Construction of an Artificial Cell Capable of Protein Expression at Low Temperatures Using a Cell Extract Derived from Pseudomonas fluorescens
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of CFPES
2.1.1. CFPES Derived from P. fluorescens
2.1.2. CFPES Derived from E. coli
2.2. Measurement of Total Protein Concentration in the Cell Extract
2.3. Luciferase Assay of the CFPES
2.4. Formation of the LBACs
2.5. Fluorescent Imaging of the LBACs
3. Results
3.1. Optimization of the Cell Extract Derived from P. fluorescens for High-Yield Protein Expression
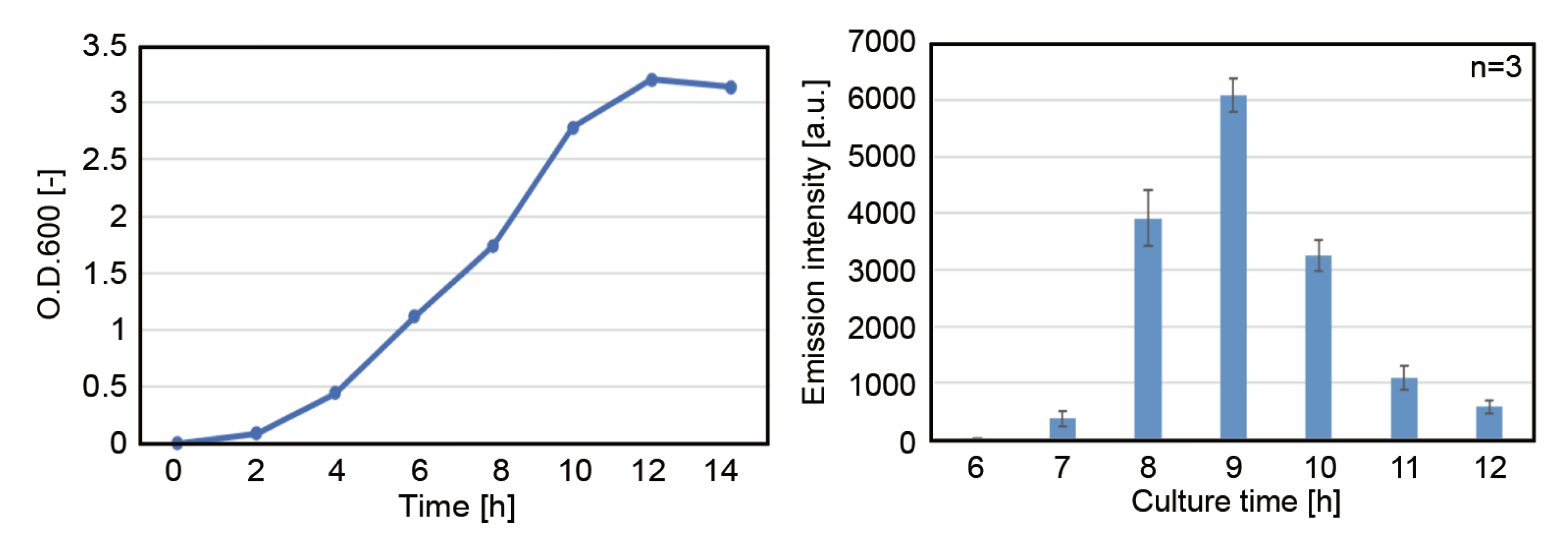
3.1.1. Effect of Incubation Period before Cell Harvest
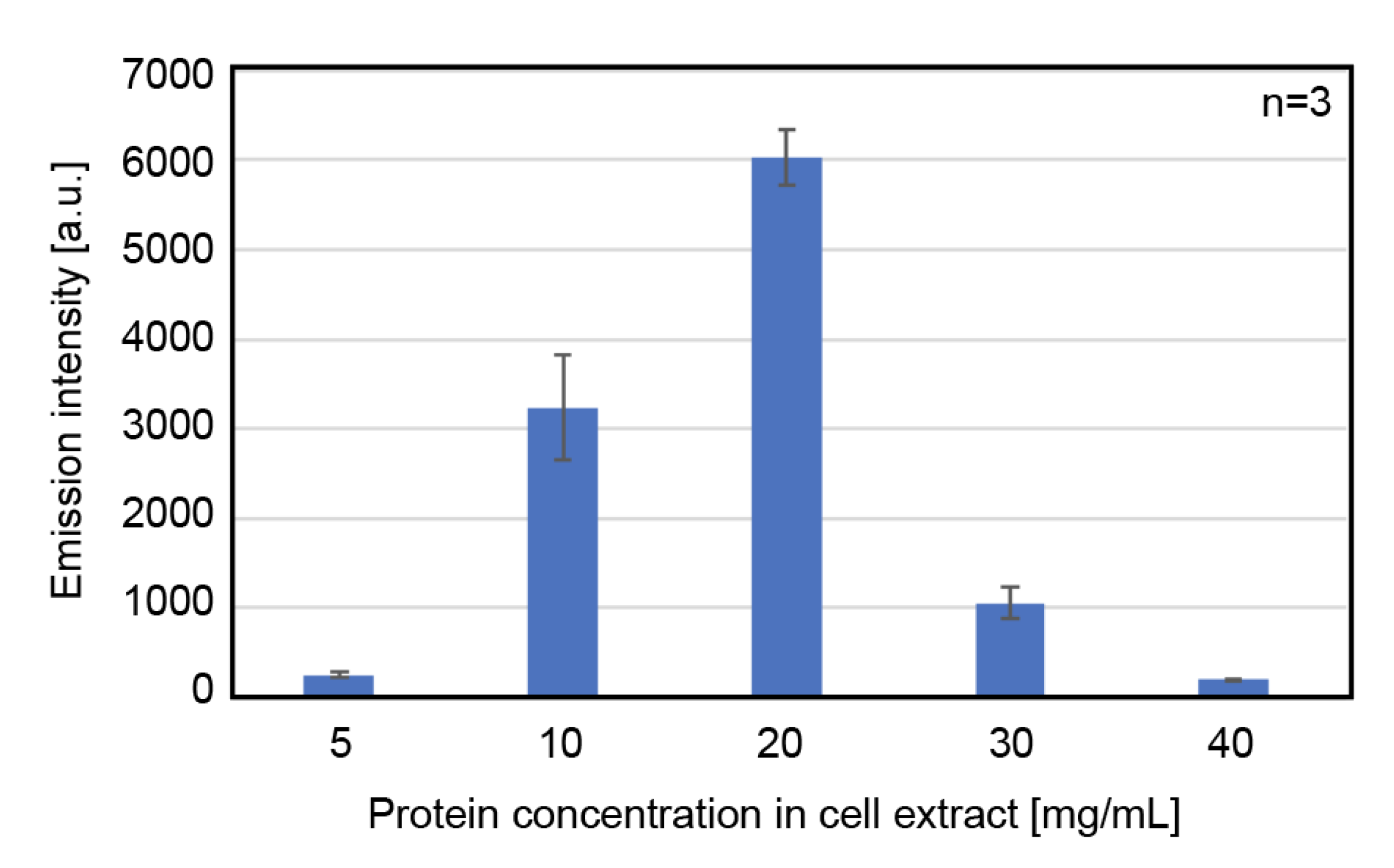
3.1.2. Effect of Total Protein Concentration of the Cell Extract on Protein Expression Activity
3.2. Optimization of Other Components in the CFPES for High-yield Protein Expression
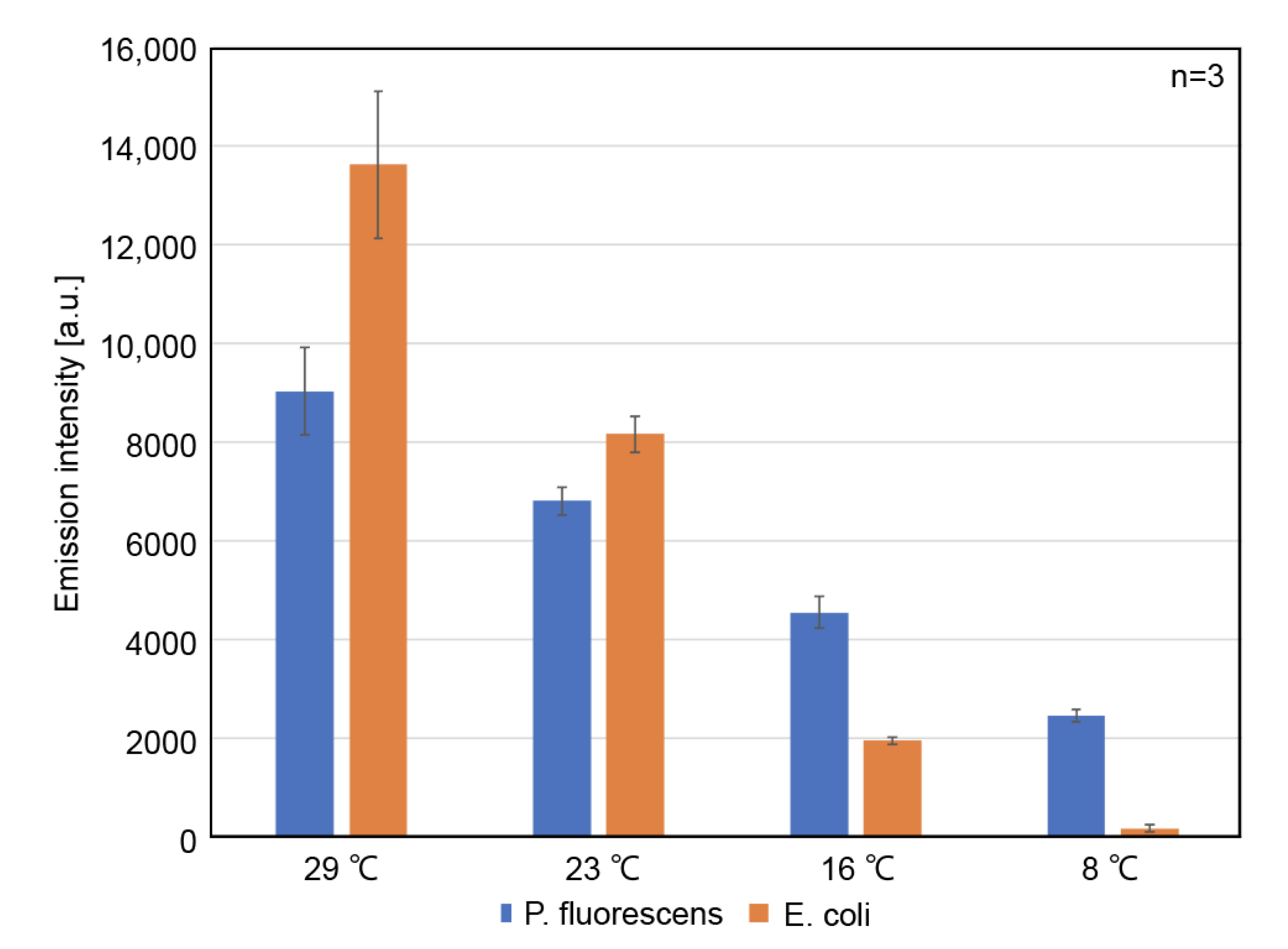
3.3. Protein Expression Capability of the Optimized CFPES at Various Temperatures
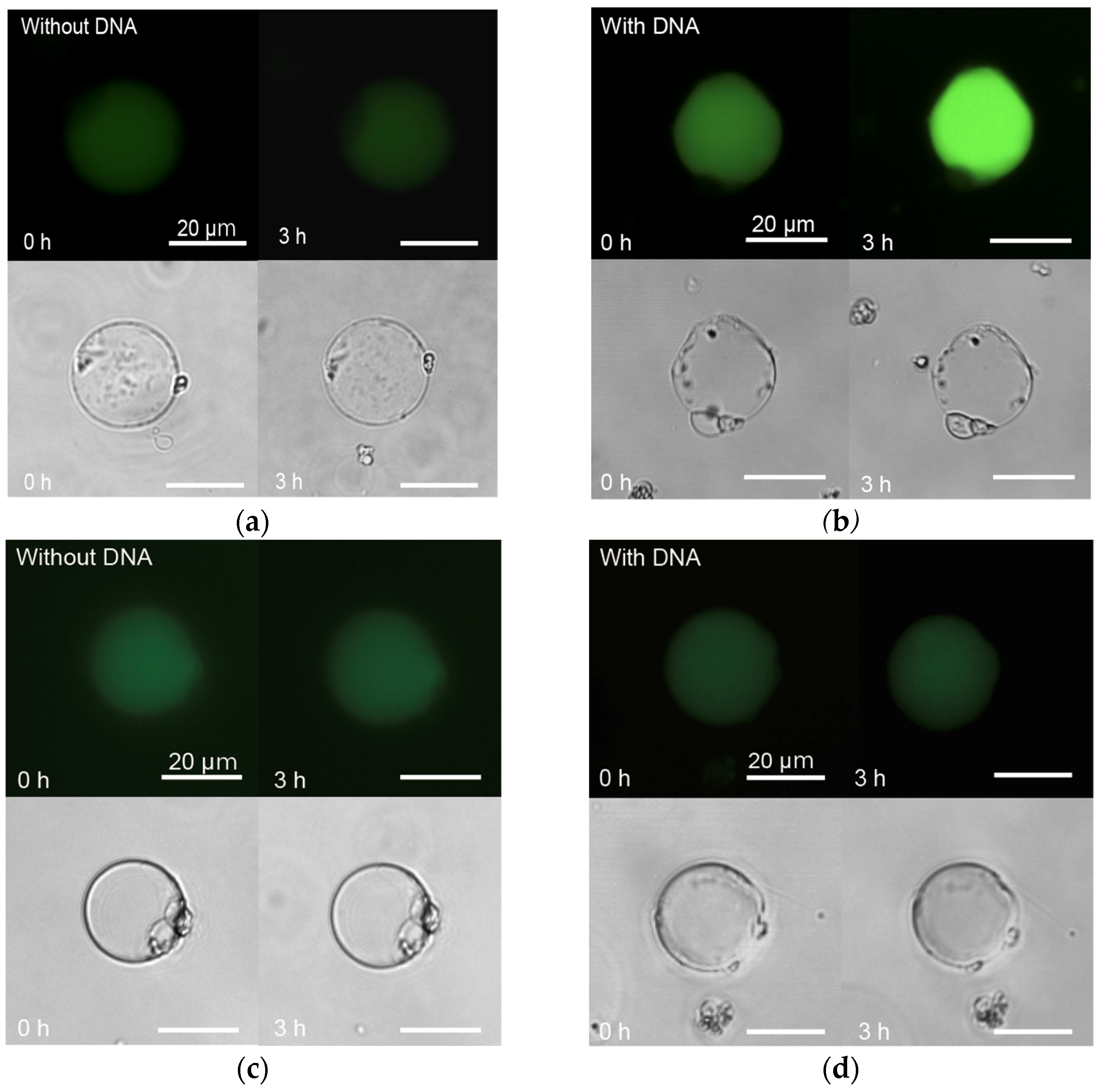
3.4. Protein Expression at 8 °C, in the LBAC Derived from P. fluorescens
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garamella, J.; Garenne, D.; Noireaux, V. TXTL-based approach to synthetic cells. Methods Enzymol. 2019, 617, 217–239. [Google Scholar] [PubMed]
- Lentini, R.; Martín, Y.N.; Forlin, M.; Belmonte, L.; Fontana, J.; Cornella, M.; Martini, L.; Tamburini, S.; Bentley, E.W.; Jousson, O.; et al. Two-way chemical communication between artificial and natural cells. ACS Cent. Sci. 2017, 3, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Rampioni, G.; D’Angelo, F.; Messina, M.; Zennaro, A.; Kuruma, Y.; Tofani, D.; Leoni, T.; Stano, P. Synthetic cells produce a quorum sensing chemical signal perceived by Pseudomonas aeruginosa. Chem. Commun. 2018, 54, 2090–2093. [Google Scholar] [CrossRef] [PubMed]
- Berhanu, S.; Ueda, T.; Kuruma, Y. Artificial photosynthetic cell producing energy for protein synthesis. Nat. Commun. 2019, 10, 1325. [Google Scholar] [CrossRef] [PubMed]
- Seike, Y.; Kobori, S.; Whitaker, C.; Matsuura, T.; Yokobayashi, Y. Programmable artificial cells using histamine-responsive synthetic riboswitch. J. Am. Chem. Soc. 2019, 141, 11103–11114. [Google Scholar]
- Hindley, W.J.; Zheleva, G.D.; Elani, Y.; Charalambous, K.; Barter, C.L.M.; Booth, J.P.; Bevan, L.C.; Law, V.R.; Ces, O. Building a synthetic mechanosensitive signaling pathway in compartmentalized artificial cells. Proc. Natl. Acad. Sci. USA 2019, 116, 16711–16716. [Google Scholar] [CrossRef] [PubMed]
- Garamella, J.; Majumder, S.; Liu, P.A.; Noireaux, V. An adaptive synthetic cell based on mechanosensing, biosensing, and inducible gene circuits. ACS Synth. Biol. 2019, 16, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Adamala, P.K.; Martin-Alarcon, A.D.; Guthrie-Honea, R.K.; Boyden, S.E. Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem. 2017, 9, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Contreras-Llano, E.L.; Morris, E.; Mao, M.; Tan, C. Minimizing context dependency of gene networks using artificial cells. ACS Appl. Mater. Interfaces 2018, 10, 30137–30146. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.Z.; Hayes, A.C.; Shin, J.; Caschera, F.; Murray, M.R. Protocols for implementing an Escherichia Coli based TX-TL cell-free expression system for synthetic biology. J. Vis. Exp. 2013, 79, e50762. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, N.; Tamura, T. Cell-free protein synthesis using cell extract of Pseudomonas fluorescens and CspA promoter. Biochem. Biophys. Res. Commun. 2004, 319, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Noireaux, V.; Libchaber, A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl. Acad. Sci. USA 2004, 101, 17669–17674. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Copeland, E.C.; Padumane, R.S.; Yong-Chan, K. A crude extract preparation and optimization from a genomically engineered Escherichia coli for the cell-free protein synthesis system. Methods Protoc. 2019, 2, 68. [Google Scholar] [CrossRef] [PubMed]
- Caschera, F.; Noireaux, V. A cost-effective polyphosphate-based metabolism fuels an all E. coli cell-free expression system. Metab. Eng. 2015, 27, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Köhler, T.; Heida, T.; Hoefgen, S.; Weigel, N.; Valiante, V.; Thiele, J. Cell-free protein synthesis and in Situ immobilization of deGFP-MatB in polymer microgels for malonate-to-malonyl CoA conversion. RSC Adv. 2020, 10, 40588–40596. [Google Scholar] [CrossRef]
- Miyake, R.; Kawamoto, J.; Wei, L.Y.; Kitagawa, M.; Kato, I.; Kurihara, T.; Esaki, N. Construction of a low-temperature protein expression system using a cold-adapted bacterium, Shewanella sp. strain Ac10, as the host. Appl. Environ. Microbiol. 2007, 73, 4849–4856. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, J.; Jewett, C.M. Development of a Pseudomonas putida cell-free protein synthesis platform for rapid screening of gene regulatory elements. Synth. Biol. 2018, 3, ysy003. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukumoto, M.; Tonooka, T. Construction of an Artificial Cell Capable of Protein Expression at Low Temperatures Using a Cell Extract Derived from Pseudomonas fluorescens. Processes 2021, 9, 212. https://doi.org/10.3390/pr9020212
Fukumoto M, Tonooka T. Construction of an Artificial Cell Capable of Protein Expression at Low Temperatures Using a Cell Extract Derived from Pseudomonas fluorescens. Processes. 2021; 9(2):212. https://doi.org/10.3390/pr9020212
Chicago/Turabian StyleFukumoto, Mana, and Taishi Tonooka. 2021. "Construction of an Artificial Cell Capable of Protein Expression at Low Temperatures Using a Cell Extract Derived from Pseudomonas fluorescens" Processes 9, no. 2: 212. https://doi.org/10.3390/pr9020212
APA StyleFukumoto, M., & Tonooka, T. (2021). Construction of an Artificial Cell Capable of Protein Expression at Low Temperatures Using a Cell Extract Derived from Pseudomonas fluorescens. Processes, 9(2), 212. https://doi.org/10.3390/pr9020212




