Bioactive Peptides from Liquid Milk Protein Concentrate by Sequential Tryptic and Microbial Hydrolysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ultra-Heat-Treated (UHT) Skimmed Cow’s Milk
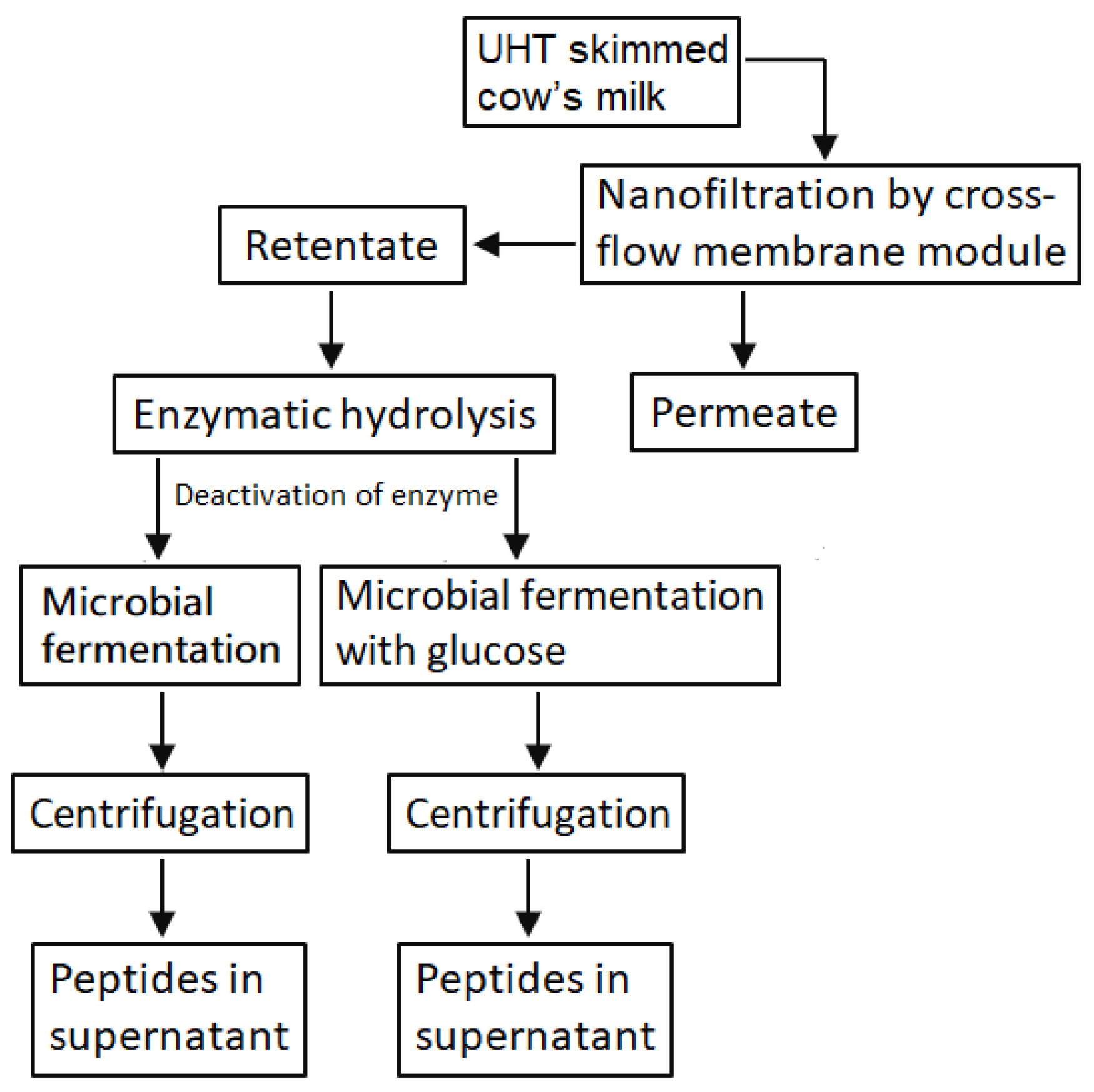
2.2. Production of Peptides with Functional Values from LMPC
2.2.1. Preparation of LMPC
2.2.2. In Vitro Hydrolysis of LMPC by Trypsin
2.2.3. Microbial Hydrolysis
2.3. Analytical Methods
2.3.1. Understanding the Molecular Weight Distribution of Proteins and Peptides
2.3.2. Immunoblotting
2.3.3. Determination of Protein Concentration
2.3.4. Determination of the Antioxidant Capacity
Ferric Reducing Ability of Plasma (FRAP) Assay
2,2-Diphenyl-1-picrylhydrazyl (DPPH) Radical-Scavenging Assay
2.3.5. Estimation of Angiotensin-Converting Enzyme Inhibitory Activity
2.3.6. Microbiological Assay
2.3.7. Near Infrared (NIR) Spectroscopy and Aquaphotomics Analysis
2.4. Statistical Analysis
3. Results and Discussions
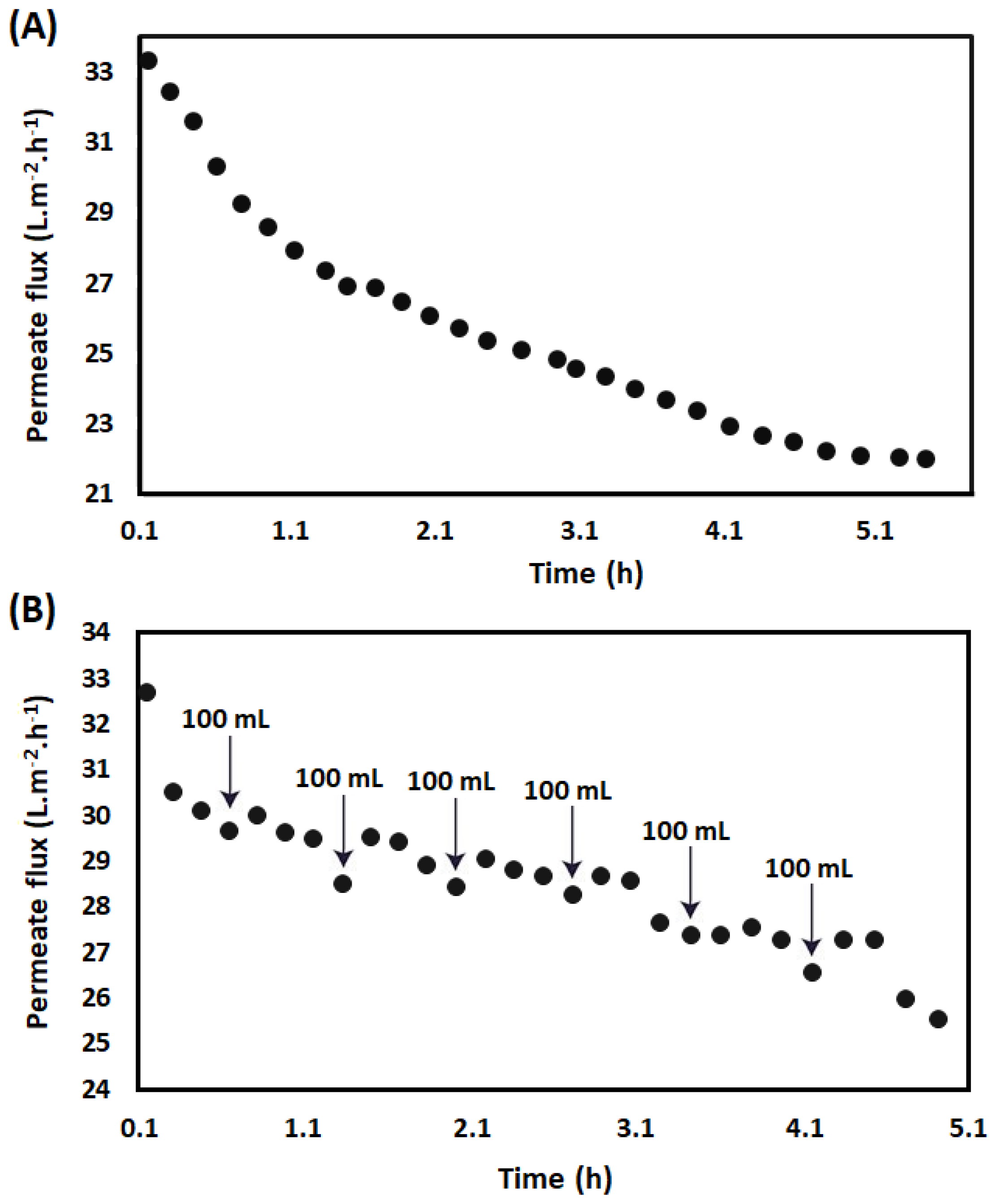
3.1. Preparation of LMPC
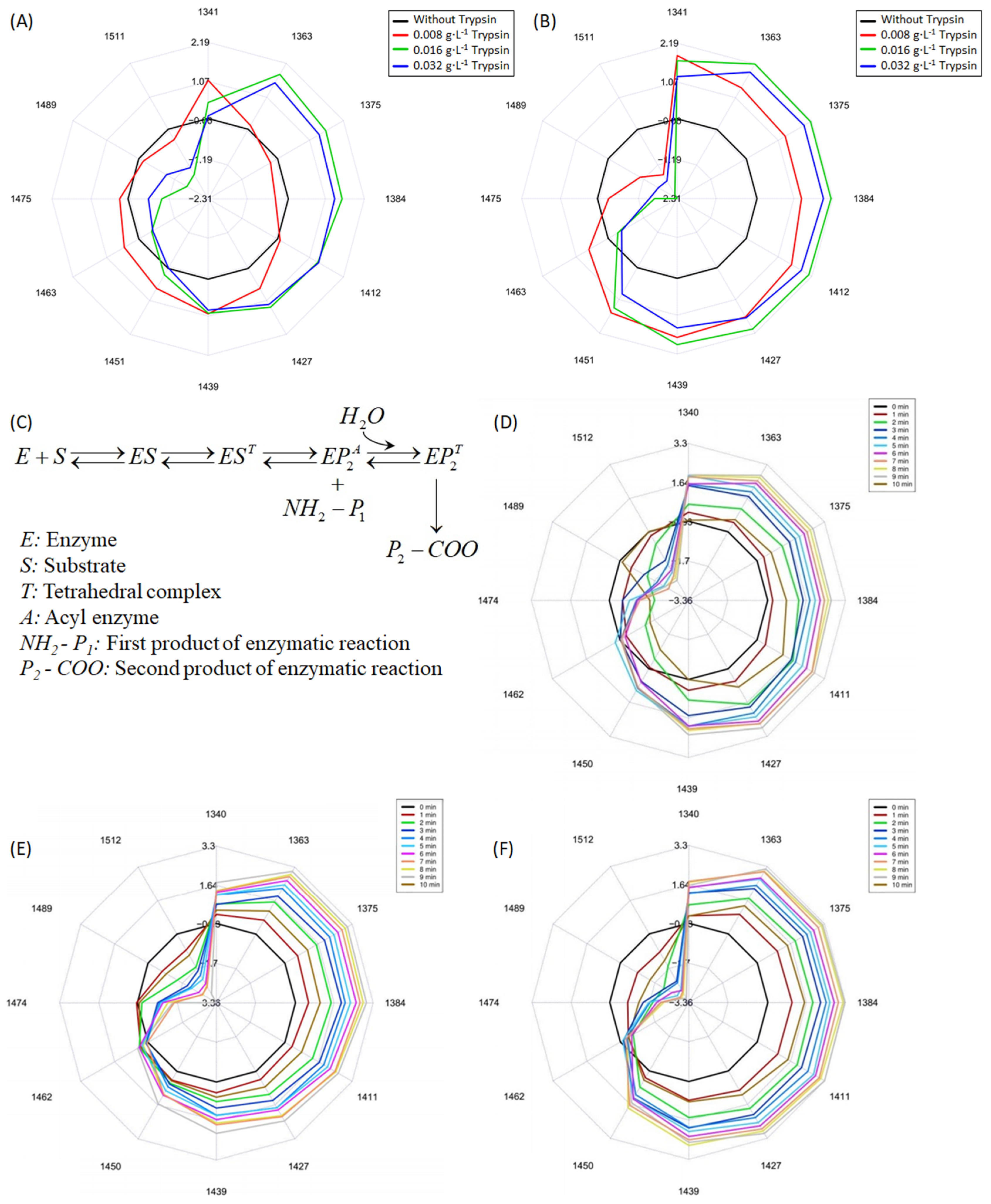
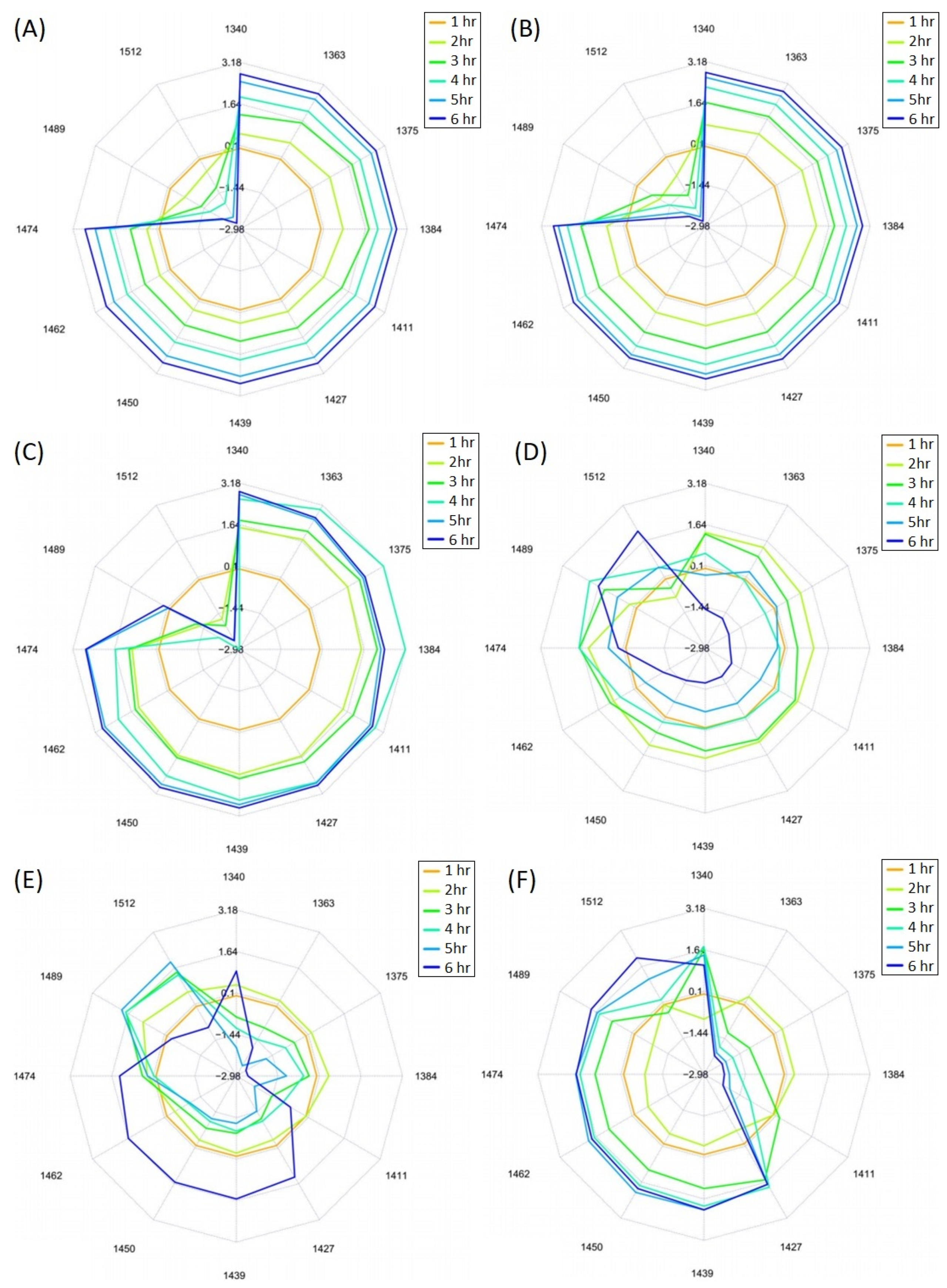
3.2. Hydrolysis of Proteins in LMPC
3.3. Aquaphotomics Analysis
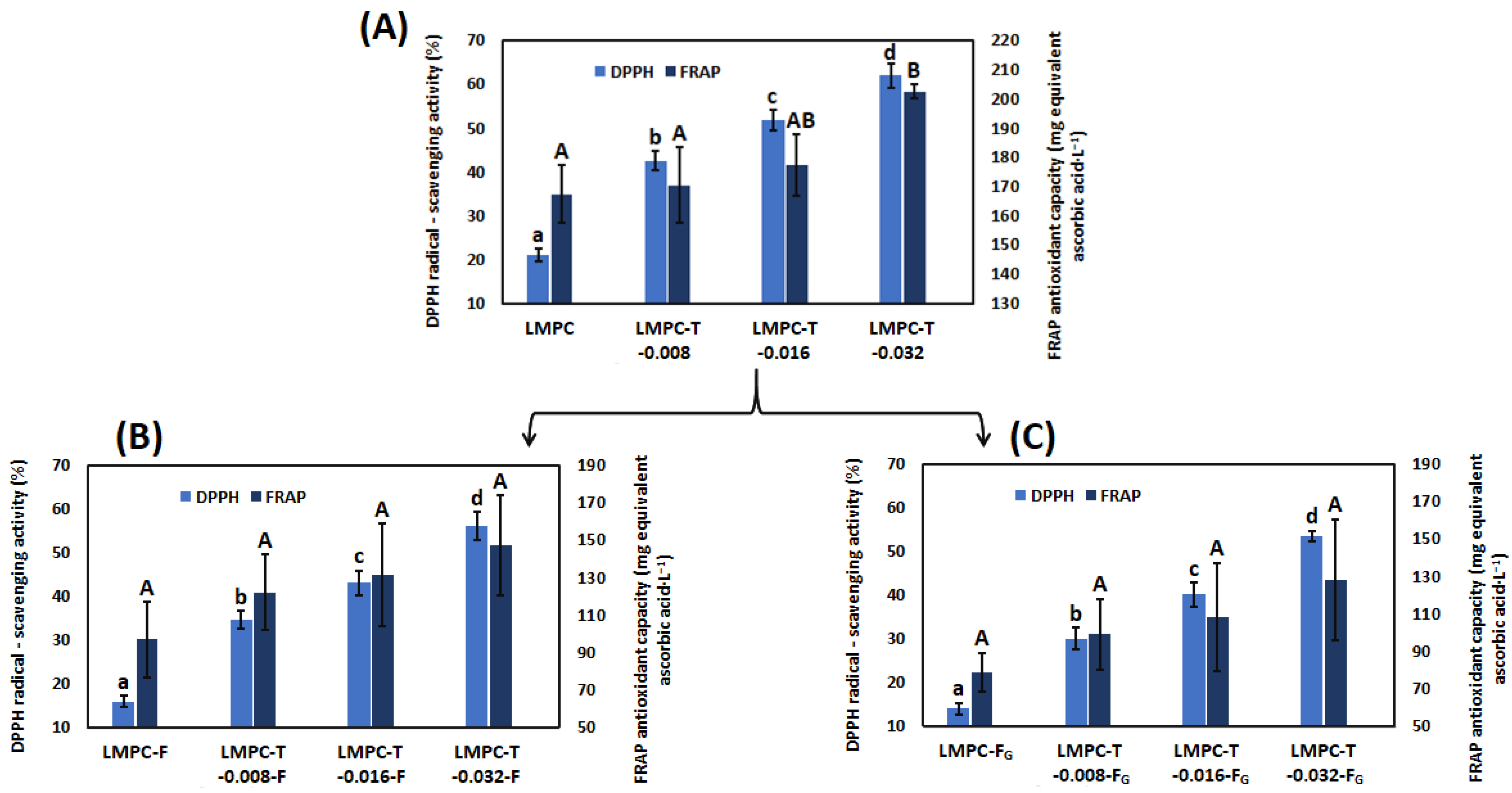
3.4. Antioxidant Capacity
3.5. Angiotensin-Converting Enzyme Inhibitory Activity
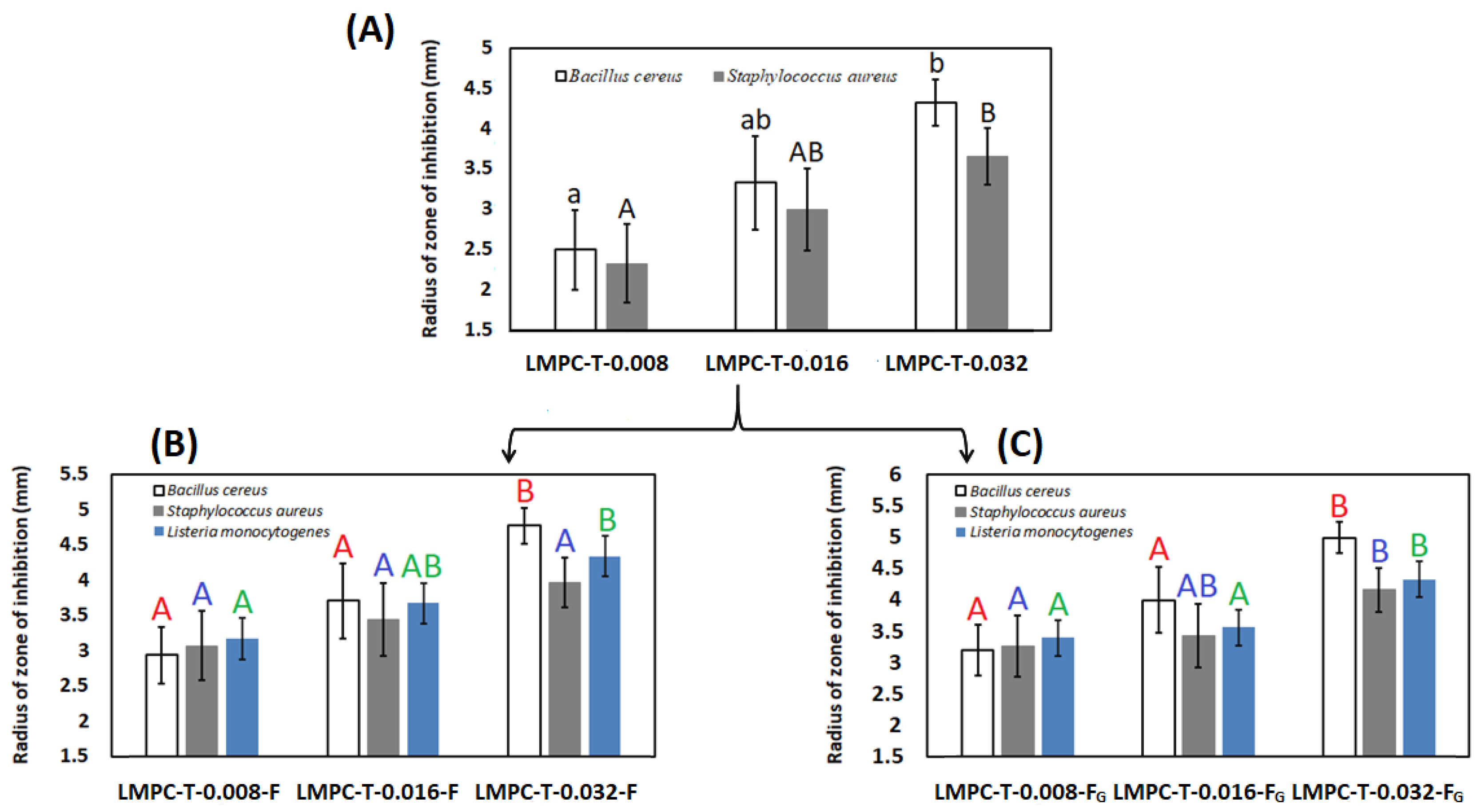
3.6. Antibacterial Activity
3.7. Immunogenicity
3.7.1. Antigenicity
3.7.2. Allergenicity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Han, T.S.; Lean, M.E. A clinical perspective of obesity, metabolic syndrome and cardiovascular disease. JRSM Cardiovasc. Dis. 2016, 5, 204800401663337. [Google Scholar] [CrossRef] [Green Version]
- Ferder, L.; Inserra, F.; Martínez-Maldonado, M. Inflammation and the metabolic syndrome: Role of angiotensin II and oxidative stress. Curr. Hypertens. Rep. 2006, 8, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Mancia, G.; Dell’Oro, R.; Quarti-Trevano, F.; Scopelliti, F.; Grassi, G. Angiotensin-sympathetic system interactions in cardiovascular and metabolic disease. J. Hypertens. 2006, 24, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Gaynor, R.B. Therapeutic potential of inhibition of the NF-κB pathway in the treatment of inflammation and cancer. J. Clin. Investig. 2001, 107, 135–142. [Google Scholar] [CrossRef] [Green Version]
- Tham, D.M.; Martin-McNulty, B.; Wang, Y.X.; Wilson, D.W.; Vergona, R.; Sullivan, M.E.; Dole, W.; Rutledge, J.C. Angiotensin II is associated with activation of NF-κB-mediated genes and downregulation of PPARs. Physiol. Genom. 2003, 11, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Luyckx, V.A.; Ots, M.; Lee, K.W.; Ziai, F.; Troy, J.L.; Brenner, B.M.; Mackenzie, H.S. Renin-angiotensin blockade lowers MCP-1 expression in diabetic rats. Kidney Int. 1999, 56, 1037–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornig, B.; Landmesser, U.; Kohler, C.; Ahlersmann, D.; Spiekermann, S.; Christoph, A.; Tatge, H.; Drexler, H. Comparative effect of ACE inhibition and angiotensin II type 1 receptor antagonism on bioavailability of nitric oxide in patients with coronary artery disease: Role of superoxide dismutase. Circulation 2001, 103, 799–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, S.; Zhang, G.X.; Nishiyama, A.; Shokoji, T.; Yao, L.; Fan, Y.Y.; Rahman, M.; Abe, Y. Mitochondria-derived reactive oxygen species and vascular MAP kinases: Comparison of angiotensin II and diazoxide. Hypertension 2005, 45, 438–444. [Google Scholar] [CrossRef] [Green Version]
- Zafari, A.M.; Ushio-fukai, M.; Akers, M.; Yin, Q.; Shah, A.; Harrison, D.G.; Taylor, W.R.; Griendling, K.K. Angiotensin II–Induced Vascular Hypertrophy. Hypertension 1998, 32, 488–495. [Google Scholar] [CrossRef] [Green Version]
- Beckman, J.S.; Beckman, T.W.; Chen, J.; Marshall, P.A.; Freeman, B.A. Apparent hydroxyl radical production by peroxynitrite: Implications for endothelial injury from nitric oxide and superoxide. Proc. Natl. Acad. Sci. USA 1990, 87, 1620–1624. [Google Scholar] [CrossRef] [Green Version]
- Wautier, M.P.; Chappey, O.; Corda, S.; Stern, D.M.; Schmidt, A.M.; Wautier, J.L. Activation of NADPH oxidase by AGE links oxidant stress to altered gene expression via RAGE. Am. J. Physiol.-Endocrinol. Metab. 2001, 280, E685–E694. [Google Scholar] [CrossRef] [PubMed]
- Rocha, R.; Funder, J.W. The pathophysiology of aldosterone in the cardiovascular system. Ann. N. Y. Acad. Sci. 2002, 970, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Marcus, Y.; Shefer, G.; Stern, N. Adipose tissue renin-angiotensin-aldosterone system (RAAS) and progression of insulin resistance. Mol. Cell. Endocrinol. 2013, 378, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.K.W.; Kam, K.K.H.; Yan, B.P.; Lam, Y.Y. Renin-angiotensin-aldosterone system blockade for cardiovascular diseases: Current status. Br. J. Pharmacol. 2010, 160, 1273–1292. [Google Scholar] [CrossRef] [PubMed]
- Jahandideh, F.; Wu, J. Perspectives on the potential benefits of antihypertensive peptides towards metabolic syndrome. Int. J. Mol. Sci. 2020, 21, 2192. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-López, F.; Tasset, I.; Agüera, E.; Feijóo, M.; Fernández-Bolaños, R.; Sánchez, F.M.; Ruiz, M.C.; Cruz, A.H.; Lix Gascón, F.; Túnez, I. Oxidative stress and inflammation biomarkers in the blood of patients with huntington’s disease. Neurol. Res. 2012, 34, 721–724. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Neuenschwander, L.C.; Bittencourt, H.; Ribeiro, A.F.T.; Teixeira, A.L.; Teixeira, M.M.; Teixeira, J.C.; Nobre, V. Plasma levels of procalcitonin and eight additional inflammatory molecules in febrile neutropenic patients. Clinics 2011, 66, 1699–1705. [Google Scholar]
- Dwyer, D.J.; Belenky, P.A.; Yang, J.H.; Cody MacDonald, I.; Martell, J.D.; Takahashi, N.; Chan, C.T.Y.; Lobritz, M.A.; Braff, D.; Schwarz, E.G.; et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA 2014, 111, E2100–E2109. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarti, S.; Jahandideh, F.; Wu, J. Food-derived bioactive peptides on inflammation and oxidative stress. BioMed Res. Int. 2014, 2014, 608979. [Google Scholar] [CrossRef] [Green Version]
- Oyinloye, B.E.; Adenowo, A.F.; Kappo, A.P. Reactive oxygen species, apoptosis, antimicrobial peptides and human inflammatory diseases. Pharmaceuticals 2015, 8, 151–175. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Aluko, R.E. Food protein-derived bioactive peptides: Production, processing, and potential health benefits. J. Food Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef]
- Nath, A.; Eren, B.A.; Csighy, A.; Pastorne-Huszar, K.; Kisko, G.; Abranko, L.; Toth, A.; Szerdahelyi, E.; Kovacs, Z.; Koris, A.; et al. Production of Liquid Milk Protein Concentrate with Antioxidant Capacity, Angiotensin Converting Enzyme Inhibitory Activity, Antibacterial Activity, and Hypoallergenic Property by Membrane Filtration and Enzymatic Modification of Proteins. Processes 2020, 8, 871. [Google Scholar] [CrossRef]
- Vila, L.; Beyer, K.; Järvinen, K.M.; Chatchatee, P.; Bardina, L.; Sampson, H.A. Role of conformational and linear epitopes in the achievement of tolerance in cow’s milk allergy. Clin. Exp. Allergy 2001, 31, 1599–1606. [Google Scholar] [CrossRef]
- Muro Urista, C.; Álvarez Fernández, R.; Riera Rodriguez, F.; Arana Cuenca, A.; Téllez Jurado, A. Review: Production and functionality of active peptides from milk. Food Sci. Technol. Int. 2011, 17, 293–317. [Google Scholar] [CrossRef]
- Crittenden, R.; Little, C.; Georgiou, G.; Forsyth, S.; Bennett, L. Cow’s Milk Allergy: A Complex Disorder. Aust. J. Dairy Technol. 2007, 62, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Haddad, Z.H.; Kalra, V.; Verma, S. IgE antibodies to peptic and peptic-tryptic digests of betalactoglobulin: Significance in food hypersensitivity. Ann. Allergy 1979, 42, 368–371. [Google Scholar] [PubMed]
- Colantuono, A.; D’Incecco, P.; Fortina, M.G.; Rosi, V.; Ricci, G.; Pellegrino, L. Milk substrates influence proteolytic activity of Pseudomonas fluorescens strains. Food Control 2020, 111, 107063. [Google Scholar] [CrossRef]
- Troise, A.D.; Bandini, E.; De Donno, R.; Meijer, G.; Trezzi, M.; Fogliano, V. The quality of low lactose milk is affected by the side proteolytic activity of the lactase used in the production process. Food Res. Int. 2016, 89, 514–525. [Google Scholar] [CrossRef] [Green Version]
- Li-Chan, E.C.Y. Bioactive peptides and protein hydrolysates: Research trends and challenges for application as nutraceuticals and functional food ingredients. Curr. Opin. Food Sci. 2015, 1, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Beshkova, D.; Simova, E.; Frengova, G.; Simov, Z. Production of flavour compounds by yogurt starter cultures. J. Ind. Microbiol. Biotechnol. 1998, 20, 180–186. [Google Scholar] [CrossRef]
- Li, S.; Tang, S.; He, Q.; Hu, J.; Zheng, J. Changes in Proteolysis in Fermented Milk Produced by Streptococcus thermophilus in Co-Culture with Lactobacillus plantarum or Bifidobacterium animalis subsp. Lactis during Refrigerated Storage. Molecules 2019, 24, 3699. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Hajós, G.; Polgár, M.; Farkas, J. High-pressure effects on IgE immunoreactivity of proteins in a sausage batter. Innov. Food Sci. Emerg. Technol. 2004, 5, 443–449. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The Ferric Reducing Ability of Plasma (FRAP) as a Measure of “Antioxidant Power”: The FRAP Assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Arfaoui, L. Total polyphenol content and radical scavenging activity of functional yogurt enriched with dates. Czech J. Food Sci. 2020, 38, 287–292. [Google Scholar] [CrossRef]
- Fagyas, M.; Úri, K.; Siket, I.M.; Daragó, A.; Boczán, J.; Bányai, E.; Édes, I.; Papp, Z.; Tóth, A. New Perspectives in the Renin-Angiotensin-Aldosterone System (RAAS) I: Endogenous Angiotensin Converting Enzyme (ACE) Inhibition. PLoS ONE 2014, 9, e87843. [Google Scholar]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Y.; Chen, Y.; Zhang, Y.; Shi, T.; Wang, J.; Hong, Y.; Fei, T.; Zhang, Y. The influence of spectral pretreatment on the selection of representative calibration samples for soil organic matter estimation using vis-NIR reflectance spectroscopy. Remote Sens. 2019, 11, 450. [Google Scholar] [CrossRef] [Green Version]
- Tsenkova, R. Aquaphotomics: Water in the biological and aqueous world scrutinised with invisible light. Spectrosc. Eur. 2010, 22, 6–10. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. 2017. Available online: https://www.R-project.org/ (accessed on 17 September 2021).
- Kovacs, Z.; Pollner, B. Dedicated Aquaphotomics-Software R-Package „aquap2“ General Introduction and Workshop. In Proceedings of the Understanding Water in Biology at the 2nd International Symposium, Kobe, Japan, 26–29 November 2021; Kobe University, Faculty of Agriculture: Kobe, Japan, 2016. [Google Scholar]
- Al-Mutwalli, S.A.; Dilaver, M.; Koseoglu-Imer, D.Y. Performance evaluation of ceramic membrane on ultrafiltration and diafiltration modes for efficient recovery of whey protein. J. Membr. Sci. Res. 2020, 6, 138–146. [Google Scholar]
- Jeswan Singh, M.; Chandrapala, J.; Udabage, P.; McKinnon, I.; Augustin, M.A. Heat-induced changes in the properties of modified skim milks with different casein to whey protein ratios. J. Dairy Res. 2015, 82, 135–142. [Google Scholar] [CrossRef]
- Galani, D.; Owusu Apenten, R.K. Heat-induced denaturation and aggregation of β-lactoglobulin: Kinetics of formation of hydrophobic and disulphide-linked aggregates. Int. J. Food Sci. Technol. 1999, 34, 467–476. [Google Scholar] [CrossRef]
- Morr, C.V. Protein Aggregation in Conventional and Ultra High-Temparature Heated Skimmilk. J. Dairy Sci. 1969, 52, 1174–1180. [Google Scholar] [CrossRef]
- Gezimati, J.; Singh, H.; Creamer, L.K. Aggregation and Gelation of Bovine β-Lactoglobulin, α-Lactalbumin, and Serum Albumin. ACS Symp. Ser. 1996, 650, 113–123. [Google Scholar]
- Jang, H.D.; Swaisgood, H.E. Disulfide Bond Formation Between Thermally Denatured β-Lactoglobulin and κ-Casein in Casein Micelles. J. Dairy Sci. 1990, 73, 900–904. [Google Scholar] [CrossRef]
- Vasbinder, A.J.; Van De Velde, F.; De Kruif, C.G. Gelation of casein-whey protein mixtures. J. Dairy Sci. 2004, 87, 1167–1176. [Google Scholar] [CrossRef]
- Li, Y.; Dalgleish, D.; Corredig, M. Influence of heating treatment and membrane concentration on the formation of soluble aggregates. Food Res. Int. 2015, 76, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Sindhu, J.S.; Arora, S. Milk|Buffalo Milk. In Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 503–511. ISBN 978-0-12-374407-4. [Google Scholar]
- Liu, E.; Zheng, H.; Shi, T.; Ye, L.; Konno, T.; Oda, M.; Shen, H.; Ji, Z.-S. Relationship between Lactobacillus bulgaricus and Streptococcus thermophilus under whey conditions: Focus on amino acid formation. Int. Dairy J. 2016, 56, 141–150. [Google Scholar] [CrossRef]
- Tzvetkova, I.; Dalgalarrondo, M.; Danova, S.; Iliev, I.; Ivanova, I.; Chobert, J.M.; Haertlé, T. Hydrolysis of major dairy proteins by lactic acid bacteria from Bulgarian yogurts. J. Food Biochem. 2007, 31, 680–702. [Google Scholar] [CrossRef]
- Tsenkova, R. Introduction: Aquaphotomics: Dynamic spectroscopy of aqueous and biological systems describes peculiarities of water. J. Near Infrared Spectrosc. 2009, 17, 303. [Google Scholar] [CrossRef]
- Muncan, J.; Tsenkova, R. Aquaphotomics—From Innovative Knowledge to Integrative Platform in Science and Technology. Molecules 2019, 24, 2742. [Google Scholar] [CrossRef] [Green Version]
- Perutka, Z.; Šebela, M. Pseudotrypsin: A Little-Known Trypsin Proteoform. Molecules 2018, 23, 2637. [Google Scholar] [CrossRef] [Green Version]
- Weiner, S.J.; Seibel, G.L.; Kollman, P.A. The nature of enzyme catalysis in trypsin. Proc. Natl. Acad. Sci. USA 1986, 83, 649–653. [Google Scholar] [CrossRef] [Green Version]
- Muncan, J.; Tei, K.; Tsenkova, R. Real-Time Monitoring of Yogurt Fermentation Process by Aquaphotomics Near-Infrared Spectroscopy. Sensors 2021, 21, 177. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, P.; MEISEL, H. Influence of trypsin action in yoghurt milk on the release of caseinophosphopeptide-rich fractions and physical properties of the fermented products. Int. J. Dairy Technol. 2005, 58, 119–124. [Google Scholar] [CrossRef]
- Okumura, M.; Yeh, L.I.; Myers, J.D.; Lee, Y.T. Infrared spectra of the solvated hydronium ion: Vibrational predissociation spectroscopy of mass-selected H3O+.cntdot.(H2O)n.cntdot.(H2)m. J. Phys. Chem. 1990, 94, 3416–3427. [Google Scholar] [CrossRef]
- Jiang, J.-C.; Wang, Y.-S.; Chang, H.-C.; Lin, S.H.; Lee, Y.T.; Niedner-Schatteburg, G.; Chang, H.-C. Infrared Spectra of H+(H2O)5–8 Clusters: Evidence for Symmetric Proton Hydration. J. Am. Chem. Soc. 2000, 122, 1398–1410. [Google Scholar] [CrossRef]
- Broyard, C.; Gaucheron, F. Modifications of structures and functions of caseins: A scientific and technological challenge. Dairy Sci. Technol. 2015, 95, 831–862. [Google Scholar] [CrossRef]
- Savijoki, K.; Ingmer, H.; Varmanen, P. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 2006, 71, 394–406. [Google Scholar] [CrossRef]
- Pescuma, M.; Hébert, E.M.; Mozzi, F.; de Valdez, G.F. Hydrolysis of whey proteins by Lactobacillus acidophilus, Streptococcus thermophilus and Lactobacillus delbrueckii ssp. bulgaricus grown in a chemically defined medium. J. Appl. Microbiol. 2007, 103, 1738–1746. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Y.; Li, H.; Liu, X. The potential of proteins, hydrolysates and peptides as growth factors for Lactobacillus and Bifidobacterium: Current research and future perspectives. Food Funct. 2020, 11, 1946–1957. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, S.; Imm, J.-Y. Buffering Capacity of Dairy Powders and Their Effect on Yoghurt Quality. Korean J. Food Sci. Anim. Resour. 2018, 38, 273–281. [Google Scholar] [PubMed]
- Bouteille, R.; Gaudet, M.; Lecanu, B.; This, H. Monitoring lactic acid production during milk fermentation by in situ quantitative proton nuclear magnetic resonance spectroscopy. J. Dairy Sci. 2013, 96, 2071–2080. [Google Scholar] [CrossRef] [PubMed]
- Chatham, J.C.; Forder, J.R. Lactic acid and protein interactions: Implications for the NMR visibility of lactate in biological systems. Biochim. Biophys. Acta–Gen. Subj. 1999, 1426, 177–184. [Google Scholar] [CrossRef]
- Kanetro, B.; Slamet, A.; Wazyka, A. Effect of various solvent on the specific amino acids of black soybean (Glycine soja) sprout. IOP Conf. Ser. Earth Environ. Sci. 2018, 102, 12002. [Google Scholar] [CrossRef]
- Kinsella, J.; Melachouris, N. Functional properties of proteins in foods: A survey. Crit. Rev. Food Sci. Nutr. 2009, 7, 219–280. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Enzymatic hydrolysis of proteins for increased solubility. J. Agric. Food Chem. 1976, 24, 1090–1093. [Google Scholar] [CrossRef]
- Biasutti, E.; Vieira, C.; Capobiango, M.; Silva, V.; Silvestre, M. Study of Some Functional Properties of Casein: Effect of pH and Tryptic Hydrolysis. Int. J. Food Prop. 2007, 10, 173–183. [Google Scholar] [CrossRef]
- Purohit, D.H.; Hassan, A.N.; Bhatia, E.; Zhang, X.; Dwivedi, C. Rheological, sensorial, and chemopreventive properties of milk fermented with exopolysaccharide-producing lactic cultures. J. Dairy Sci. 2009, 92, 847–856. [Google Scholar] [CrossRef] [Green Version]
- Gezginc, Y.; Topcal, F.; Comertpay, S.; Akyol, I. Quantitative analysis of the lactic acid and acetaldehyde produced by Streptococcus thermophilus and Lactobacillus bulgaricus strains isolated from traditional Turkish yogurts using HPLC. J. Dairy Sci. 2015, 98, 1426–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bills, D.D.; Yang, C.S.; Morgan, M.E.; Bodyfelt, F.W. Effect of Sucrose on the Production of Acetaldehyde and Acids by Yogurt Culture Bacteria. J. Dairy Sci. 1972, 55, 1570–1573. [Google Scholar] [CrossRef]
- Jamshidian, M.; Arab-Tehrany, E.; Imran, M.; Jacquot, M.; Desobry, S. Poly-Lactic Acid: Production, Applications, Nanocomposites, and Release Studies. Compr. Rev. Food Sci. Food Saf. 2010, 9, 552–571. [Google Scholar] [CrossRef] [PubMed]
- Williams, P. Influence of Water on Prediction of Composition and Quality Factors: The Aquaphotomics of Low Moisture Agricultural Materials. J. Near Infrared Spectrosc. 2009, 17, 315–328. [Google Scholar] [CrossRef]
- Neto, Y.A.A.H.; Rosa, J.C.; Cabral, H. Peptides with antioxidant properties identified from casein, whey, and egg albumin hydrolysates generated by two novel fungal proteases. Prep. Biochem. Biotechnol. 2019, 49, 639–648. [Google Scholar] [CrossRef]
- Bamdad, F.; Shin, S.H.; Suh, J.-W.; Nimalaratne, C.; Sunwoo, H. Anti-Inflammatory and Antioxidant Properties of Casein Hydrolysate Produced Using High Hydrostatic Pressure Combined with Proteolytic Enzymes. Molecules 2017, 22, 609. [Google Scholar]
- Abd El-Fattah, A.; Sakr, S.; El-Dieb, S.; Elkashef, H. Bioactive peptides with ACE-I and antioxidant activity produced from milk proteolysis. Int. J. Food Prop. 2017, 20, 3033–3042. [Google Scholar] [CrossRef] [Green Version]
- Sabeena Farvin, K.H.; Baron, C.P.; Nielsen, N.S.; Jacobsen, C. Antioxidant activity of yoghurt peptides: Part 1-in vitro assays and evaluation in ω-3 enriched milk. Food Chem. 2010, 123, 1081–1089. [Google Scholar] [CrossRef]
- Sabeena Farvin, K.H.; Baron, C.P.; Nielsen, N.S.; Otte, J.; Jacobsen, C. Antioxidant activity of yoghurt peptides: Part 2–Characterisation of peptide fractions. Food Chem. 2010, 123, 1090–1097. [Google Scholar] [CrossRef]
- Yilmaz-Ersan, L.; Ozcan, T.; Akpinar-Bayizit, A.; Sahin, S. Comparison of antioxidant capacity of cow and ewe milk kefirs. J. Dairy Sci. 2018, 101, 3788–3798. [Google Scholar]
- Aloğlu, H.; Oner, Z. Determination of antioxidant activity of bioactive peptide fractions obtained from yogurt. J. Dairy Sci. 2011, 94, 5305–5314. [Google Scholar] [CrossRef] [Green Version]
- Courtin, P.; Rul, F. Interactions between microorganisms in a simple ecosystem: Yogurt bacteria as a study model. Le Lait 2003, 84, 125–134. [Google Scholar] [CrossRef]
- Schieber, A.; Brückner, H. Characterization of oligo- and polypeptides isolated from yoghurt. Eur. Food Res. Technol. 2000, 210, 310–313. [Google Scholar]
- López-Fandiño, R.; Otte, J.; van Camp, J. Physiological, chemical and technological aspects of milk-protein-derived peptides with antihypertensive and ACE-inhibitory activity. Int. Dairy J. 2006, 16, 1277–1293. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xu, Z.; Lu, S.; Xin, M.; Kong, J. Effects of glutathione on acid stress resistance and symbiosis between Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus. Int. Dairy J. 2016, 61, 22–28. [Google Scholar] [CrossRef]
- Pophaly, S.D.; Poonam, S.; Pophaly, S.D.; Kapila, S.; Nanda, D.K.; Tomar, S.K.; Singh, R. Glutathione biosynthesis and activity of dependent enzymes in food-grade lactic acid bacteria harbouring multidomain bifunctional fusion gene (gshF). J. Appl. Microbiol. 2017, 123, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Ramchandran, L.; Shah, N.P. Characterization of functional, biochemical and textural properties of synbiotic low-fat yogurts during refrigerated storage. LWT–Food Sci. Technol. 2010, 43, 819–827. [Google Scholar] [CrossRef]
- Ramchandran, L.; Shah, N.P. Effect of exopolysaccharides on the proteolytic and angiotensin-I converting enzyme-inhibitory activities and textural and rheological properties of low-fat yogurt during refrigerated storage. J. Dairy Sci. 2009, 92, 895–906. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Tu, M.; Wu, D.; Chen, H.; Chen, C.; Wang, Z.; Jiang, L. Identification of an ACE-Inhibitory Peptide from Walnut Protein and Its Evaluation of the Inhibitory Mechanism. Int. J. Mol. Sci. 2018, 19, 1156. [Google Scholar] [CrossRef] [Green Version]
- Vermeirssen, V.; Van Camp, J.; Decroos, K.; Van Wijmelbeke, L.; Verstraete, W. The impact of fermentation and in vitro digestion on the formation of angiotensin-I-converting enzyme inhibitory activity from pea and whey protein. J. Dairy Sci. 2003, 86, 429–438. [Google Scholar] [CrossRef]
- Manso, M.A.; López-Fandiño, R. Angiotensin I converting enzyme-inhibitory activity of bovine, ovine, and caprine kappa-casein macropeptides and their tryptic hydrolysates. J. Food Prot. 2003, 66, 1686–1692. [Google Scholar] [CrossRef] [PubMed]
- Pihlanto-Leppälä, A.; Koskinen, P.; Piilola, K.; Tupasela, T.; Korhonen, H. Angiotensin I-converting enzyme inhibitory properties of whey protein digests: Concentration and characterization of active peptides. J. Dairy Res. 2000, 67, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Mullally, M.M.; Meisel, H.; FitzGerald, R.J. Synthetic peptides corresponding to alpha-lactalbumin and beta-lactoglobulin sequences with angiotensin-I-converting enzyme inhibitory activity. Biol. Chem.-Hoppe Seyler 1996, 377, 259–260. [Google Scholar]
- Asoodeh, A.; Homayouni-Tabrizi, M.; Shabestarian, H.; Emtenani, S.; Emtenani, S. Biochemical characterization of a novel antioxidant and angiotensin I-converting enzyme inhibitory peptide from Struthio camelus egg white protein hydrolysis. J. Food Drug Anal. 2016, 24, 332–342. [Google Scholar] [CrossRef] [Green Version]
- Cheung, H.S.; Wang, F.L.; Ondetti, M.A.; Sabo, E.F.; Cushman, D.W. Binding of peptide substrates and inhibitors of angiotensin-converting enzyme. Importance of the COOH-terminal dipeptide sequence. J. Biol. Chem. 1980, 255, 401–407. [Google Scholar] [CrossRef]
- Hernández-Ledesma, B.; Amigo, L.; Ramos, M.; Recio, I. Angiotensin converting enzyme inhibitory activity in commercial fermented products. Formation of peptides under simulated gastrointestinal digestion. J. Agric. Food Chem. 2004, 52, 1504–1510. [Google Scholar] [CrossRef] [PubMed]
- Minervini, F.; Algaron, F.; Rizzello, C.G.; Fox, P.F.; Monnet, V.; Gobbetti, M. Angiotensin I-converting-enzyme-inhibitory and antibacterial peptides from Lactobacillus helveticus PR4 proteinase-hydrolyzed caseins of milk from six species. Appl. Environ. Microbiol. 2003, 69, 5297–5305. [Google Scholar] [CrossRef] [Green Version]
- Shakerian, M.; Razavi, S.H.; Ziai, S.A.; Khodaiyan, F.; Yarmand, M.S.; Moayedi, A. Proteolytic and ACE-inhibitory activities of probiotic yogurt containing non-viable bacteria as affected by different levels of fat, inulin and starter culture. J. Food Sci. Technol. 2015, 52, 2428–2433. [Google Scholar] [CrossRef] [Green Version]
- Fagyas, M.; Úri, K.; Siket, I.M.; Fülöp, G.Á.; Csató, V.; Daragó, A.; Boczán, J.; Bányai, E.; Szentkirályi, I.E.; Maros, T.M.; et al. New perspectives in the renin-angiotensin-aldosterone system (RAAS) II: Albumin suppresses angiotensin converting enzyme (ACE) activity in human. PLoS ONE 2014, 9, e87844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guang, C.; Phillips, R.D. Plant food-derived Angiotensin I converting enzyme inhibitory peptides. J. Agric. Food Chem. 2009, 57, 5113–5120. [Google Scholar] [CrossRef] [PubMed]
- Vallabha, V.; Kaultiku, P. Antihypertensive Peptides Derived from Soy Protein by Fermentation. Int. J. Pept. Res. Ther. 2014, 20, 161. [Google Scholar] [CrossRef]
- Medeiros, V.; Rainha, N.; Paiva, L.; Lima, E.; Baptista, J. Bovine Milk Formula Based on Partial Hydrolysis of Caseins by Bromelain Enzyme: Better Digestibility and Angiotensin-Converting Enzyme-Inhibitory Properties. Int. J. Food Prop. 2014, 17, 806–817. [Google Scholar] [CrossRef]
- Wang, R.; Han, Z.; Ji, R.; Xiao, Y.; Si, R.; Guo, F.; He, J.; Hai, L.; Ming, L.; Yi, L. Antibacterial Activity of Trypsin-Hydrolyzed Camel and Cow Whey and Their Fractions. Animals 2020, 10, 337. [Google Scholar] [CrossRef] [Green Version]
- Bruni, N.; Capucchio, M.T.; Biasibetti, E.; Pessione, E.; Cirrincione, S.; Giraudo, L.; Corona, A.; Dosio, F. Antimicrobial Activity of Lactoferrin-Related Peptides and Applications in Human and Veterinary Medicine. Molecules 2016, 21, 752. [Google Scholar] [CrossRef]
- Silva, A.; Honjoya, E.; Cardoso, S.; Souza, C.; Costa, M.; Santana, E.; Aragon-Alegro, L. Antimicrobial action of lactoferrin on Staphylococcus aureus inoculated in Minas frescal cheese. Arch. Latinoam. Nutr. 2012, 62, 68–72. [Google Scholar]
- Seifu, E.; Buys, E.M.; Donkin, E.F. Significance of the lactoperoxidase system in the dairy industry and its potential applications: A review. Trends Food Sci. Technol. 2005, 16, 137–154. [Google Scholar] [CrossRef]
- Davidson, P.M.; Taylor, T.M.; Schmidt, S.E. Chemical Preservatives and Natural Antimicrobial Compounds. In Food Microbiology: Fundamentals and Frontiers; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2012; pp. 765–801. ISBN 9781683670582. [Google Scholar]
- Travkova, O.G.; Moehwald, H.; Brezesinski, G. The interaction of antimicrobial peptides with membranes. Adv. Colloid Interface Sci. 2017, 247, 521–532. [Google Scholar] [CrossRef]
- Patrzykat, A.; Douglas, S.E. Antimicrobial peptides: Cooperative approaches to protection. Protein Pept. Lett. 2005, 12, 19–25. [Google Scholar] [CrossRef]
- Matin, M.; Monnai, M.; Otani, H. Isolation and Characterization of a Cytotoxic Pentapeptide, κ-casecidin, from Bovine κ-casein Digested with Bovine Trypsin. Nihon Chikusan Gakkaiho 2000, 71, 197–207. [Google Scholar] [CrossRef]
- Pellegrini, A.; Thomas, U.; Bramaz, N.; Hunziker, P.; von Fellenberg, R. Isolation and identification of three bactericidal domains in the bovine α-lactalbumin molecule. Biochim. Biophys. Acta–Gen. Subj. 1999, 1426, 439–448. [Google Scholar] [CrossRef]
- Pellegrini, A.; Dettling, C.; Thomas, U.; Hunziker, P. Isolation and characterization of four bactericidal domains in the bovine β-lactoglobulin. Biochim. Biophys. Acta–Gen. Subj. 2001, 1526, 131–140. [Google Scholar] [CrossRef]
- Zucht, H.D.; Raida, M.; Adermann, K.; Mägert, H.J.; Forssmann, W.G. Casocidin-I: A casein-αs2 derived peptide exhibits antibacterial activity. FEBS Lett. 1995, 372, 185–188. [Google Scholar] [CrossRef] [Green Version]
- Akpinar, A.; Yerlikaya, O.; Kiliç, S. Antimicrobial activity and antibiotic resistance of Lactobacillus delbrueckii ssp bulgaricus and Streptococcus thermophilus strain isolated from Turkish homemade yoghurts. Afr. J. Microbiol. Res. 2011, 5, 675–682. [Google Scholar]
- Yerlikaya, O.; Saygili, D.; Akpinar, A. Evaluation of antimicrobial activity and antibiotic susceptibility profiles of Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus strains isolated from commercial yoghurt starter cultures. Ciênc. Tecnol. Aliment. 2020, 41, 418–425. [Google Scholar] [CrossRef]
- Al-Nabulsi, A.A.; Olaimat, A.N.; Osaili, T.M.; Ayyash, M.M.; Abushelaibi, A.; Jaradat, Z.W.; Shaker, R.; Al-Taani, M.; Holley, R.A. Behavior of Escherichia coli O157:H7 and Listeria monocytogenes during fermentation and storage of camel yogurt. J. Dairy Sci. 2016, 99, 1802–1811. [Google Scholar] [CrossRef] [Green Version]
- Gomez, S.; Cosson, C.; Deschamps, A.M. Evidence for a bacteriocin-like substance produced by a new strain of Streptococcus sp., inhibitory to gram-positive food-borne pathogens. Res. Microbiol. 1997, 148, 757–766. [Google Scholar] [CrossRef]
- Massa, S.; Trovatelli, L.D.; Canganella, F. Survival of Listeria monocytogenes in yogurt during storage at 4 °C. Lett. Appl. Microbiol. 1991, 13, 112–114. [Google Scholar] [CrossRef]
- Benkerroum, N.; Oubel, H.; Mimoun, L. Behavior of Listeria monocytogenes and Staphylococcus aureus in Yogurt Fermented with a Bacteriocin-Producing Thermophilic Starter. J. Food Prot. 2002, 65, 799–805. [Google Scholar] [CrossRef]
- Miteva, V.; Ivanova, I.; Budakov, I.; Pantev, A.; Stefanova, T.; Danova, S.; Moncheva, P.; Mitev, V.; Dousset, X.; Boyaval, P. Detection and characterization of a novel antibacterial substance produced by a Lactobacillus delbrueckii strain 1043. J. Appl. Microbiol. 1998, 85, 603–614. [Google Scholar] [CrossRef]
- Wal, J.-M. Bovine milk allergenicity. Ann. Allergy Asthma Immunol. 2004, 93, S2–S11. [Google Scholar] [CrossRef]
- Fritsché, R. Role for technology in dairy allergy. Aust. J. Dairy Technol. 2003, 58, 89–91. [Google Scholar]
- Bu, G.; Luo, Y.; Zheng, Z.; Zheng, H. Effect of heat treatment on the antigenicity of bovine α-lactalbumin and β-lactoglobulin in whey protein isolate. Food Agric. Immunol. 2009, 20, 195–206. [Google Scholar] [CrossRef]
- Xu, Q.; Shi, J.; Yao, M.; Jiang, M.; Luo, Y. Effects of heat treatment on the antigenicity of four milk proteins in milk protein concentrates. Food Agric. Immunol. 2015, 27, 401–413. [Google Scholar] [CrossRef]
- Liu, F.; Teodorowicz, M.; van Boekel, M.A.J.S.; Wichers, H.J.; Hettinga, K.A. The decrease in the IgG-binding capacity of intensively dry heated whey proteins is associated with intense Maillard reaction, structural changes of the proteins and formation of RAGE-ligands. Food Funct. 2016, 7, 239–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antoine, E.; Souza, C. Study by Differential Scanning Calorimetry of the Thermal Stability of Whey Proteins Concentrate. Biotechnology 2007, 6, 431–435. [Google Scholar] [CrossRef]
- Deeth, H. Optimum Thermal Processing for Extended Shelf-Life (ESL) Milk. Foods 2017, 6, 102. [Google Scholar] [CrossRef] [Green Version]
- Natale, M.; Bisson, C.; Monti, G.; Peltran, A.; Garoffo, L.P.; Valentini, S.; Fabris, C.; Bertino, E.; Coscia, A.; Conti, A. Cow’s milk allergens identification by two-dimensional immunoblotting and mass spectrometry. Mol. Nutr. Food Res. 2004, 48, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Wróblewska, B.; Kaliszewska, A. Cow’s milk proteins immunoreactivity and allergenicity in processed food. Czech J. Food Sci. 2018, 30, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Pescuma, M.; Hébert, E.M.; Rabesona, H.; Drouet, M.; Choiset, Y.; Haertlé, T.; Mozzi, F.; de Valdez, G.F.; Chobert, J.-M. Proteolytic action of Lactobacillus delbrueckii subsp. bulgaricus CRL 656 reduces antigenic response to bovine β-lactoglobulin. Food Chem. 2011, 127, 487–492. [Google Scholar] [CrossRef]
- Bu, G.; Luo, Y.; Zhang, Y.; Chen, F. Effects of fermentation by lactic acid bacteria on the antigenicity of bovine whey proteins. J. Sci. Food Agric. 2010, 90, 2015–2020. [Google Scholar] [CrossRef] [PubMed]
- Kleber, N.; Weyrich, U.; Hinrichs, J. Screening of lactic acid bacteria with potential to reduce antigenic response of β-lactoglobulin bovine skim milk and sweet whey. Innov. Food Sci. Emerg. Technol. 2006, 7, 233–238. [Google Scholar] [CrossRef]
- Abd El-Salam, M.; El-Shibiny, S. Reduction of Milk Protein Antigenicity by Enzymatic Hydrolysis and Fermentation. A Review. Food Rev. Int. 2019, 37, 276–295. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nath, A.; Csighy, A.; Eren, B.A.; Tjandra Nugraha, D.; Pásztorné-Huszár, K.; Tóth, A.; Takács, K.; Szerdahelyi, E.; Kiskó, G.; Kovács, Z.; et al. Bioactive Peptides from Liquid Milk Protein Concentrate by Sequential Tryptic and Microbial Hydrolysis. Processes 2021, 9, 1688. https://doi.org/10.3390/pr9101688
Nath A, Csighy A, Eren BA, Tjandra Nugraha D, Pásztorné-Huszár K, Tóth A, Takács K, Szerdahelyi E, Kiskó G, Kovács Z, et al. Bioactive Peptides from Liquid Milk Protein Concentrate by Sequential Tryptic and Microbial Hydrolysis. Processes. 2021; 9(10):1688. https://doi.org/10.3390/pr9101688
Chicago/Turabian StyleNath, Arijit, Attila Csighy, Burak Attila Eren, David Tjandra Nugraha, Klára Pásztorné-Huszár, Attila Tóth, Krisztina Takács, Emőke Szerdahelyi, Gabriella Kiskó, Zoltán Kovács, and et al. 2021. "Bioactive Peptides from Liquid Milk Protein Concentrate by Sequential Tryptic and Microbial Hydrolysis" Processes 9, no. 10: 1688. https://doi.org/10.3390/pr9101688
APA StyleNath, A., Csighy, A., Eren, B. A., Tjandra Nugraha, D., Pásztorné-Huszár, K., Tóth, A., Takács, K., Szerdahelyi, E., Kiskó, G., Kovács, Z., Koris, A., & Vatai, G. (2021). Bioactive Peptides from Liquid Milk Protein Concentrate by Sequential Tryptic and Microbial Hydrolysis. Processes, 9(10), 1688. https://doi.org/10.3390/pr9101688








