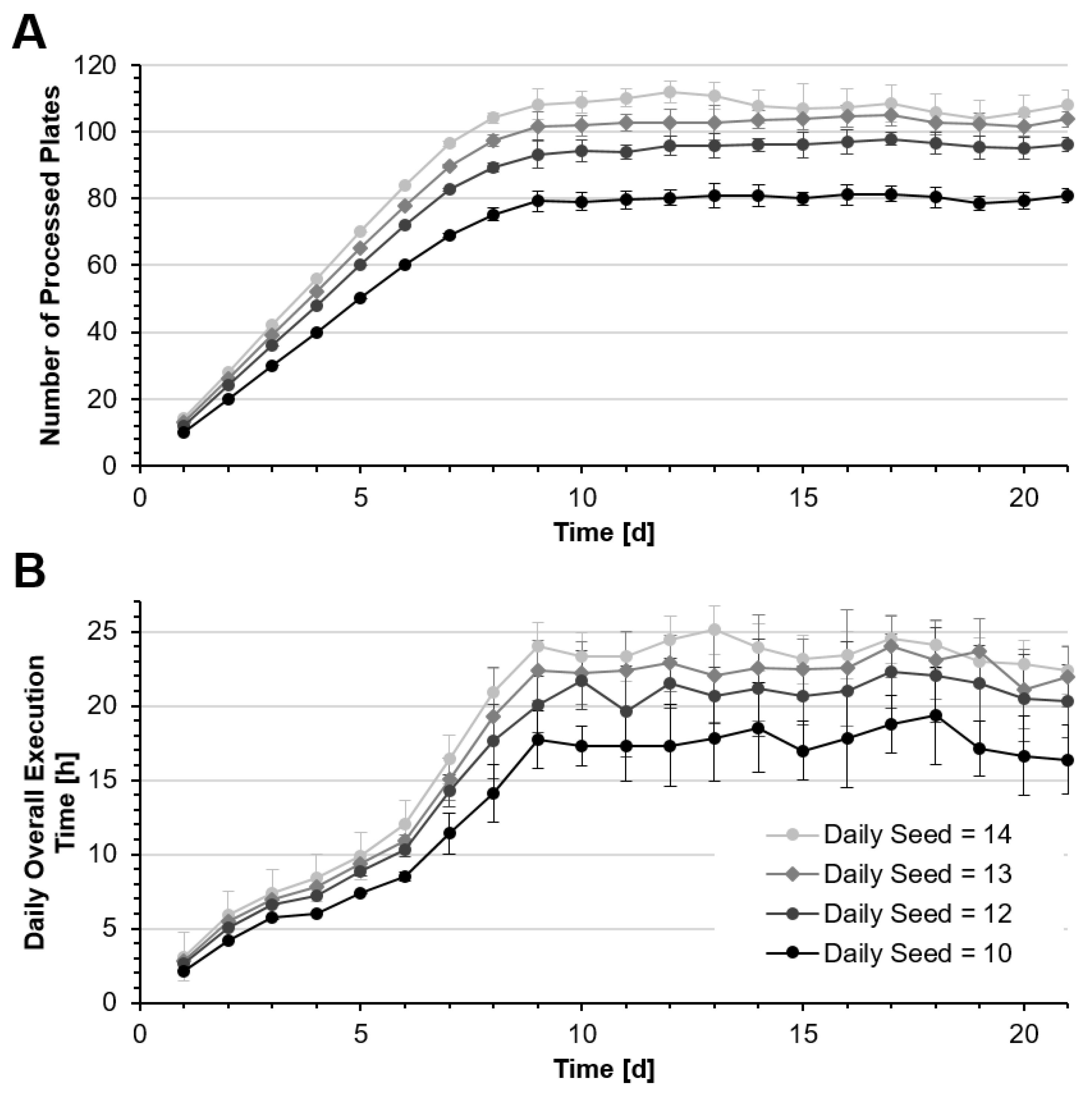
Figure 8A shows the numbers of culture plates processed per day while
Figure 8B shows corresponding overall system utilization. For all seeding rates, the number of cultures processed increases over the first nine days and then reaches a steady state, when the same number of cultures are harvested and removed from the system as new ones are seeded.
Table 3 shows the device and system utilization as well as the numbers of plates processed. If 14 new cultures are seeded every day, about 108 plates can be processed on average within 24 h. In this case, the daily process time is 23.6 ± 2.6 h. Due to the biological variability, the actual daily harvesting yield can vary considerably, as some cultures require longer incubation than others.
Since the harvesting process requires an about five-fold longer time than a medium change, the number of harvests per day impacts the system utilization per day. As a result, the daily processing time can vary and, in some cases, will be longer than 24 h. In addition, it has to be taken into account that stocking and maintenance may require additional time every day. Therefore, a daily seed of twelve new cultures may represent the system capacity more realistically, with 95.6 ± 3.0 plates processed per day and a daily system utilization of 21.0 ± 2.5 h. This still means that on 11.5% of the days, the system may require longer than 24 h to process the batch of cells. It is likely, however, that this deviation can be absorbed by the schedule of the subsequent days.
The system utilization also shows that the processes performed by the liquid handling unit are the bottleneck in the system, which is in operation 98% of the time.
4. Conclusions
Fully automated, robot-assisted technologies can assume labor-intensive tasks, especially in areas where highly parallelized generation of cells in small scale is required. In this paper, we presented the StemCellDiscovery for automating the cultivation of adherent cells. The system is designed as a fully equipped robotic laboratory that can host all the equipment required for automated cell production and monitoring. In contrast to many commercially available automated systems, the StemCellDiscovery integrates an industrial six-axis robot for complex handling and manipulation steps and provides room for storage spaces and the different laboratory devices. Furthermore, a user-friendly control software allows protocol definition and operation of the system via an intuitive graphical user interface with minimal training required.
Another key element is the in-line quality control through visual assessment. This does not only require performant imaging systems such as the high-speed microscope described by Schenk et al. [
19] but also image processing algorithms capable of translating the image into meaningful information. Such an approach was first introduced into the StemCellFactory, a fully robotic laboratory for the production of iPSCs [
31]. Here, we introduced a deep learning-based image processing approach for detection of hMSCs. We demonstrated the capability of the trained algorithm to correctly detect cells, background and detached cells in a phase contrast microscope image and calculate confluences in cultures with low to high density. Thereby, the algorithm performed even better than the conventional algorithm that was used to segment the ground truth [
23]. The deep learning-based algorithm is a vital element for automated decision making in robot-assisted, autonomous production. However, the field of application reaches beyond automation as the algorithm may also provide a useful tool for standardized, non-invasive culture and growth assessment in manual cell culture.
Using this algorithm, we showed successful automated cultivation of adipose-derived hMSCs on the StemCellDiscovery from two donors in six-well plates. These results should be interpreted as a first proof of concept for robot-assisted production of hMSCs on the StemCellDiscovery. Future investigations will include more sophisticated evaluation of the system by benchmarking the results against manual culture.
While the system has a storage capacity of more than 360 cell culture plates, which is comparable to other commercial systems [
14], it does not yet allow any conclusions to be drawn about the workload that can actually be processed within 24 h. In order to investigate the workload capacity of such a robotic cultivation system, we modeled the production environment in a simulation software, taking into account the inherent biological variability. The results suggest that the system can process over 95 cell culture plates per day, with an average harvesting rate of twelve culture plates per day. Since the system can be operated day and night, it can process a significantly higher number of culture plates than what a human operator would be able to process within a working shift. The main tasks of the operator thus shift towards system operation, restocking of consumables, process supervision and evaluation. In consequence, the throughput of cell culture laboratories can be increased significantly while allowing allocation of the highly trained personnel to more important tasks.
The simulation also showed that the main bottleneck of the system is the liquid handling processes, which are also most affected by day-to-day process variations. This also affects material consumption, requiring foresighted material supply. In the future, these challenges may be addressed by intelligent process supervision and anticipation. The confluence detection algorithm presented in this paper provides insight on the current cell density and allows quantification of the growth over time. A combination of this confluence detection algorithm with a modeling of the cell growth may allow anticipation of the processes. Thus, predictive process scheduling and intelligent resource management will become feasible, paving the way for fully autonomous cell production.