Salicylate or Phthalate: The Main Intermediates in the Bacterial Degradation of Naphthalene
Abstract
1. Introduction
2. Some Representatives of Naphthalene Destructors
3. Degradation of Naphthalene by Gram-Negative Bacteria
4. Degradation of Naphthalene by Gram-Positive Bacteria
5. Peculiarities of Econiches for Naphthalene-Degrading Bacteria
- Bioaugmentation is a bioremediation strategy that involves introducing indigenous microorganisms to the contaminated site to detoxify and degrade environmental contaminants. This strategy is used in both anaerobic [94] and aerobic conditions. Several successful bioaugmentation cases have been documented. Decomposing naphthalene Streptomyces sp. strain QWE-35, isolated from activated sludge, was introduced into a membrane bioreactor, which significantly increased the efficiency of naphthalene decomposition [95]. The effect of microbial inoculation on naphthalene mineralization in biosolid treatment was evaluated in soil manure microcosms. Inoculation by Pseudomonas putida G7 carrying the naphthalene dioxygenase (nahA) gene resulted in rapid mineralization of naphthalene, whereas indigenous microorganisms in the PAH-contaminated soil required a 28 h adaptation period before significant mineralization occurred [96].
- Biostimulation concerns the activation of native oil-oxidizing microflora by creating optimal conditions for its development. This strategy also has many successful examples. For example, this is demonstrated in [97]. Thus, in the research of [98], it was shown that under laboratory conditions, the introduction of organic fertilizers increases the decomposition rate of naphthalene in soil samples by more than two times within 28 days.
- The use of bacterial cultures in ex situ conditions in bioreactors of various configurations. Thus, a good example of the biodegradation of naphthalene using the bacterium Pseudomonas putida M8, isolated from soil in the suspension phase, shows the advantage of increased availability of pollutants for bacteria. The experiments were carried out in a reactor with a stirrer and oxygen. The results obtained confirmed the success of the selected bioremediation technology in the treatment of contaminated soils [99]. The use of a Batch Bioreactor with a Moving Bed (MBBR) with mixed microbial consortia operating under anaerobic, anoxic, and aerobic conditions resulted in the degradation of 90–94.8% of naphthalene at a concentration of 10 to 100 mg/L in 24 h (RT) [100]. Highly active strains degrading resistant compounds isolated from contaminated sites can be effectively used in bioreactors. Additionally, strains adapted to the decomposition of pollutants in bioreactors are a source of microflora activity for remediation by bioaugmentation [101].
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Polycyclic Aromatic Hydrocarbons (PAHs). In Air Quality Guidelines, 2nd ed.; WHO Regional Office for Europe: Copenhagen, Denmark, 2000; Chapter 5.9; 24p.
- Fonger, G.C.; Hakkinen, P.; Jordan, S.; Publicker, S. The National Library of Medicine’s (NLM) Hazardous Substances Data Bank (HSDB): Background, recent enhancements and future plans. Toxicology 2014, 325, 209–216. [Google Scholar] [CrossRef]
- Kanaly, R.A.; Harayama, S. Biodegradation of High-Molecular-Weight Polycyclic Aromatic Hydrocarbons by Bacteria. J. Bacteriol. 2000, 182, 2059–2067. [Google Scholar] [CrossRef]
- Peng, R.-H.; Xiong, A.-S.; Xue, Y.; Fu, X.-Y.; Gao, F.; Zhao, W.; Tian, Y.-S.; Yao, Q.-H. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol. Rev. 2008, 32, 927–955. [Google Scholar] [CrossRef]
- Lee, H.J.; Shim, W.J.; Lee, J.; Kim, G.B. Temporal and geographical trends in the genotoxic effects of marine sediments after accidental oil spill on the blood cells of striped beakperch (Oplegnathus fasciatus). Mar. Pollut. Bull. 2011, 62, 2264–2268. [Google Scholar] [CrossRef] [PubMed]
- Bekki, K.; Toriba, A.; Tang, N.; Kameda, T.; Hayakawa, K. Biological effects of polycyclic aromatic hydrocarbon derivatives. J. UOEH 2013, 35, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Haritash, A.; Kaushik, C. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): A review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Krzyszczak, A.; Czech, B. Occurrence and toxicity of polycyclic aromatic hydrocarbons derivatives in environmental matrices. Sci. Total. Environ. 2021, 788, 147738. [Google Scholar] [CrossRef] [PubMed]
- Låg, M.; Øvrevik, J.; Refsnes, M.; Holme, J.A. Potential role of polycyclic aromatic hydrocarbons in air pollution-induced non-malignant respiratory diseases. Respir. Res. 2020, 21, 1–22. [Google Scholar] [CrossRef]
- Ren, B. Research on polycyclic aromatic hydrocarbons in environment from 2005 to 2020: A bibliometric analysis. IOP Conf. Ser. Earth Environ. Sci. 2021, 760, 012050. [Google Scholar] [CrossRef]
- Honda, M.; Suzuki, N. Toxicities of Polycyclic Aromatic Hydrocarbons for Aquatic Animals. Int. J. Environ. Res. Public Health 2020, 17, 1363. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. Toxicological Review of Naphthalene (CAS No. 91-20-3) in Support of Summary Information on the Integrated Risk Information System (IRIS); National Center for Environmental Assessment: Cincinnati, OH, USA, 1998.
- Atlas, R.M.; Hazen, T.C. Oil Biodegradation and Bioremediation: A Tale of the Two Worst Spills in U.S. History. Environ. Sci. Technol. 2011, 45, 6709–6715. [Google Scholar] [CrossRef] [PubMed]
- Reddy, C.M.; Arey, J.S.; Seewald, J.S.; Sylva, S.P.; Lemkau, K.L.; Nelson, R.; Carmichael, C.A.; McIntyre, C.; Fenwick, J.; Ventura, G.T.; et al. Composition and fate of gas and oil released to the water column during the Deepwater Horizon oil spill. Proc. Natl. Acad. Sci. USA 2011, 109, 20229–20234. [Google Scholar] [CrossRef] [PubMed]
- James, M.O.; Kleinow, K.M. Trophic transfer of chemicals in the aquatic environment. In Aquatic Toxicology: Molecular, Biochemical, and Cellular Perspectives; Malins, D.C., Ostrander, G.K., Eds.; Lewis: Boca Raton, FL, USA, 1994; pp. 1–36. [Google Scholar]
- Pulster, E.L.; Main, K.; Wetzel, D.; Murawski, S. Species-specific metabolism of naphthalene and phenanthrene in 3 species of marine teleosts exposed to Deepwater Horizon crude oil. Environ. Toxicol. Chem. 2017, 36, 3168–3176. [Google Scholar] [CrossRef] [PubMed]
- Murawski, S.A.; Hogarth, W.T.; Peebles, E.B.; Barbeiri, L. Prevalence of external skin lesions and polycyclic aromatic hydrocarbon concentrations in Gulf of Mexico fishes, Post-Deepwater Horizon. Trans. Am. Fish. Soc. 2014, 143, 1084–1097. [Google Scholar] [CrossRef]
- Brown-Peterson, N.J.; Krasnec, M.; Takeshita, R.; Ryan, C.N.; Griffitt, K.J.; Lay, C.; Mayer, G.D.; Bayha, K.M.; Hawkins, W.E.; Lipton, I.; et al. A multiple endpoint analysis of the effects of chronic exposure to sediment contaminated with Deepwater Horizon oil on juvenile Southern flounder and their associated microbiomes. Aquat. Toxicol. 2015, 165, 197–209. [Google Scholar] [CrossRef]
- Klinger, D.H.; Dale, J.J.; Machado, B.E.; Incardona, J.P.; Farwell, C.J.; Block, B.A. Exposure to Deepwater Horizon weathered crude oil increases routine metabolic demand in chub mackerel, Scomber japonicus. Mar. Pollut. Bull. 2015, 98, 259–266. [Google Scholar] [CrossRef]
- Esbaugh, A.J.; Mager, E.M.; Stieglitz, J.D.; Hoenig, R.; Brown, T.L.; French, B.L.; Linbo, T.L.; Lay, C.; Forth, H.; Scholz, N.; et al. The effects of weathering and chemical dispersion on Deepwater Horizon crude oil toxicity to mahi-mahi (Coryphaena hippurus) early life stages. Sci. Total. Environ. 2015, 543, 644–651. [Google Scholar] [CrossRef]
- Hedgpeth, B.M.; Griffitt, R.J. Simultaneous exposure to chronic hypoxia and dissolved polycyclic aromatic hydrocarbons results in reduced egg production and larval survival in the sheepshead minnow (Cyprinodon variegatus). Environ. Toxicol. Chem. 2016, 35, 645–651. [Google Scholar] [CrossRef]
- Olson, G.M.; Meyer, B.M.; Portier, R.J. Assessment of the toxic potential of polycyclic aromatic hydrocarbons (PAHs) affecting Gulf menhaden (Brevoortia patronus) harvested from waters impacted by the BP Deepwater Horizon Spill. Chemosphere 2016, 145, 322–328. [Google Scholar] [CrossRef]
- DeGraeve, G.M.; Elder, R.G.; Woods, D.C.; Bergman, H.L. Effects of naphthalene and benzene on fathead minnows and rainbow trout. Arch. Environ. Contam. Toxicol. 1982, 11, 487–490. [Google Scholar] [CrossRef] [PubMed]
- Broderius, S.J.; Kah, M.D.; Hoglund, M.D. Use of joint toxic response to define the primary-mode of toxic action for diverse in-dustrial organic-chemicals. Environ. Toxicol. Chem. 1995, 14, 1591–1605. [Google Scholar] [CrossRef]
- Irwin, R.J. Phenanthrene. In Environmental Contaminants Encyclopedia; Irwin, R.J., Ed.; National Park Service, Water Resources Division: Fort Collins, CO, USA, 1997. [Google Scholar]
- Pollino, C.A.; Holdway, D.A. Toxicity Testing of Crude Oil and Related Compounds Using Early Life Stages of the Crimson-Spotted Rainbowfish (Melanotaenia fluviatilis). Ecotoxicol. Environ. Saf. 2002, 52, 180–189. [Google Scholar] [CrossRef]
- dos Santos, T.d.C.A.; Van Ngan, P.; Passos, M.J.D.A.C.R.; Gomes, V. Effects of naphthalene on metabolic rate and ammonia excretion of juvenile Florida pompano, Trachinotus carolinus. J. Exp. Mar. Biol. Ecol. 2006, 335, 82–90. [Google Scholar] [CrossRef]
- Collier, T.K.; Anulacion, B.F.; Arkoosh, M.R.; Dietrich, J.P.; Incardona, J.P.; Johnson, L.L.; Ylitalo, G.M.; Myers, M.S. Effects on fish of polycyclic aromatic hydrocarbonS (PAHS) and naphthenic acid exposures. Fish Physiol. 2013, 195–255. [Google Scholar] [CrossRef]
- Neff, J.M.; Anderson, J.W.; Cox, B.A.; Laughlin, R.B., Jr.; Rossi, S.S.; Tatem, H.E. Effects of petroleum on survival, res-piration and growth of marine animals. In Sources, Effects and Sinks of Hydrocarbons in the Aquatic Environment; American Institute of Biological Sciences: Washington, DC, USA, 1976; pp. 516–539. [Google Scholar]
- Gao, W.; Yin, X.; Mi, T.; Zhang, Y.; Lin, F.; Han, B.; Zhao, X.; Luan, X.; Cui, Z.; Zheng, L. Microbial diversity and ecotoxicity of sediments 3 years after the Jiaozhou Bay oil spill. AMB Express 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bihari, Z.; Szvetnik, A.; Szabó, Z.; Blastyák, A.; Zombori, Z.; Balázs, M.; Kiss, I. Functional analysis of long-chain n-alkane de-gradation by Dietzia spp. FEMS Microbiol. Lett. 2011, 316, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Das, N.; Chandran, P. Microbial degradation of petroleum hydrocarbon contaminants: An overview. Biotechnol. Res. Int. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.-Y.; Zhang, T.; Fang, H.H.-P. Bacteria-mediated PAH degradation in soil and sediment. Appl. Microbiol. Biotechnol. 2011, 89, 1357–1371. [Google Scholar] [CrossRef] [PubMed]
- Khanna, P.; Goyal, D.; Khanna, S. Characterization of pyrene utilizing Bacillus spp. from crude oil contaminated soil. Braz. J. Microbiol. 2012, 43, 606–617. [Google Scholar] [CrossRef][Green Version]
- Stibal, M.; Bælum, J.; Holben, W.E.; Sørensen, S.R.; Jensen, A.; Jacobsen, C.S. Microbial Degradation of 2,4-Dichlorophenoxyacetic Acid on the Greenland Ice Sheet. Appl. Environ. Microbiol. 2012, 78, 5070–5076. [Google Scholar] [CrossRef] [PubMed]
- Adebusoye, S.A.; Ilori, M.; Amund, O.O.; Teniola, O.D.; Olatope, S.O. Microbial degradation of petroleum hydrocarbons in a polluted tropical stream. World J. Microbiol. Biotechnol. 2007, 23, 1149–1159. [Google Scholar] [CrossRef]
- Ikuma, Y.K.; Gunsch, C.K. Genetic bioaugmentation as an effective method for in situ bioremediation: Functionality of catabolic plasmids following conjugal transfers. Bioengineered 2012, 3, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.; Niqui-Arroyo, J.L.; Conde, S.; Ramos, J.L.; Duque, E. Enhanced tolerance to naphthalene and enhanced rhizore-mediation performance for Pseudomonas putida KT2440 via the NAH7 catabolic plasmid. Appl. Environ. Microbiol. 2012, 78, 5104–5110. [Google Scholar] [CrossRef]
- McGenity, T.J.; Folwell, B.D.; McKew, B.A.; Sanni, G.O. Marine crude-oil biodegradation: A central role for interspecies interac-tions. Aquat. Biosyst. 2012, 8, 10. [Google Scholar] [CrossRef]
- Mihasan, M.; Brandsch, R. pAO1 of Arthrobacter nicotinovorans and the spread of catabolic traits by horizontal gene transfer in Gram-positive soil bacteria. J. Mol. Evol. 2013, 77, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Nojiri, H. Impact of catabolic plasmids on host cell physiology. Curr. Opin. Biotechnol. 2013, 24, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhao, H.; Li, Y.; Niu, S.; Cai, B. Complete nucleotide sequence of plasmid pND6-2 from Pseudomonas putida ND6 and characterization of conjugative genes. Gene 2013, 512, 148–156. [Google Scholar] [CrossRef]
- Sutton, N.B.; Maphosa, F.; Morillo, J.A.; Abu Al-Soud, W.; Langenhoff, A.A.M.; Grotenhuis, T.; Rijnaarts, H.H.M.; Smidt, H. Impact of long-term diesel contamination on soil microbial community structure. Appl. Environ. Microbiol. 2013, 79, 619–630. [Google Scholar] [CrossRef]
- Yergeau, E.; Sanschagrin, S.; Beaumier, D.; Greer, C.W. Metagenomic analysis of the bioremediation of diesel-contaminated canadian high arctic soils. PLoS ONE 2012, 7, e30058. [Google Scholar] [CrossRef]
- Guo, W.; Li, D.; Tao, Y.; Gao, P.; Hu, J. Isolation and description of a stable carbazole-degrading microbial consortium consisting of Chryseobacterium sp. NCY and Achromobacter sp. NCW. Curr. Microbiol. 2008, 57, 251–257. [Google Scholar] [CrossRef]
- Eaton, R.W.; Chapman, P.J. Bacterial metabolism of naphthalene: Construction and use of recombinant bacteria to study ring cleavage of 1,2-dihydroxynaphthalene and subsequent reactions. J. Bacteriol. 1992, 174, 7542–7554. [Google Scholar] [CrossRef]
- Chavez, F.P.; Lunsdor, F.H.; Jerez, C.A. Growth of polychlorinatedbiphenyl-degrading bacteria in the presence of biphenyl and chlorobiphenyls generates oxidative stress and massive accumulation of inorganic polyphosphate. Appl. Environ. Microbiol. 2004, 70, 3064–3072. [Google Scholar] [CrossRef]
- Dean-Ross, D.; Moody, J.; Cerniglia, C.E. Utilization of mixtures of polycyclic aromatic hydrocarbons by bacteria isolated from contaminated sediment. FEMS Microbiol. Ecol. 2002, 41, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Madueno, L.; Coppotelli, B.M.; Alvarez, H.M.; Morelli, I.S. Isolation and characterization of indigenous soil bacteria for bioaug-mentation of PAH contaminated soil of semiarid Patagonia, Argentina. Int. Biodeterior. Biodegrad. 2011, 65, 345–351. [Google Scholar] [CrossRef]
- Hedlund, B.P.; Geiselbrecht, A.D.; Bair, T.J.; Staley, J.T. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium, Neptunomonas naphthovorans gen. nov., sp. nov. Appl. Environ. Microbiol. 1999, 65, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Uz, I.; Duan, Y.; Ogram, A. Characterization of the naphthalene-degrading bacterium, Rhodococcus opacus M213. FEMS Microbiol. Lett. 2000, 185, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Annweiler, E.; Richnow, H.H.; Antranikian, G.; Hebenbrock, S.; Garms, C.; Franke, S.; Francke, W.; Michaelis, W. Naphthalene degradation and incorporation of naphthalene-derived carbon into biomass by the thermophile Bacillus thermoleovorans. Appl. Environ. Microbiol. 2000, 66, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Poswal, V.; Kaur, S.; Mahajan, A.; Begum, Z. Isolation and identification of naphthalene degradation bacteria. Intern. J. Inn. Sci. Res. Technol. 2018, 3, 682–688. [Google Scholar]
- Nair, D.; Fernández-Acero, F.J.; García-Luque, E.; Riba, I.; Del Valls, T.A. Isolation and characterization of naphthalene-degrading bacteria from sediments of Cadiz area (SW Spain). Environ. Toxicol. 2008, 23, 576–582. [Google Scholar] [CrossRef]
- Häggblom, M.M.; Daane, L. Paenibacillus Naphthalenovorans that Degrades Naphthalene. U.S. Patent 6905864, 29 May 2005. [Google Scholar]
- Yen, K.M.; Gunsalus, I.C. Plasmid gene organization: Naphthalene/salicylate oxidation. Proc. Natl. Acad. Sci. USA 1982, 79, 874–878. [Google Scholar] [CrossRef]
- Burt, D.; Ensley, T.; Gibson, D.T. Naphthalene dioxygenase: Purification and properties of a terminal oxygenase component. J. Bacteriol. 1983, 155, 505–511. [Google Scholar]
- Cane, P.A.; Williams, P.A. A Restriction map of naphthalene catabolic plasmid pWW60-1 and the location of some of its catabolic genes. Microbiology 1986, 132, 2919–2929. [Google Scholar] [CrossRef]
- Schell, M.A. Homology between nucleotide sequences of promoter regions of nah and sal operons of NAH7 plasmid of Pseu-domonas putida. Proc. Natl. Acad. Sci. USA 1986, 83, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Schell, M.A.; Wender, P.E. Identification of the nahR gene product and nucleotide sequences required for its activation of the sal operon. J. Bacteriol. 1986, 166, 9–14. [Google Scholar] [CrossRef]
- Harayama, S.; Rekik, M.; Wasserfallen, A.; Bairoch, A. Evolutionary relationships between catabolic pathways for aromatics: Conservation of gene order and nucleotide sequences of catechol oxidation genes of pWW0 and NAH7 plasmids. Mol. Genet. Genom. 1987, 210, 241–247. [Google Scholar] [CrossRef]
- Simon, M.J.; Osslund, T.D.; Saunders, R.; Ensley, B.D.; Suggs, S.; Harcourt, A.; Wen-Chen, S.; Cruder, D.L.; Gibson, D.T.; Zylstra, G.J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene 1993, 127, 31–37. [Google Scholar] [CrossRef]
- Platt, A.; Shingler, V.; Taylor, S.C.; Williams, P.A. The 4-hydroxy-2-oxovalerate aldolase and acetaldehyde dehydrogenase (acylating) encoded by the nahM and nahO genes of the naphthalene catabolic plasmid pWW60-22 provide further evidence of conservation of meta-cleavage pathway gene sequences. Microbiology 1995, 141, 2223–2233. [Google Scholar] [CrossRef]
- Davies, J.I.; Evans, W.C. Oxidative metabolism of naphthalene nucleus by soil pseudomonads—Ring fission mechanism. Biochem. J. 1964, 91, 251–261. [Google Scholar] [CrossRef]
- Zylstra, G.J.; Wang, X.P.; Kim, E.; Didolcar, V.A. Cloning and analysis of the genes for polycyclic aromatic hydrocarbon degradation. In: Recombinant DNA technology II. Ann. N. Y. Acad. Sci. 1994, 721, 386–398. [Google Scholar] [CrossRef]
- Sutherland, J.B.; Rafil, F.; Khan, A.A.; Cerniglia, C.E. Mechanisms of polycyclic aromatic hydrocarbon degradation. In Microbial Transformation and Degradation of Toxic Organic Chemicals; Willey-Liss, Inc.: Hoboken, NJ, USA, 1995; pp. 269–306. [Google Scholar]
- Miyazawa, D.; Thanh, L.; Tani, A.; Shintani, M.; Loc, N.H.; Hatta, T.; Kimbara, K. Isolation and characterization of genes res-ponsible for naphthalene degradation from thermophilic naphthalene degrader, Geobacillus sp. JF8. Microorganisms 2019, 8, 44. [Google Scholar] [CrossRef]
- Jeffrey, A.M.; Yeh, H.J.C.; Jerina, D.M.; Patel, T.R.; Davey, J.F.; Gibson, D.T. Initial reactions in the oxidation of naphthalene by Pseudomonas putida. Biochemistry 1975, 14, 575–584. [Google Scholar] [CrossRef]
- Rossello-Mora, R.A.; Lalucat, J.; Garcia-Valdes, E. Comparative biochemical and genetic analysis of naphthalene degradation among Pseudomonas stutzeri strains. Appl. Environ. Microbiol. 1994, 60, 966–972. [Google Scholar] [CrossRef]
- Meyer, S.; Moser, R.; Neef, A.; Stahl, U.; Kampfer, P. Differential detection of key enzymes of polyaromatic hydrocarbon de-grading bacteria using PCR and gene probes. Microbiology 1999, 145, 1731–1741. [Google Scholar] [CrossRef]
- Moser, R.; Stahl, U. Insights into the genetic diversity of initial dioxygenases from PAH-degrading bacteria. Appl. Microbiol. Biotechnol. 2001, 55, 609–618. [Google Scholar] [CrossRef]
- Tay, M.; Roizman, D.; Cohen, Y.; Tolker-Nielsen, T.; Givskov, M.; Yang, L. Draft genome sequence of the model naphthalene utilizing organism Pseudomonas putida OUS82. Genome Announc. 2014, 2, e01161-13. [Google Scholar] [CrossRef]
- Yen, K.-M.; Serdar, C.M.; Gunsalus, I.C. Genetics of naphthalene catabolism in Pseudomonads. CRC Crit. Rev. Microbiol. 1988, 15, 247–268. [Google Scholar] [CrossRef] [PubMed]
- Kochetkov, V.V.; Balakshina, V.V.; Mordukhova, E.A.; Boronin, A.M. Plasmids encoding naphthalene biodegradation in rhizos-phere bacteria of the genus Pseudomonas. Microbiology 1997, 66, 173–177. [Google Scholar]
- Ghosal, D.; In-Soon, Y.; Gunsalus, I.C. Nucleotide sequence and expression of gene nahH of plasmid NAH7 and homology with gene xylE of TOL pWWO. Gene 1987, 55, 19–28. [Google Scholar] [CrossRef]
- You, I.S.; Ghosal, D.; Gunsalus, I.C. Nucleotide sequence of plasmid NAH7 gene nahR and DNA binding of the nahR product. J. Bacteriol. 1988, 170, 5409–5415. [Google Scholar] [CrossRef] [PubMed]
- Grimm, A.C.; Harwood, C.S. NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J. Bacteriol. 1999, 181, 3310–3316. [Google Scholar] [CrossRef]
- Kim, E.; Zylstra, G.J. Molecular and biochemical characterization of two meta-cleavage dioxygenases involved in biphenyl and m-xylene degradation by Beijerinckia sp. strain B1. J. Bacteriol. 1995, 177, 3095–3103. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Finnerty, W.R. The biology and genetics of the genus Rhodococcus. Annu. Rev. Microbiol. 1992, 46, 193–218. [Google Scholar] [CrossRef]
- Grund, E.; Denecke, B.; Eichenlaub, R. Naphthalene degradation via salicylate and gentisate by Rhodococcus sp. strain B4. Appl. Environ. Microbiol. 1992, 58, 1874–1877. [Google Scholar] [CrossRef]
- Warhurst, A.M.; Clarke, K.F.; Hill, R.A.; Holt, R.A.; Fewson, C.A. Production of catechols and muconic acids from various aromatics by the styrene-degrader Rhodococcus rhodochrous NCIMB 13259. Biotechnol. Lett. 1994, 16, 513–516. [Google Scholar] [CrossRef]
- McLeod, M.P.; Warren, R.; Hsiao, W.W.L.; Araki, N.; Myhre, M.; Fernandes, C.; Miyazawa, D.; Wong, W.; Lillquist, A.L.; Wang, D.; et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 2006, 103, 15582–15587. [Google Scholar] [CrossRef] [PubMed]
- Martínková, L.; Uhnáková, B.; Pátek, M.; Nesvera, J.; Kren, V. Biodegradation potential of the genus Rhodococcus. Environ. Int. 2009, 35, 162–177. [Google Scholar] [CrossRef]
- Pathak, A.; Green, S.; Ogram, A.; Chauhan, A. Draft genome sequence of Rhodococcus opacus strain M213 shows a diverse catabolic potential. Genome Announc. 2013, 1, e00144-12. [Google Scholar] [CrossRef] [PubMed]
- Pathak, A.; Chauhan, A.; Blom, J.; Indest, K.J.; Jung, C.; Stothard, P.; Bera, G.; Green, S.; Ogram, A. Comparative genomics and metabolic analysis reveals peculiar characteristics of Rhodococcus opacus strain M213 particularly for naphthalene degradation. PLoS ONE 2016, 11, e0161032. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, C.; Song, G.; Liu, H.; Sheng, G.; Ding, Z.; Wang, Z.; Sun, Y.; Xu, Y.; Chen, J. Characterization of a protocatechuate catabolic gene cluster in Rhodococcus ruber OA1 involved in naphthalene degradation. Ann. Microbiol. 2015, 66, 469–478. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Z.; Zhang, S.; Li, X.; Gao, H.; Zhang, C. Characterization of a naphthalene catabolic gene cluster and hetero-logous expression of naphthalene dioxygenase genes from Rhodococcus ruber OA1. Biotechnology 2017, 16, 165–173. [Google Scholar] [CrossRef][Green Version]
- Maruyama, T.; Ishikura, M.; Taki, H.; Shindo, K.; Kasai, H.; Haga, M.; Inomata, Y.; Misawa, N. Isolation and characterization of o-xylene oxygenase genes from Rhodococcus opacus TKN14. Appl. Environ. Microbiol. 2005, 71, 7705–7715. [Google Scholar] [CrossRef]
- Anokhina, T.O.; Esikova, T.Z.; Gafarov, A.B.; Polivtseva, V.N.; Baskunov, B.P.; Solyanikova, I.P. Alternative naphthalene metabolic pathway includes formation of ortho-phthalic acid and cinnamic acid derivatives in the Rhodococcus opacus strain 3D. Biokhimiya 2020, 85, 355–368. [Google Scholar] [CrossRef]
- Kulakova, A.N.; Reid, K.A.; Larkin, M.J.; Allen, C.C.; Kulakov, L.A. Isolation of Rhodococcus rhodochrous NCIMB 13064 de-rivatives with new biodegradative abilities. FEMS Microbiol. Lett. 1996, 145, 227–231. [Google Scholar] [CrossRef]
- Kulakov, L.A.; Chen, S.; Allen, C.C.R.; Larkin, M.J. Web-type evolution of Rhodococcus gene clusters associated with utilization of naphthalene. Appl. Environ. Microbiol. 2005, 71, 1754–1764. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.J.; De Mot, R.; Kulakov, L.A.; Nagy, I. Applied aspects of Rhodococcus genetics. Antonie Leeuwenhoek 1998, 74, 133–153. [Google Scholar] [CrossRef]
- Larkin, M.J.; Kulakov, L.A.; Allen, C. Biodegradation and Rhodococcus—masters of catabolic versatility. Curr. Opin. Biotechnol. 2005, 16, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, A.; Massini, G.; Miritana, V.M.; Panico, A.; Pontoni, L.; Race, M.; Rosa, S.; Signorini, A.; Fabbricino, M.; Pirozzi, F. Bioaugmentation strategy to enhance polycyclic aromatic hydrocarbons anaerobic biodegradation in contaminated soils. Chemosphere 2021, 275, 130091. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Ma, W.; Han, H.; Jia, S.; Hou, B. Isolation of a naphthalene-degrading strain from activated sludge and bioaugmentation with it in a MBR treating coal gasification wastewater. Bull. Environ. Contam. Toxicol. 2014, 94, 358–364. [Google Scholar] [CrossRef]
- Piskonen, R.; Nyyssönen, M.; Rajamäki, T.; Itävaara, M. Monitoring of accelerated naphthalene-biodegradation in a bioaugmented soil slurry. Biogeochemistry 2005, 16, 127–134. [Google Scholar] [CrossRef]
- Giovanella, P.; de Azevedo Duarte, L.; Kita, D.M.; de Oliveira, V.M.; Sette, L.D. Effect of biostimulation and bioaugmentation on hydrocarbon degradation and detoxification of diesel-contaminated soil: A microcosm study. J. Microbiol. 2021, 59, 634–643. [Google Scholar] [CrossRef]
- Olu, A.A. Effectiveness of organic fertilizer as a biostimulating agent for the removal of naphthalene in soil. Appl. J. Environ. Eng. Sci. 2017, 3, 177–189. [Google Scholar]
- Collina, E.; Bestetti, G.; Di Gennaro, P.; Franzetti, A.; Gugliersi, F.; Lasagni, M.; Pitea, D. Naphthalene biodegradation kinetics in an aerobic slurry-phase bioreactor. Environ. Int. 2005, 31, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Yadu, A.; Sahariah, B.P.; Anandkumar, J. Process optimization and comparative study on naphthalene biodegradation in anaerobic, anoxic, and aerobic moving bed bioreactors. Eng. Rep. 2020, 2, e12127. [Google Scholar] [CrossRef]
- Pawar, A.N.; Ugale, S.S.; More, M.G.; Kokani, N.F.; Khandelwal, S.R. Biological degradation of naphthalene: A new era. J. Bioremediat. Biodegrad. 2013, 4. [Google Scholar] [CrossRef]
- Loginov, O.N.; Silishchev, N.N.; Churaev, R.N.; Boyko, T.F.; Galimzyanova, N.F.; Danilova, E.A.; Podcepikhin, A.K.; Sultanov, L.O.R.; Chetverikov, S.P. Consortium of Microbial Strains Bacillus brevis and Arthrobacter sp., Used to Clean-up Water and Soil from Crude Oil and Oil Products. RU Patent 2232806, 20 July 2004. [Google Scholar]
- Loginov, O.N.; Nurtdinova, L.A.; Boyko, T.F.; Chetverikov, S.P.; Silishchev, N.N. Efficiency of a novel preparation Lenoil in oil-polluted soil remediation. Biotechnol. Rus. 2004, 1, 77–82. [Google Scholar]
- Nigam, P.; Banat, I.; Marchant, R.; Singh, D. Degradation of naphthalene by bacterial cultures. Environ. Int. 1998, 24, 671–677. [Google Scholar] [CrossRef]
- Pelaez, A.I.; Lores, I.; Sotres, A.; Mendez-Garcia, C.; Fernandez-Velarde, C.; Santos, J.A.; Gallego, J.L.R.; Sanchez, J. Design and feld-scale implementation of an “on site” bioremediation treatment in PAH-polluted soil. Environ. Pollut. 2013, 181, 190–199. [Google Scholar] [CrossRef]
- Sakshi; Haritash, A.K. A comprehensive review of metabolic and genomic aspects of PAH-degradation. Arch. Microbiol. 2020, 202, 2033–2058. [Google Scholar] [CrossRef]
- Kumar, M.; Leon, V.; Materano, A.D.S.; Ilzins, O.A.; Galindo-Castro, I.; Fuenmayor, S.L. Polycyclic aromatic hydrocarbon degradation by biosurfactant-producing Pseudomonas sp. IR1. Z. Nat. C 2006, 61, 203–212. [Google Scholar] [CrossRef]
- Zhu, X.; Ni, X.; Waigi, M.G.; Liu, J.; Sun, K.; Gao, Y. Biodegradation of mixed PAHs by PAH-degrading endophytic bacteria. Int. J. Environ. Res. Public Health 2016, 13, 805. [Google Scholar] [CrossRef]
- Hesham, A.E.-L.; Mawad, A.M.M.; Mostafa, Y.M.; Shoreit, A. Biodegradation ability and catabolic genes of petroleum-degrading Sphingomonas koreensis strain ASU-06 isolated from egyptian oily soil. BioMed Res. Int. 2014, 2014, 1–10. [Google Scholar] [CrossRef]
- Kim, T.J.; Lee, E.Y.; Kim, Y.J.; Cho, K.-S.; Ryu, H.W. Degradation of polyaromatic hydrocarbons by Burkholderia cepacia 2A-12. World J. Microbiol. Biotechnol. 2003, 19, 411–417. [Google Scholar] [CrossRef]
- Di Gennaro, P.; Rescalli, E.; Galli, E.; Sello, G.; Bestetti, G. Characterization of Rhodococcus opacus R7, a strain able to degrade naphthalene and o-xylene isolated from a polycyclic aromatic hydrocarbon-contaminated soil. Res. Microbiol. 2001, 152, 641–651. [Google Scholar] [CrossRef]
- Di Gennaro, P.; Terreni, P.; Masi, G.; Botti, S.; De Ferra, F.; Bestetti, G. Identification and characterization of genes involved in naphthalene degradation in Rhodococcus opacus R7. Appl. Microbiol. Biotechnol. 2010, 87, 297–308. [Google Scholar] [CrossRef]
- Di Gennaro, P.; Zampolli, J.; Presti, I.; Cappelletti, M.; D’Ursi, P.; Orro, A.; Mezzelani, A.; Milanesi, L. Genome sequence of Rhodococcus opacus strain R7, a biodegrader of mono- and polycyclic aromatic hydrocarbons. Genome Announc. 2014, 2, e00827-14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-C.; Mörtelmaier, C.; Margesin, R. Characterization of the bacterial archaeal diversity in hydrocarbon-contaminated soil. Sci. Total. Environ. 2012, 421–422, 184–196. [Google Scholar] [CrossRef]
- Zhang, D.; Margesin, R. Characterization of culturable heterotrophic bacteria in hydrocarbon-contaminated soil from an alpine former military site. World J. Microbiol. Biotechnol. 2014, 30, 1717–1724. [Google Scholar] [CrossRef]
- Schumann, P.; Zhang, D.-C.; França, L.; Albuquerque, L.; da Costa, M.; Margesin, R. Psychromicrobium silvestre gen. nov., sp. nov., an actinobacterium isolated from alpine forest soils. Int. J. Syst. Evol. Microbiol. 2017, 67, 640–645. [Google Scholar] [CrossRef]
- Elster, J.; Margesin, R.; Wagner, D.; Häggblom, M. Editorial: Polar and Alpine Microbiology—Earth’s cryobiosphere. FEMS Microbiol. Ecol. 2017, 93, fiw221. [Google Scholar] [CrossRef]
- Siles, J.A.; Margesin, R. Seasonal soil microbial responses are limited to changes in functionality at two Alpine forest sites differing in altitude and vegetation. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Siles, J.A.; Margesin, R. Insights into microbial communities mediating the bioremediation of hydrocarbon-contaminated soil from an Alpine former military site. Appl. Microbiol. Biotechnol. 2018, 102, 4409–4421. [Google Scholar] [CrossRef]
- Schumann, P.; Zhang, D.-C.; França, L.; Margesin, R. Marmoricola silvestris sp. nov., a novel actinobacterium isolated from alpine forest soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 1313–1318. [Google Scholar] [CrossRef]
- Siles, J.A.; Öhlinger, B.; Cajthaml, T.; Kistler, E.; Margesin, R. Characterization of soil bacterial, archaeal and fungal communities inhabiting archaeological human-impacted layers at Monte Iato settlement (Sicily, Italy). Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Molina, M.C.; González, N.; Bautista, L.F.; Sanz, R.; Simarro, R.; Sanchez-Andrea, I.; Sanz, J.L. Isolation and genetic identification of PAH degrading bacteria from a microbial consortium. Biogeochemistry 2009, 20, 789–800. [Google Scholar] [CrossRef]
- Simarro, R.; González, N.; Bautista, L.F.; Molina, M.C. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by a wood-degrading consortium at low temperatures. FEMS Microbiol. Ecol. 2012, 83, 438–449. [Google Scholar] [CrossRef] [PubMed][Green Version]
- González-Benítez, N.; Bautista, L.F.; Simarro, R.; Vargas, C.; Salmerón, A.; Murillo, Y.; Molina, M.C. Bacterial diversity in aqueous/sludge phases within diesel fuel storage tanks. World J. Microbiol. Biotechnol. 2020, 36, 180. [Google Scholar] [CrossRef]
- Lin, X.; Yang, B.; Shen, J.; Du, N. Biodegradation of crude oil by an arctic psychrotrophic bacterium Pseudoalteromonas sp. P29. Curr. Microbiol. 2009, 59, 341–345. [Google Scholar] [CrossRef]
- Xuezheng, L.; Shuoshuo, C.; Guoying, X.; Shuai, W.; Ning, D.; Jihong, S. Cloning and heterologous expression of two cold-active lipases from the Antarctic bacterium Psychrobacter sp. G. Polar Res. 2010, 29, 421–429. [Google Scholar] [CrossRef]
- Mooney, T.J.; King, C.K.; Wasley, J.; Andrew, N.R. Toxicity of diesel contaminated soils to the subantarctic earthworm Microscolex macquariensis. Environ. Toxicol. Chem. 2012, 32, 370–377. [Google Scholar] [CrossRef]
- Bramley-Alves, J.; Wasley, J.; King, C.K.; Powell, S.; Robinson, S.A. Phytoremediation of hydrocarbon contaminants in subantarctic soils: An effective management option. J. Environ. Manag. 2014, 142, 60–69. [Google Scholar] [CrossRef]
- Arbel, J.; King, C.K.; Raymond, B.; Winsley, T.; Mengersen, K.L. Application of a Bayesian nonparametric model to derive toxicity estimates based on the response of Antarctic microbial communities to fuel-contaminated soil. Ecol. Evol. 2015, 5, 2633–2645. [Google Scholar] [CrossRef]
- Macoustra, G.K.; King, C.K.; Wasley, J.; Robinson, S.A.; Jolley, D.F. Impact of hydrocarbons from a diesel fuel on the germination and early growth of subantarctic plants. Environ. Sci. Process. Impacts 2015, 17, 1238–1248. [Google Scholar] [CrossRef][Green Version]
- Nydahl, A.C.; King, C.K.; Wasley, J.; Jolley, D.F.; Robinson, S.A. Toxicity of fuel-contaminated soil to Antarctic moss and terrestrial algae. Environ. Toxicol. Chem. 2015, 34, 2004–2012. [Google Scholar] [CrossRef]
- Ferrari, B.C.; Zhang, C.; van Dorst, J. Recovering greater fungal diversity from pristine and diesel fuel contaminated sub-antarctic soil through cultivation using both a high and a low nutrient media approach. Front. Microbiol. 2011, 2, 217. [Google Scholar] [CrossRef] [PubMed]
- Van Dorst, J.; Siciliano, S.D.; Winsley, T.; Snape, I.; Ferrari, B.C. Bacterial targets as potential indicators of diesel fuel toxicity in subantarctic soils. Appl. Environ. Microbiol. 2014, 80, 4021–4033. [Google Scholar] [CrossRef]
- Chang, W.; Akbari, A.; Snelgrove, J.; Frigon, D.; Ghoshal, S. Biodegradation of petroleum hydrocarbons in contaminated clayey soils from a subarctic site: The role of aggregate size and microstructure. Chemosphere 2013, 91, 1620–1626. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Dyen, M.; Spagnuolo, L.; Simon, P.; Whyte, L.; Ghoshal, S. Biodegradation of semi- and non-volatile petroleum hydrocarbons in aged, contaminated soils from a sub-Arctic site: Laboratory pilot-scale experiments at site temperatures. Chemosphere 2010, 80, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Akbari, A.; Ghoshal, S. Pilot-scale bioremediation of a petroleum hydrocarbon-contaminated clayey soil from a sub-Arctic site. J. Hazard. Mater. 2014, 280, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Akbari, A.; Ghoshal, S. Effects of diurnal temperature variation on microbial community and petroleum hydrocarbon biodegradation in contaminated soils from a sub-Arctic site. Environ. Microbiol. 2015, 17, 4916–4928. [Google Scholar] [CrossRef]
- Bell, T.H.; Yergeau, E.; Maynard, C.; Juck, D.; Whyte, L.G.; Greer, C.W. Predictable bacterial composition and hydrocarbon de-gradation in Arctic soils following diesel and nutrient disturbance. ISME J. 2013, 7, 1200–1210. [Google Scholar] [CrossRef]
- Bell, T.H.; Yergeau, E.; Juck, D.; Whyte, L.G.; Greer, C.W. Alteration of microbial community structure affects diesel biodegradation in an Arctic soil. FEMS Microbiol. Ecol. 2013, 85, 51–61. [Google Scholar] [CrossRef]
- Abulencia, C.B.; Wyborski, D.L.; Garcia, J.A.; Podar, M.; Chen, W.; Chang, S.H.; Chang, H.W.; Watson, D.; Brodie, E.L.; Hazen, T.; et al. Environmental Whole-Genome Amplification to Access Microbial Populations in Contaminated Sediments. Appl. Environ. Microbiol. 2006, 72, 3291–3301. [Google Scholar] [CrossRef][Green Version]
- Liu, R.; Zhang, Y.; Ding, R.; Li, D.; Gao, Y.; Yang, M. Comparison of archaeal and bacterial community structures in heavily oil-contaminated and pristine soils. J. Biosci. Bioeng. 2009, 108, 400–407. [Google Scholar] [CrossRef]
- Boronin, A.M.; Kosheleva, I.A. Biodiversity of naphthalene biodegradation systems in soil bacteria. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1155–1163. [Google Scholar]
- Guazzaroni, M.-E.; Herbst, F.-A.; Lores, I.; Tamames, J.; Peláez, A.I.; López-Cortés, N.; Alcaide, M.; Del Pozo, M.V.; Vieites, J.M.; Von Bergen, M.; et al. Metaproteogenomic insights beyond bacterial response to naphthalene exposure and biostimulation. ISME J. 2012, 7, 122–136. [Google Scholar] [CrossRef]
- Tobalina, L.; Bargiela, R.; Pey, J.; Herbst, F.-A.; Lores, I.; Rojo, D.; Barbas, C.; Peláez, A.I.; Sánchez, J.; Von Bergen, M.; et al. Context-specific metabolic network reconstruction of a naphthalene-degrading bacterial community guided by metaproteomic data. Bioinformatics 2015, 31, 1771–1779. [Google Scholar] [CrossRef]
- Hidalgo, K.J.; Sierra-Garcia, I.N.; Dellagnezze, B.M.; de Oliveira, V.M. Metagenomic insights into the mechanisms for biodegradation of polycyclic aromatic hydrocarbons in the oil supply chain. Front. Microbiol. 2020, 11, 561506. [Google Scholar] [CrossRef]
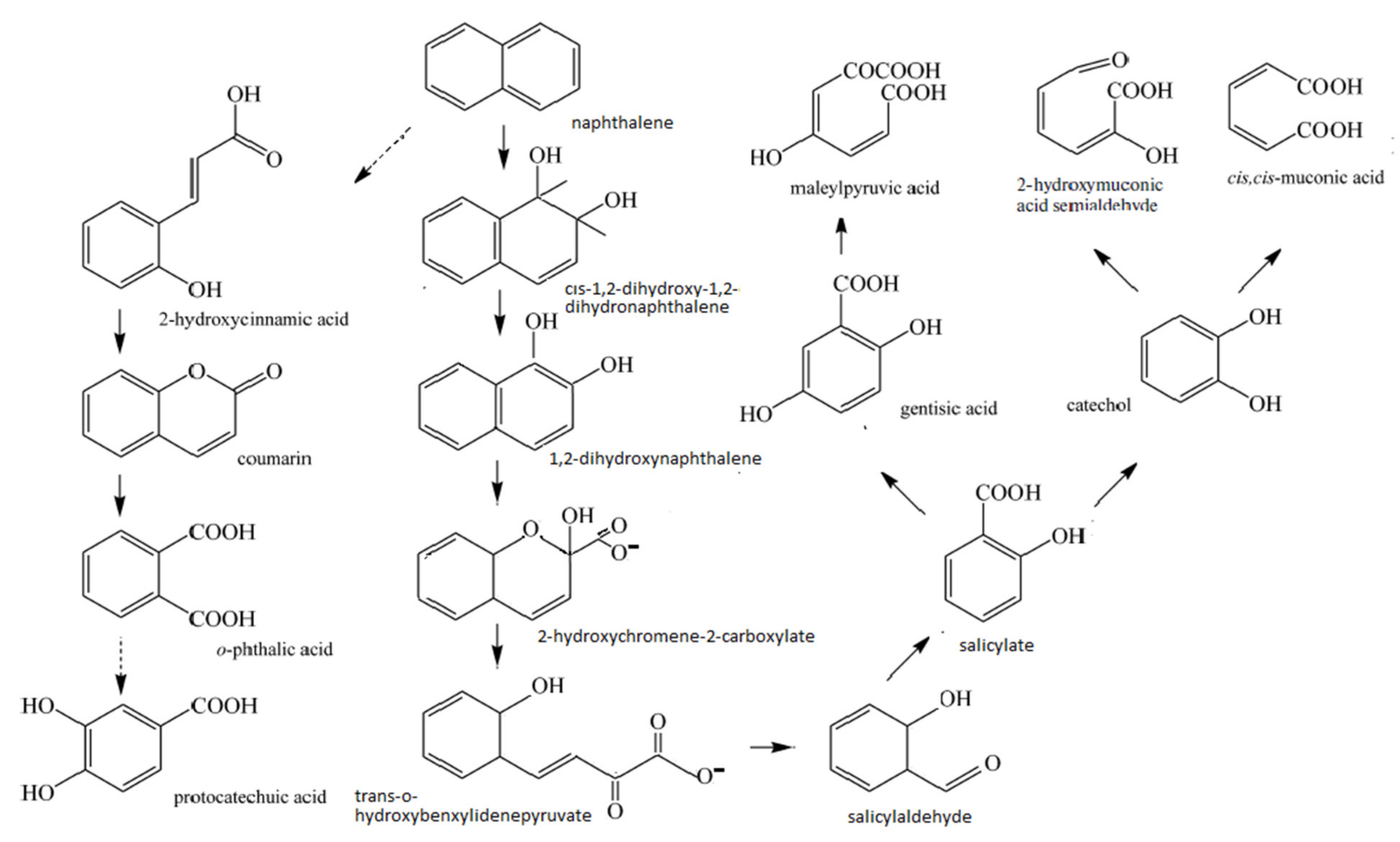
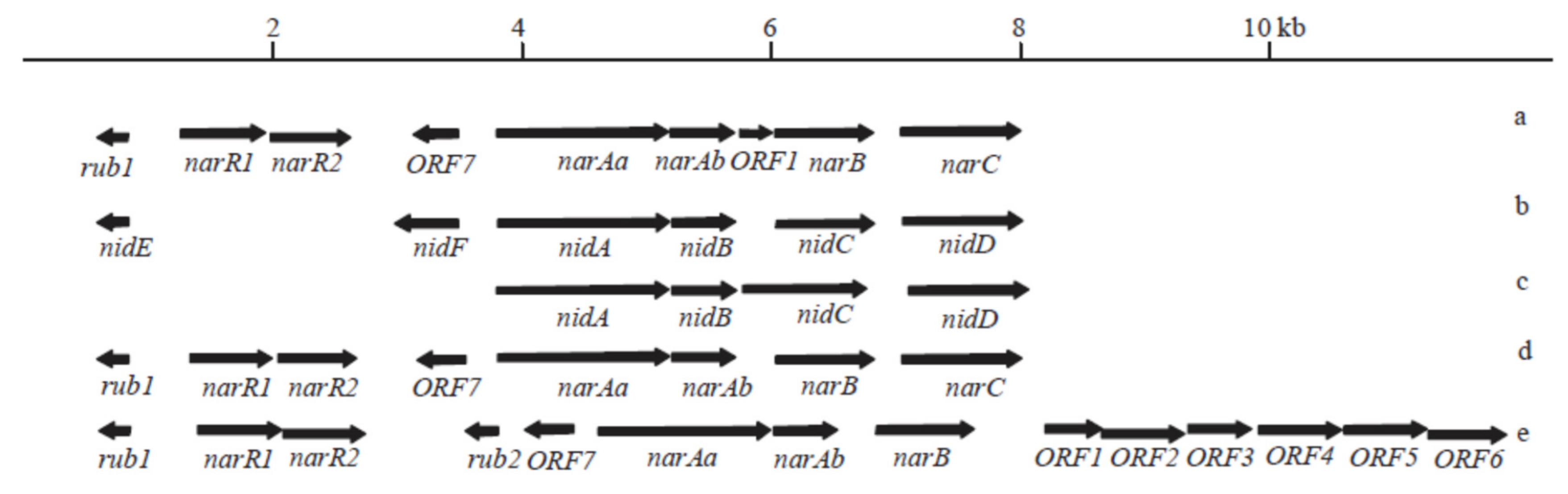
| Microbial Species | Source | Condition; Temperature (°C); Period (Days) | PAH Compound (Ci—ppm) | Degradation Efficiency (%) | Genes Coding Enzymes | Peculiarities | Reference |
|---|---|---|---|---|---|---|---|
| Pseudomonas putida IR1 | Hydrocarbon-contaminated soil | Aerobic; 30; 7 | NAP (200) PHE (200) PYR (200) | 72 ± 2 60 ± 4 69 ± 3 | nahA (naphthalene dioxygenase) and nahE (trans-o-hydroxybenzylidene pyruvate hydratase-aldolase) | Biosurfactants were synthesized. The surface tension decreased from 54.9 dN cm−1 to 35.4 dN cm−1. An emulsifying activity of 74% with diesel oil, when grown on dextrose, was present. | [107] |
| Pseudomonas sp. | Plants growing at PAH-contaminated sites | Aerobic; 30; 7 | NAP (100) FLU (100) PHE (100) PYR (100) | 95.3 87.9 90.4 6.9 | No information | Strain P3 has more potential for use in the removal of PAHs from plant tissues. | [108] |
| Stenotrophomonas sp. | Plants growing at PAH-contaminated sites | Aerobic; 30; 7 | NAP (100) FLU (100) PHE (100) PYR (100) B(a)P (10) | 98.0 83.1 87.8 14.4 1.6 | No information | [108] | |
| Neptunomonas naphthovorans NAG-2N-126 | Creosote-contaminated sediment | Aerobic; 20; 7 | NAP (5) PHE (1) | 100 100 | Naphthalene dioxygenase iron–sulfur protein (ISP) gene fragments | A naphthalene dioxygenase iron–sulfur protein (ISP) gene deduced amino acid sequences and showed that the genes were distantly related to the genes encoding naphthalene dioxygenases of Pseudomonas and Burkholderia strains. | [50] |
| Sphingomonas koreensis ASU-06 | Oil-contaminated soil | Aerobic; 30; 15 | NAP (100) PHE (100) ANT (100) | 100 99 98 | Catabolic genes alkB, alkB1, nahAc, C12O, and C23O | Degradation of 15 various PAH and production of the extracellular biosurfactant. | [109] |
| Burkholderia cepacia 2A-12 | Oil-contaminated soil | Aerobic; 30; 2 | NAP (215) PHE (215) PYR (215) | 100 100 0 | No information | The PAH degradation rate of the strain was enhanced by the addition of other organic materials such as YE, peptone, glucose, and sucrose. | [110] |
| Rhodococcus ruber OA1 | Pharmaceutical wastewater treatment plant | Aerobic; 6 | NAP (500 mg) | 100 | The genes encoding NDO large and small subunits (narAaAb/nidAB), cis-naphthalene dihydrodioldehydrogenase(narB/nidC), and putative aldolase (narC/nidD) Regulated narR1 and narR2 genes | Heterologous expression of narAaAb and rub1 genes. | [86,87] |
| Rhodococcus opacus R7 | Polycyclic aromatic hydrocarbon-contaminated soil | Aerobic; 30; 15 | NAP (1 g) | 100 | High-quality draft genome sequence of strain R7, consisting of 10, 118, and 052 bp | 1,2-Dihydro-1,2-dihydroxynaphthalene as well as salicylic and gentisic acids were identified as metabolites. | [111,112,113] |
| Rhodococcus sp. strain B4 | Polycyclic aromatic hydrocarbon-contaminated soil | Aerobic, 1 | NAP (0.5 g) | 100 | Unusual cofactor requirements: 1,2-dihydroxynaphthalene oxygenase activity depends on NADH and the salicylate 5-hydroxylase requires NADPH, ATP, and coenzyme A. | [80] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Travkin, V.M.; Solyanikova, I.P. Salicylate or Phthalate: The Main Intermediates in the Bacterial Degradation of Naphthalene. Processes 2021, 9, 1862. https://doi.org/10.3390/pr9111862
Travkin VM, Solyanikova IP. Salicylate or Phthalate: The Main Intermediates in the Bacterial Degradation of Naphthalene. Processes. 2021; 9(11):1862. https://doi.org/10.3390/pr9111862
Chicago/Turabian StyleTravkin, Vasili M., and Inna P. Solyanikova. 2021. "Salicylate or Phthalate: The Main Intermediates in the Bacterial Degradation of Naphthalene" Processes 9, no. 11: 1862. https://doi.org/10.3390/pr9111862
APA StyleTravkin, V. M., & Solyanikova, I. P. (2021). Salicylate or Phthalate: The Main Intermediates in the Bacterial Degradation of Naphthalene. Processes, 9(11), 1862. https://doi.org/10.3390/pr9111862






