1. Introduction
It is estimated that every year, at least two million women are diagnosed with breast or cervical cancer [
1]. Cervical cancer is the fourth most common women’s cancer worldwide, both in incidence and mortality, while it is the most common cancer in 38 countries [
1]. Global inequities (both geographical and socio-economical) in cervical cancer incidence and mortality have long been observed [
2], and they persist to this day. Consequently, women’s cancers are still a major challenge for global healthcare, especially in low and middle-income countries (LMIC) where approximately 90% of deaths from cervical cancer occur, according to the World Health Organization (WHO). In [
1], it was estimated that approximately 85% of women diagnosed and 88% of women who die from cervical cancer live in an LMIC. For example, a large proportion of cervical cancers in Ethiopia are diagnosed at an advanced stage [
3].
For 2020, 1,806,590 new cancer cases and 606,520 cancer deaths are projected to occur in the United States alone [
4]. The three most common cancers for women in the United States are breast, lung, and colorectal. The cancer death rate in the United States rose until 1991. It then fell continuously through 2017 [
4] for all common cancer types, excluding uterine cervix and uterine corpus cancers. These stagnant rates for these two types of women’s cancers can be attributed to the lack of major treatment advances [
5,
6]. This critical aspect, together with the fact that cervical cancer is a largely preventable disease with a known causative agent (i.e., human papillomavirus (HPV) with several major oncogenic subtypes) [
7] call for the development of the early reliable detection of cervical precancerous lesions and cancer and for age-appropriate screening strategies [
8].
WHO indicates that global mortality from cervical cancer can be reduced by actively applying a number of measures, including prevention, vaccination, early diagnosis, effective screening, and treatment. The screening tests to detect precancerous lesions include HPV DNA testing, cytology (Papanicolaou (Pap) testing), and visual inspection of the cervix. HPV testing is more frequently used as the primary cervical cancer screening test [
9], while cytology-based screening is expensive and requires complicated training [
10]. The Bethesda system (which was originally proposed in 1988 [
11] and further revised in 1991, 2001, and 2014) is the most commonly accepted nomenclature for Pap smears [
7].
Abnormal cells can be found after a routine Pap smear. CIN (cervical intraepithelial neoplasia, also called cervical dysplasia) is a precancerous lesion characterized by abnormal growth of the cells on the surface of the cervix. According to how much epithelial tissue is affected, CIN may be characterized [
12] as:
CIN1 (low-grade neoplasia), which is about one-third of the thickness of the epithelium.
CIN2, which is one-third to two-thirds of the thickness of the epithelium.
CIN3, which affects more than two-thirds of the thickness of the epithelium.
One of the goals of clinical diagnosis is to discern between normal/CIN1, and CIN2/CIN3 (or CIN2+). In [
13], the authors proposed to change the terminology for HPV-associated squamous lesions of the anogenital tract to LSIL (low-grade squamous intraepithelial lesion) or HSIL (high-grade squamous intraepithelial lesion), as follows:
CIN1 is referred to as LSIL.
CIN2 (p16 negative, which is a high-risk HPV marker) is referred to as LSIL.
CIN2 (p16 positive) is referred to as HSIL.
CIN3 is referred to as HSIL.
The visual inspections of the cervix with acetic acid (VIA) and Lugol’s iodine (VILI) are an alternative to cytology, especially in LMIC. VIA is a visual inspection of the cervix after the application of 3–5% acetic acid [
10]. Following the application, a precancerous lesion becomes white (acetowhite). However, an acetowhite area may also be a benign lesion, which needs to be excluded before considering a VIA-positive lesion as precancerous or cancer. VILI follows VIA and the precancerous lesions (tissues with low glycogen concentration), and cancer turn yellow after the application of Lugol’s iodine. Benign lesions need to be excluded, such as in the case of VIA. Colposcopy involves the examination of the cervix and surrounding areas under a microscope (colposcope), which is the most common procedure for visualization of the cervix before and after the application of acetic acid.
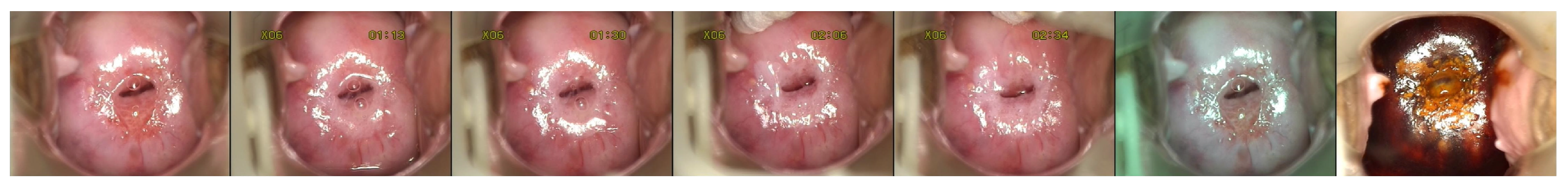
The present study applies the deep learning methodology to a cervical image dataset of 477 colposcopy cases (each case is composed of five pictures after the VIA procedure, one taken using a green lens and one after VILI) and proposes a precancerous lesion diagnosis method that is based on an ensemble of MobileNetV2 networks, which represents the first use of these networks for the automated classification of precancerous lesions. After a section dedicated to the evolution and advances in the computer-aided diagnosis of colposcopy images, this paper continues with details on the actual implementation of the proposed method and the obtained results. It ends with a section on future possible developments.
2. Related Work
In the early years of computer-aided diagnosis applied in medical imaging, there were presented several studies on the use of traditional image analysis techniques for cervical cancer diagnosis. A statistical texture analysis approach focusing on six different vascular patterns that relate to cervical lesions was shown to demonstrate a classification accuracy of over 95% [
14]. A multispectral imaging system for the in vivo detection of cervical precancer and cancer, after the application of acetic acid (to enhance the reflectance differences between normal and pathologic epithelium), was developed and presented in [
15] where improved sensitivity and specificity was demonstrated. ColpoCAD (a Computer-Aided-Diagnosis (CAD) system for colposcopy) was introduced in [
16] as a multi-sensor, multi-data, and multi-feature colposcopy images analysis tool. For example, ColpoCAD features algorithms to assess the smoothness of a lesion margin and to detect mosaic and punctuation vessel patterns. Segmentation of macro-regions in colposcopy images was at the basis of a unified model-based image analysis framework reported in [
17]. Again, the classification of vascular abnormalities was modeled as a problem of texture classification, and the major contribution of that paper was to address the detection of all vascular abnormalities in a unified framework.
In the age of data-driven machine learning techniques, biomedical imaging and analysis have been attracting a largely increasing interest from the scientific community. The application areas are very diverse, and they target various organs. For example, the use of 3D convolutional neural networks (CNNs) to discriminate between primary and metastatic liver tumors from MRI data was proposed and evaluated in [
18]. In [
19]; a deep CNN-based method and transfer learning were proposed for breast MRI tumor classification; and in [
20], transfer learning was used for the diagnosis of lung diseases, including cancer and tuberculosis. Recently, an entire issue of the Proceedings of the IEEE was dedicated to diverse computational strategies based on deep learning for the analysis of medical images [
21]. In [
22], a survey on using deep learning techniques (mainly CNNs) for medical images analysis was given and presented over 300 contributions to the field (mainly in image classification, but also in object detection, segmentation, and registration). Cervical dysplasia diagnosis was included in the application areas.
Multimodal deep learning for cervical dysplasia diagnosis was presented in [
23] where the results of using a CNN indicated 87.83% sensitivity at 90% specificity on a large dataset. Another CNN-based method for localized classification of uterine cervical cancer histology images was presented in [
24], where the accuracy of 77.25% was reported.
In [
25], from a total of 485 images in the dataset (from 157 patients), a total of 233 were captured with a green filter, and the remaining 252 were captured without a green filter. Of the total number of images, 142 images were of severe dysplasia, 257 of CIS (carcinoma in situ), and 86 of invasive cancer. The training was performed using Keras and TensorFlow. The accuracy obtained was approximately 50%.
In [
26], images from 330 patients who underwent colposcopy were analyzed (97 patients were diagnosed with LSIL and 213 with HSIL). A CNN with 11 layers was used, and the accuracy and sensitivity for diagnosing HSIL were 0.823 and 0.797, respectively.
A mobile phone application to be used in low-income settings, dedicated to the preliminary analysis of the digital images of the cervix (after VIA), was presented in [
27].
Cell morphology was combined with cell image appearance for the classification of cervical cells in Pap smear in a CNN-based approach in [
28]. The dataset used was the Herlev benchmark Pap smear on which four CNN models (AlexNet, GoogLeNet, ResNet, and DenseNet) pretrained on the ImageNet dataset were fine-tuned. For the two-class and the seven-class classification tasks, GoogLeNet obtained the best accuracies, 94.5% and 64.5%, respectively.
The first study to use transfer learning with the DenseNet model (ImageNet and Kaggle) for the classification of colposcopy images was presented in [
29] where the accuracy of 73.08% in 600 test images was reported.
Other machine learning techniques have been used for cervical disease diagnosis as well. In [
30], several cervigram images from the U.S. National Cancer Institute were collected and further enhanced to evaluate image-based cervical disease classification algorithms. Further, to differentiate between high-risk and low-risk patients, seven classic machine learning algorithms (random forest (RF), gradient boosting decision tree (GBDT), AdaBoost, support vector machines (SVM), logistic regression (LR), multilayer perceptron (MLP), and k-nearest neighbors (kNN)) were trained and tested on the same datasets on a ten-round ten-fold cross-validation. The RF algorithm was shown to yield the best results with accuracy, sensitivity, and specificity of 80%, 84.06%, and 75.94%, respectively. RF was also used in [
31] where image segmentation for cervigrams was the first step of the approach. The method’s next steps were the extraction of color and texture features for the interpretation of uterine cervix images. Next, the Boruta algorithm was applied for feature selection. The final step involved the use of RF.
The present paper is the first to propose the use of MobileNetV2 [
32] networks for the automatic classification of colposcopy images. We used a diverse input composed of three different types of images: from VIA, from a green lens, and from VILI. Another contribution of this paper is the modeling of the ensemble, which is built from several MobileNetV2 sub-networks and an inverted residual unit (a stack of convolutional layers specific to the MobileNetV2 architecture). The sub-networks transform the different images into three-dimensional embeddings that capture the discriminative features for the problem of CIN classification. The inverted residual unit processes the embeddings from multiple images in order to find distinctive patterns. Apart from the network modeling, we directed our research towards handling class imbalance, which is common in this domain, and manipulated the loss function, to be more robust to this phenomenon. Last, to better understand the functioning of the network, we visualized its activations and identified problem areas.
5. Discussion
In deep learning, there is an infinite set of variations to be chosen from when it comes to network structure, parameters, and training procedures. For this reason, you might hear from some people that deep learning is like “art”; this expresses the freedom and lack of constraints in designing a solution. We approached this problem with an incremental strategy similar to gradient descent. It was impossible to find the best solution; therefore, we searched for a setup that worked well among a predefined set of experiments, then we explored additional aspects while keeping the tested ones fixed. For choosing the correct set of experiments at each step, the researcher relies on intuition and prior knowledge. In this section, we describe the journey we took for arriving at the final solution and highlight the most important ideas.
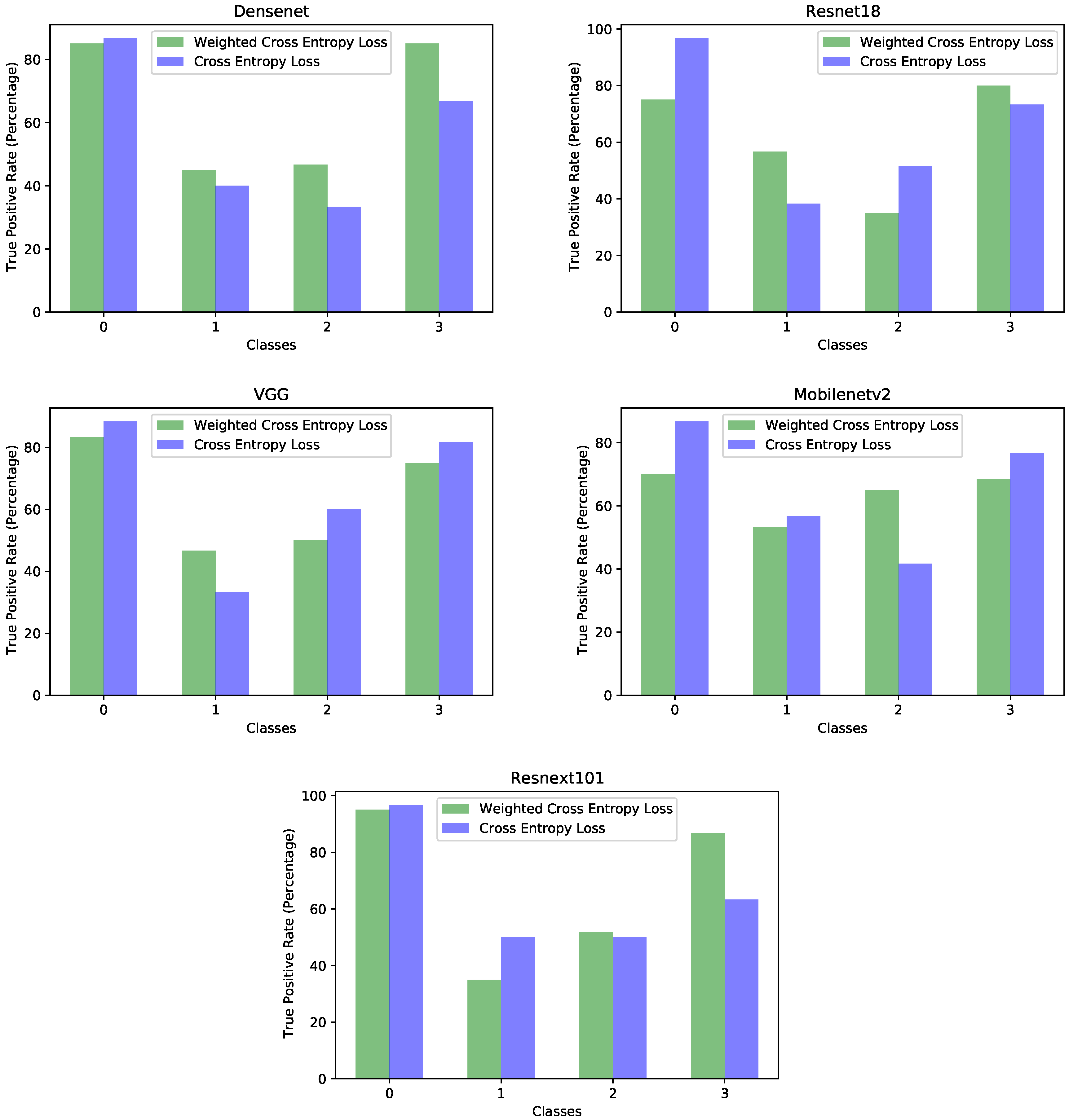
We started with a very simplistic approach, which used a single neural network pretrained on ImageNet [
40]. As was noted in the results displayed in
Table 5, despite the networks being pretrained, the accuracies were low, most models reporting approximately 64% on the test set. Upon analysis of the results, we concluded that the main reason for the low accuracy was overfitting; thus, we chose a model having a lower capacity, MobileNetV2 [
32], and applied to it as much regularization as possible. The most important and less intuitive modification was to change the size, in terms of the number of feature maps (channels), of the last convolutional layer. This modification constrained the capacity of the network while preserving the already learned structure from the ImageNet pretraining. Furthermore, as explained in
Section 3.2.2, the last convolutional layer of the MobileNetV2 acts as an adapter, transforming the internal representation of the network to the output representation; thus, it makes sense to change this adapter to better fit the required task. Another regularization technique was the two dimensional dropout layer inserted at the end of the convolution. As you can see, even though the MobileNetV2 network had a smaller capacity compared to other deeper networks, it still needed additional regularization to be effective on this task. If the dataset were larger and more diverse, we could decrease the regularization and take advantage of the full capacity of the network or use more complex models such as the ResNext101 [
44] model, which provided the highest capacity and was pretrained on the greatest amount of images [
44]. Therefore, the aspects related to regularization could change from one scenario to another, but it is useful to have the knowledge or methodology to better regularize a model, depending on the task. This paper presented such an example. Moreover, datasets on specific medical topics can be much more scarce and reduced in volume compared to others. For this reason, it is important to be able to adapt the existing models to specific tasks that are constrained by a lower volume of data. Our tuning of the MobileNetV2 network resulted in an accuracy of approximately 71%.
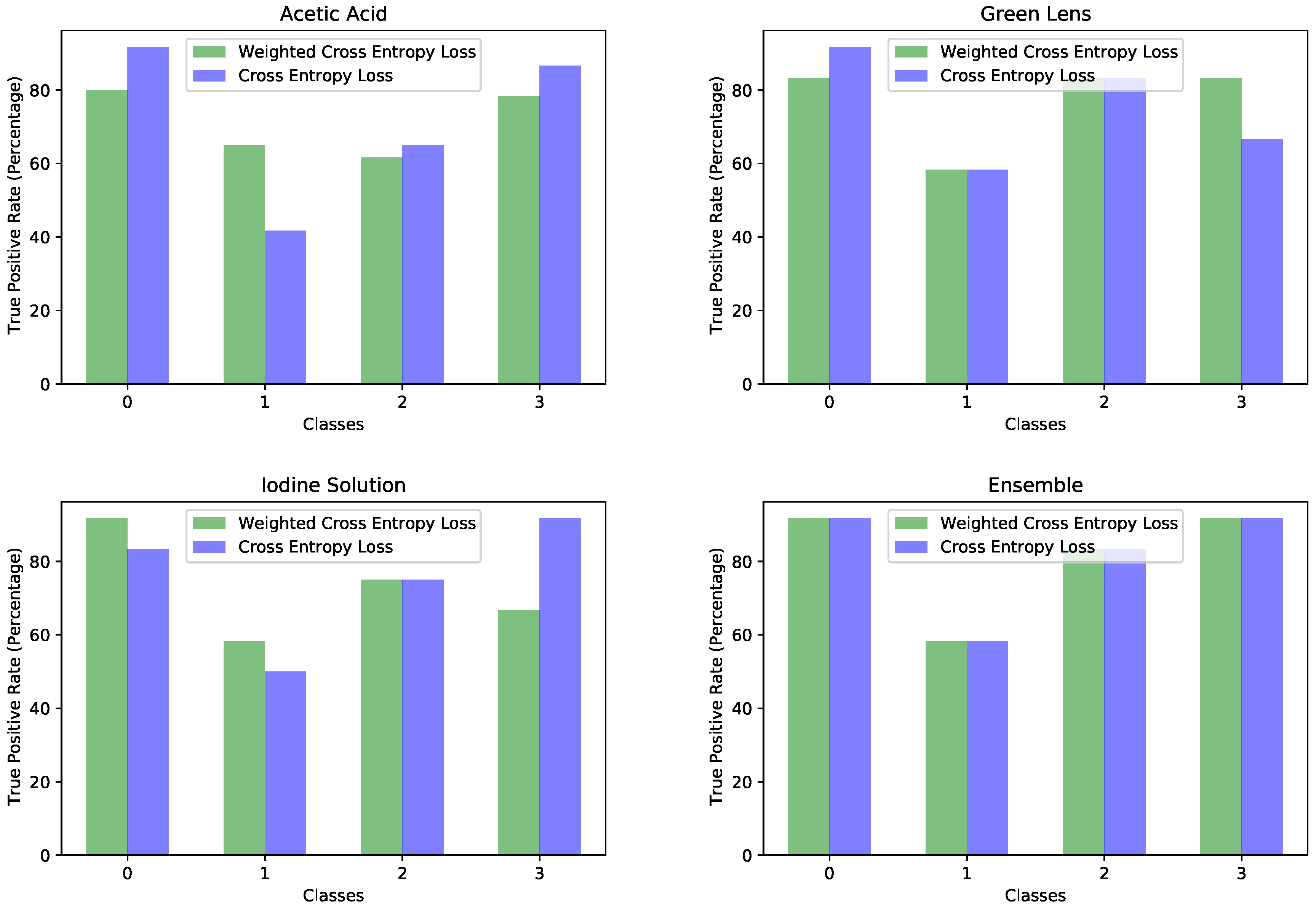
Once we decided on the MobileNetV2 network using the experiments made on the images having the acetic acid solution, we extended the solution to the rest of the dataset input, the images through the green lens and with the iodine solution. On those types of images, we reported accuracies of 75% for both image types. The increase in accuracy was most probably due to the distinctive features of these types of images. The green lens helped in creating a stronger contrast against the red color of the cervical lesions. Upon the application of the iodine solution, the abnormal areas changed color, becoming yellow or orange, which made a strong contrast with the dark red color of the iodine solution. These types of images were easier to visualize, even for humans.
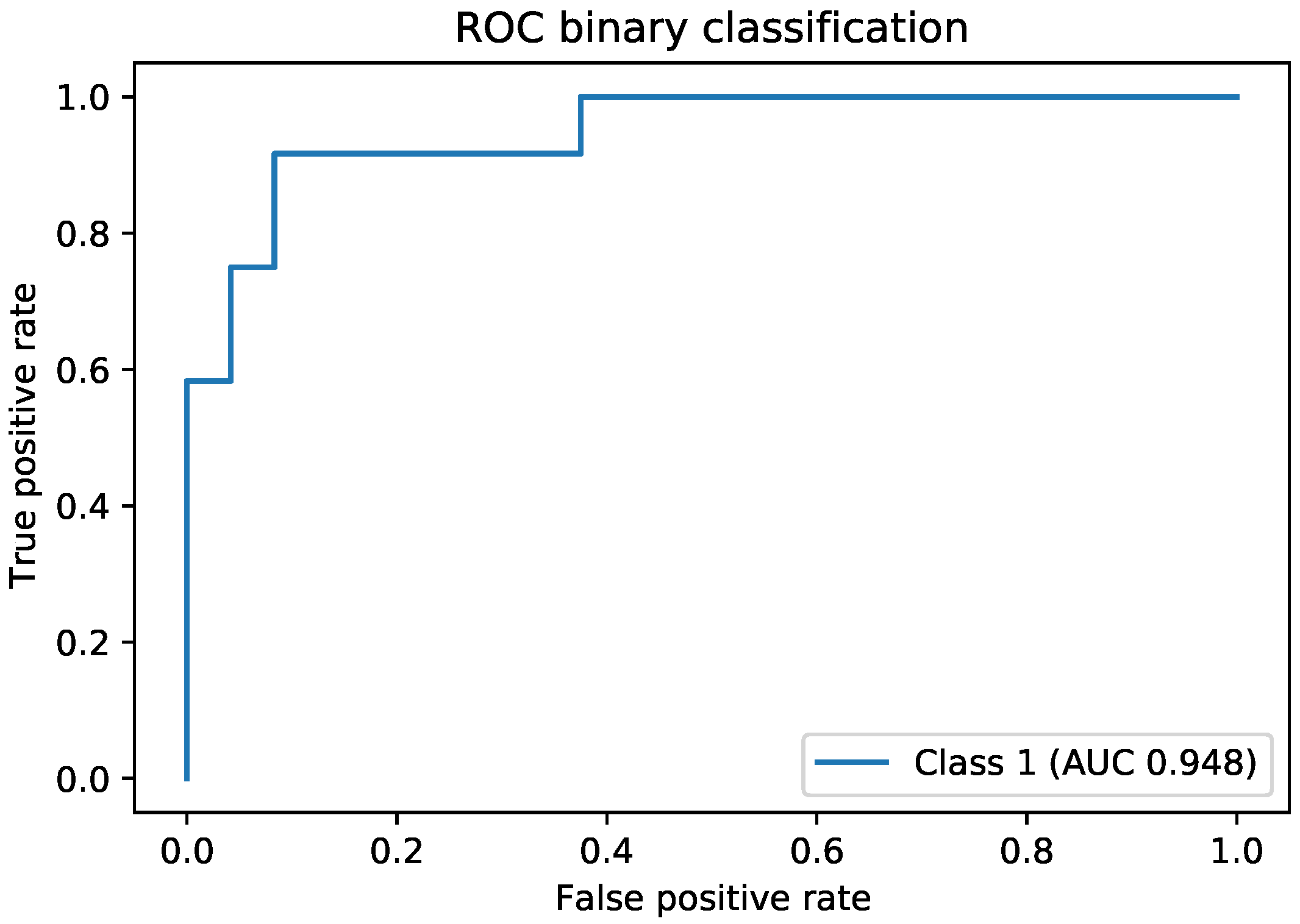
Up to this point, we explored all the types of images available; however, we did not take advantage of the existing relations between them and of the temporal sequence displayed by the first five images with the acetic acid solution. This was achieved using the ensemble, which provided the best accuracy reported in this paper of 83% for the four-class problem and 91% for binary classification. A very important aspect to note about the design of the ensemble was the use of representation learning. In the ensemble, we did not use the final output of the MobileNetV2 networks; instead, we used an internal network representation that preserved the spatial information and the relevant features for this classification task. We chose as image representation the output of the last convolutional layer from the MobileNetV2 network. This was a tensor of size
for acetic acid images and iodine solution images and of size
for green lens images. Thus, when constraining the size of the last convolutional layer in the MobileNetV2 layer, we also had in mind the computation of image representations. If the size of the representation was too large, the network might overfit instead of capturing the generic and useful features. The representations of the images were not learned by the ensemble; they were learned in the previous steps when training the single networks. We tried fine-tuning the ensemble as a whole so that the ensemble could adjust the computation of the representations, but the accuracy was lower, probably due to overfitting issues. For analyzing and relating the seven images together, we used an additional convolutional network made out of a single inverted residual unit, whose structure is described in
Table 4. An alternative to this is to use a recurrent neural network; this can be the subject of future research. In our case, we chose the convolutional network over the recurrent version because it does not pose the vanishing and exploding gradient problem and because the sizes of the image representations and the sequence length were both small and made the training through a convolutional network feasible.
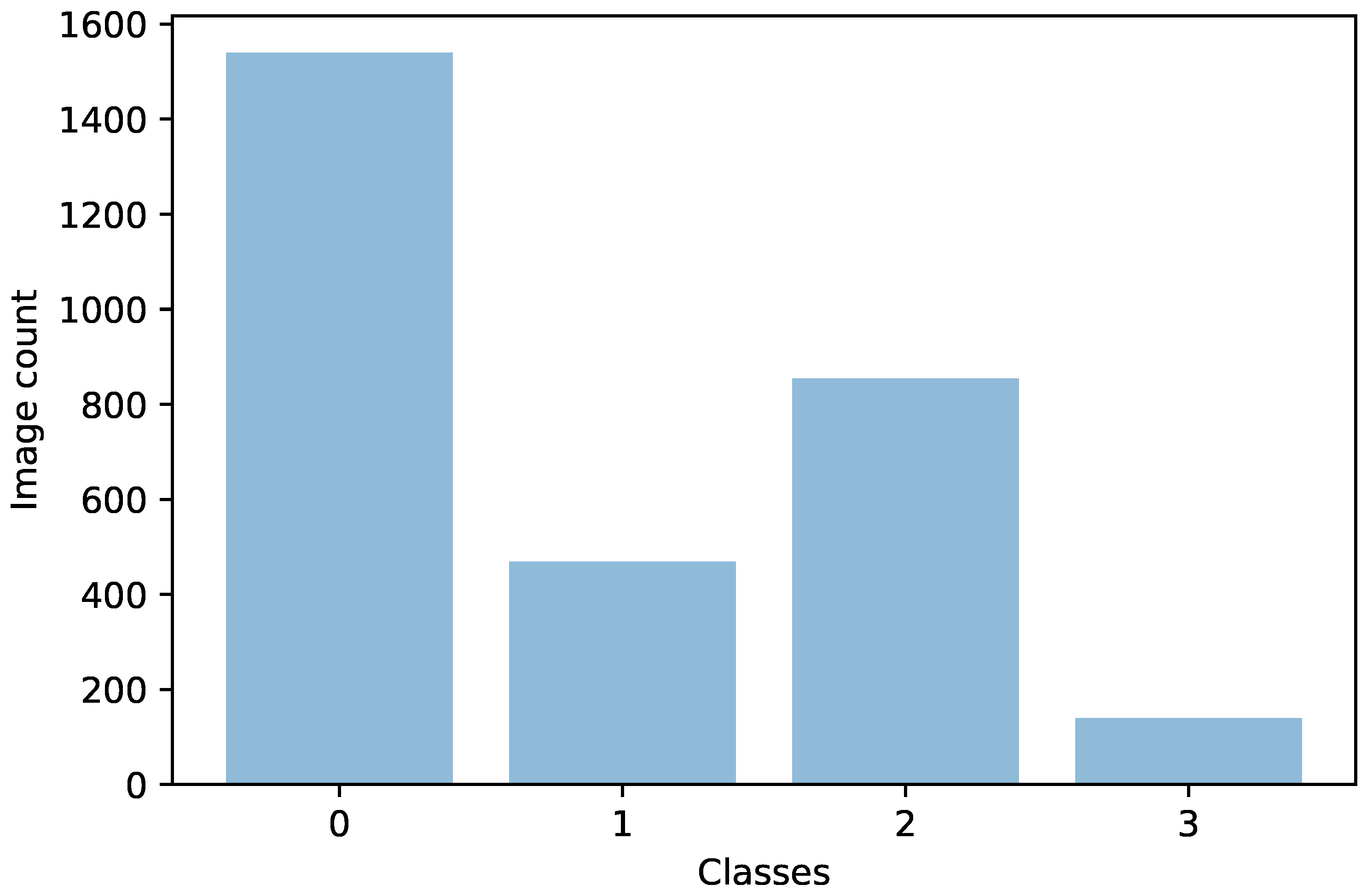
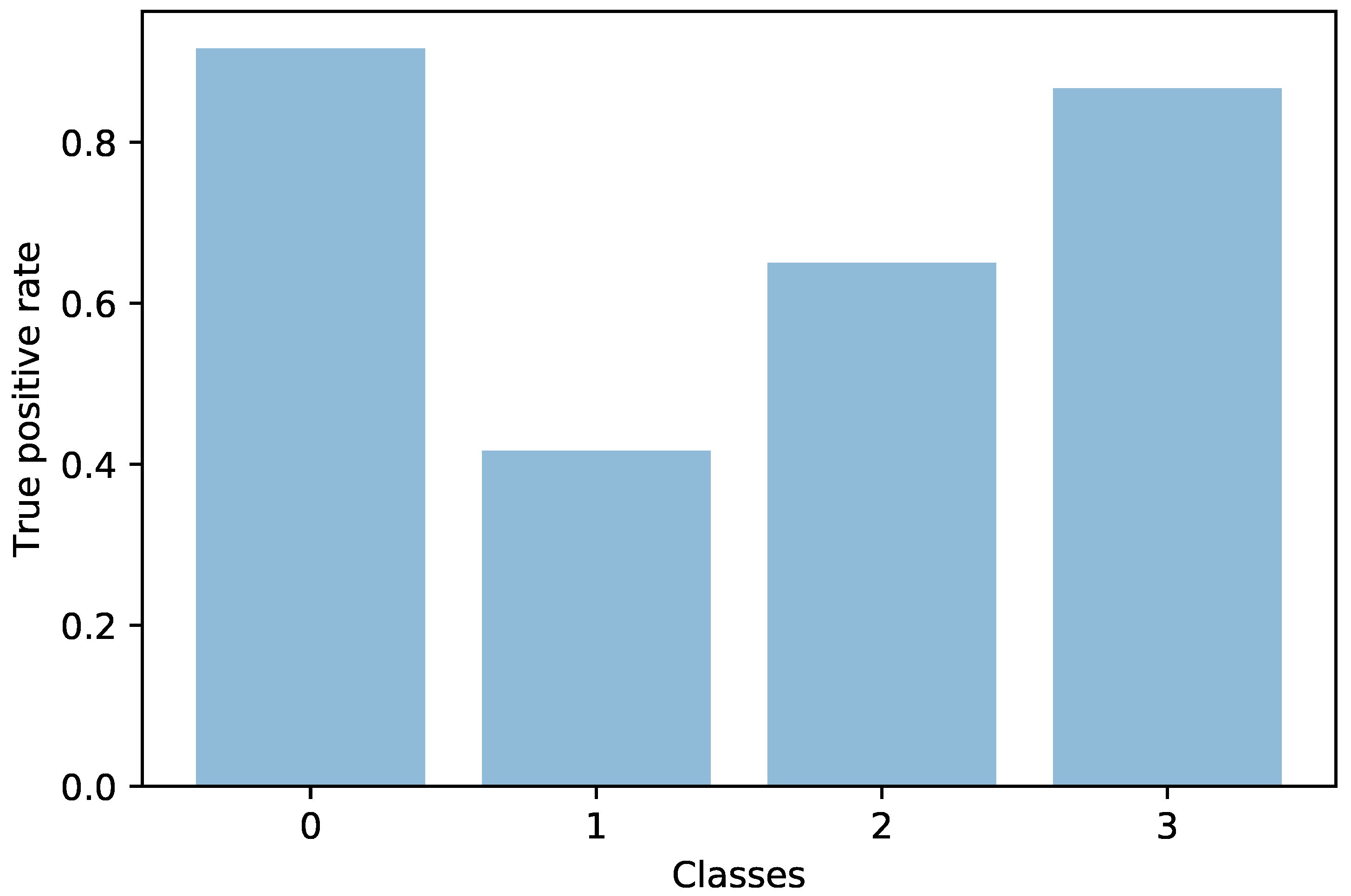
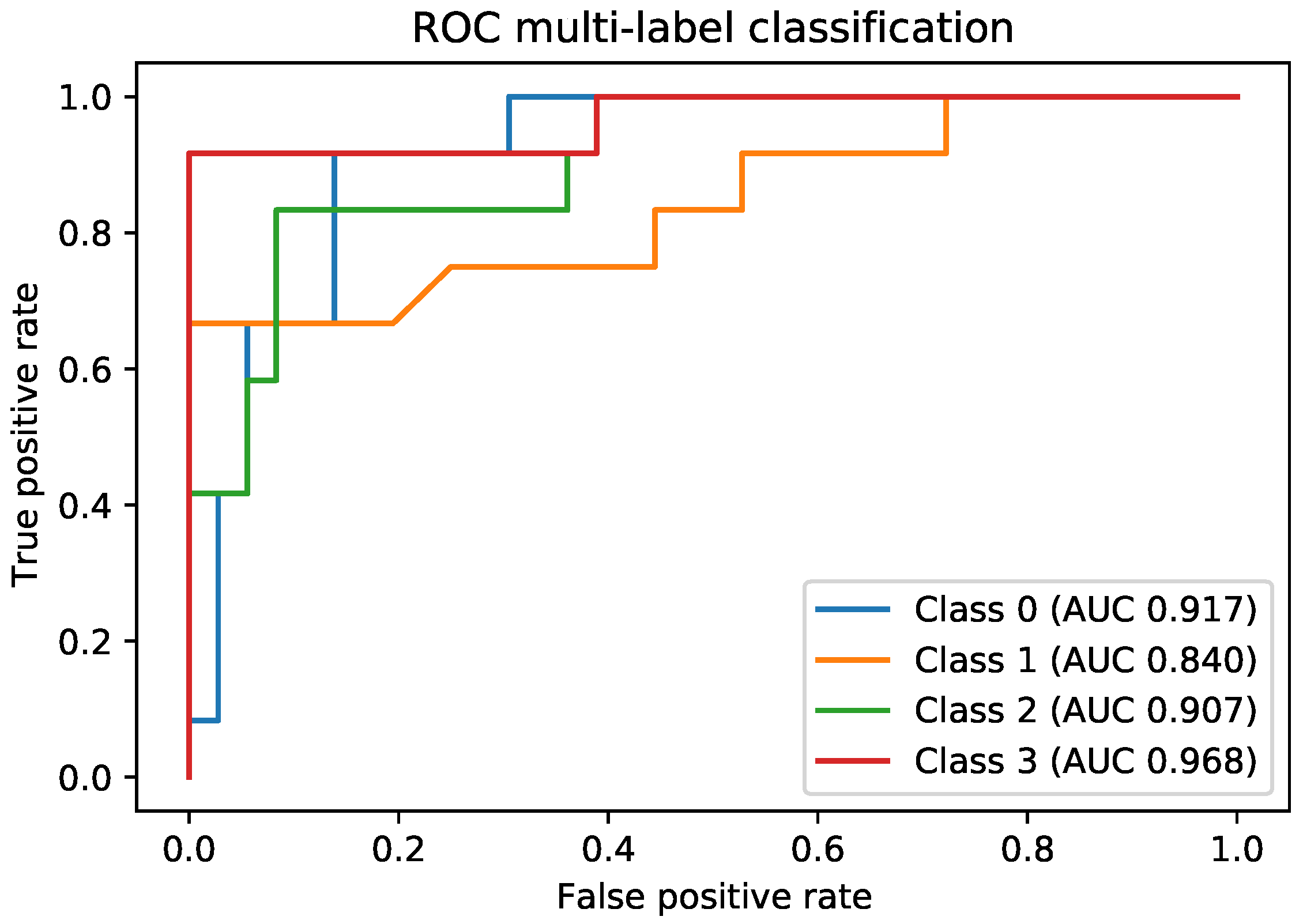
Another direction of our study was the handling of the dataset imbalance in the context of the current problem. The first step was to analyze the already existing model (trained with classic cross entropy) to assess the effects of this phenomenon. We used the true positive rate recorded per each class as a measure of classification bias. In an ideal scenario, all classes should present a 100% true positive detection rate; however, in practice, the classes having a higher number of samples performed much better than those with a reduced number of samples. This can be noticed by viewing
Figure 3 and
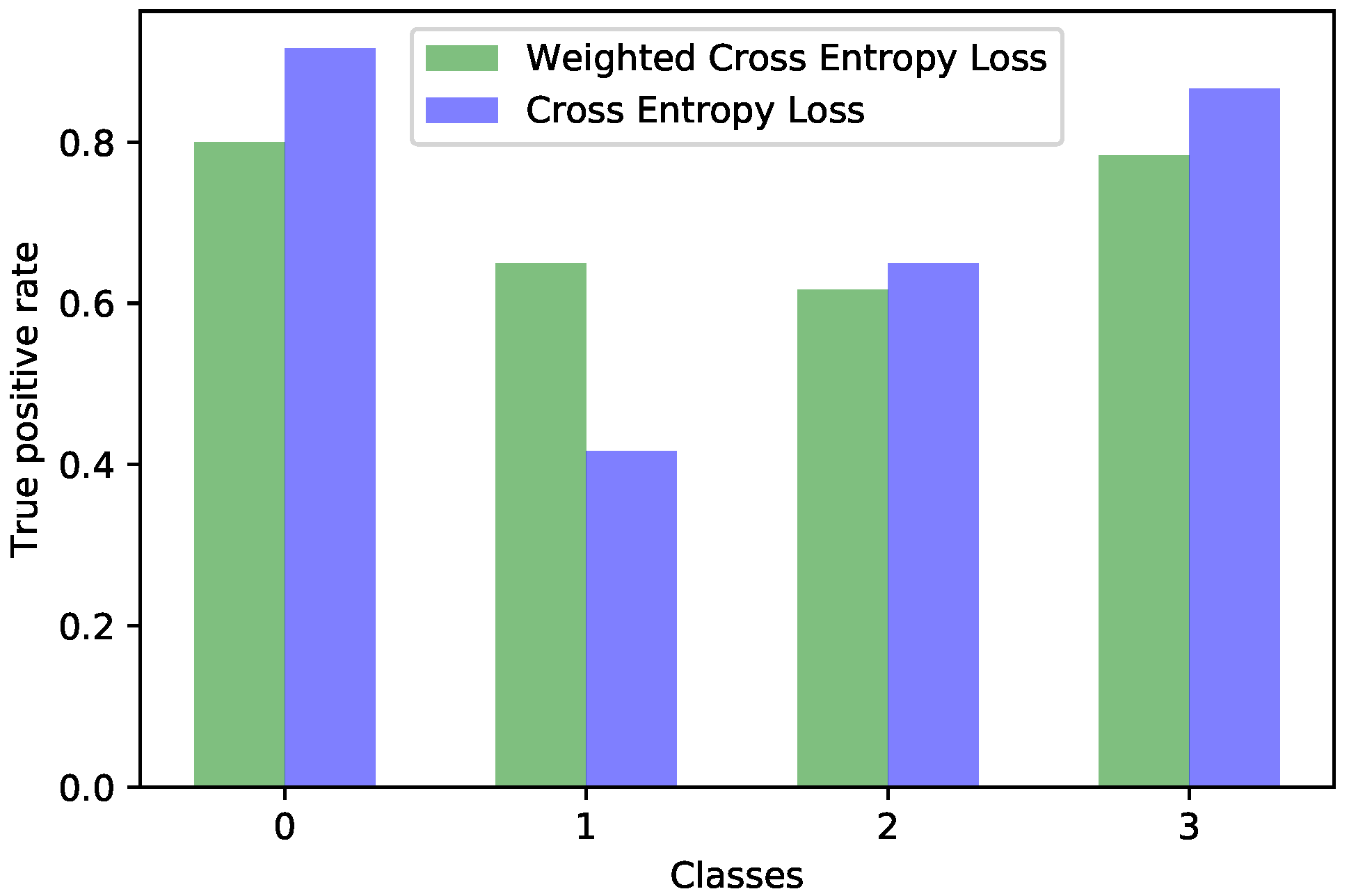
Figure 4 in parallel. It also meant that the model could perform better if presented with more data. We should also know that sometimes a higher true positive rate for one class could be attributed to biased learning, and while the model tended to better recognize one class, the rest of the classes would have a lower detection rate; this is especially true for classes that are harder to discriminate. Through our experiments, we tried to reduce this classification bias; ideally, the model should classify samples as if there would be an even distribution between the classes. Apart from the cross entropy loss, we experimented with another two standard losses for imbalanced datasets, the weighted cross entropy and focal loss [
41]. For the current problem, we obtained the best results with the weighted cross entropy having a dynamic weight calculation based on the training time reported errors per each epoch. The idea of the training algorithm was to work constantly towards recognizing the harder classes. This method levels the classification bias among the classes; however, it does not guarantee keeping the overall accuracy the same. From our experiments, the overall accuracy stayed approximately the same for the acetic acid solution images, increased for the green lens, because it worked on the problem areas, while for the iodine solution, the overall accuracy dropped. Looking at the true positive rate for each class provided a transparent and accurate view on the behavior of the model; nevertheless, comparing the performance of multiple models this way is hard. Which criteria should be used? Are some classes more important than others? What if a model performs very well on most of the classes, but very poorly on a certain one? For our task, we tried to encourage a model that had a good overall accuracy while distributing its classification bias as evenly as possible. Being inspired by the F1 measure score, which computes the harmonic mean between precision and recall, we computed the harmonic mean of the true positive rates of the classes. The harmonic mean penalizes the smaller values more then the arithmetic mean; thus, imbalanced models would get penalized more, but the overall accuracy is also taken into account. We used this measure for evaluating the effectiveness of the tested strategies. After performing these experiments on each type of image separately, we applied them on the ensemble by training each of the composing models with the established strategy and then the ensemble network as well. Unfortunately, this method did not yield any improvement for the ensemble; the true positive rate per each class was the same as those reported by simple training through cross entropy. The most probable explanation is that the ensemble network already made the best out of the available data, and any training variation around the same model was not able to bring additional advantages. Even though the ensemble could not be improved by these methods, we consider that the results obtained on the single networks and the analysis of the model behavior concerning the class imbalance are important pieces of information. Additionally, it is necessary to raise the awareness of the practitioners about these problems and about the traps of measuring the performance of a model in the context of an imbalanced dataset. Most of the time, the practitioner might not even realize that the dataset being used is imbalanced; an investigation to find this needs to be done.
Unfortunately, the method used (dynamically weighted cross entropy based on training time errors) has some disadvantages as well. The most notable one, which we also emphasize in
Section 4.4, is that it does not always work. In certain cases, it even makes the training diverge. However, with proper parameter selection, we expect that it provides good results most of the time. Another notable problem is that the parameter selection could be a complicated process, which requires more experiments. To make matters worse, the training time increases greatly for each experiment, because a smaller learning rate and slow progression across more epochs are required for this method to be effective. In this paper, we employed a transformation to the errors
, which was specific to this problem; for other cases, the researchers would need to find the appropriate transformation through experimentation. This is both an advantage since it allows for greater flexibility, but can also be seen as a disadvantage, as it is not generic enough. Nevertheless, in our opinion, this method is worth being taken into account when dealing with imbalanced datasets.
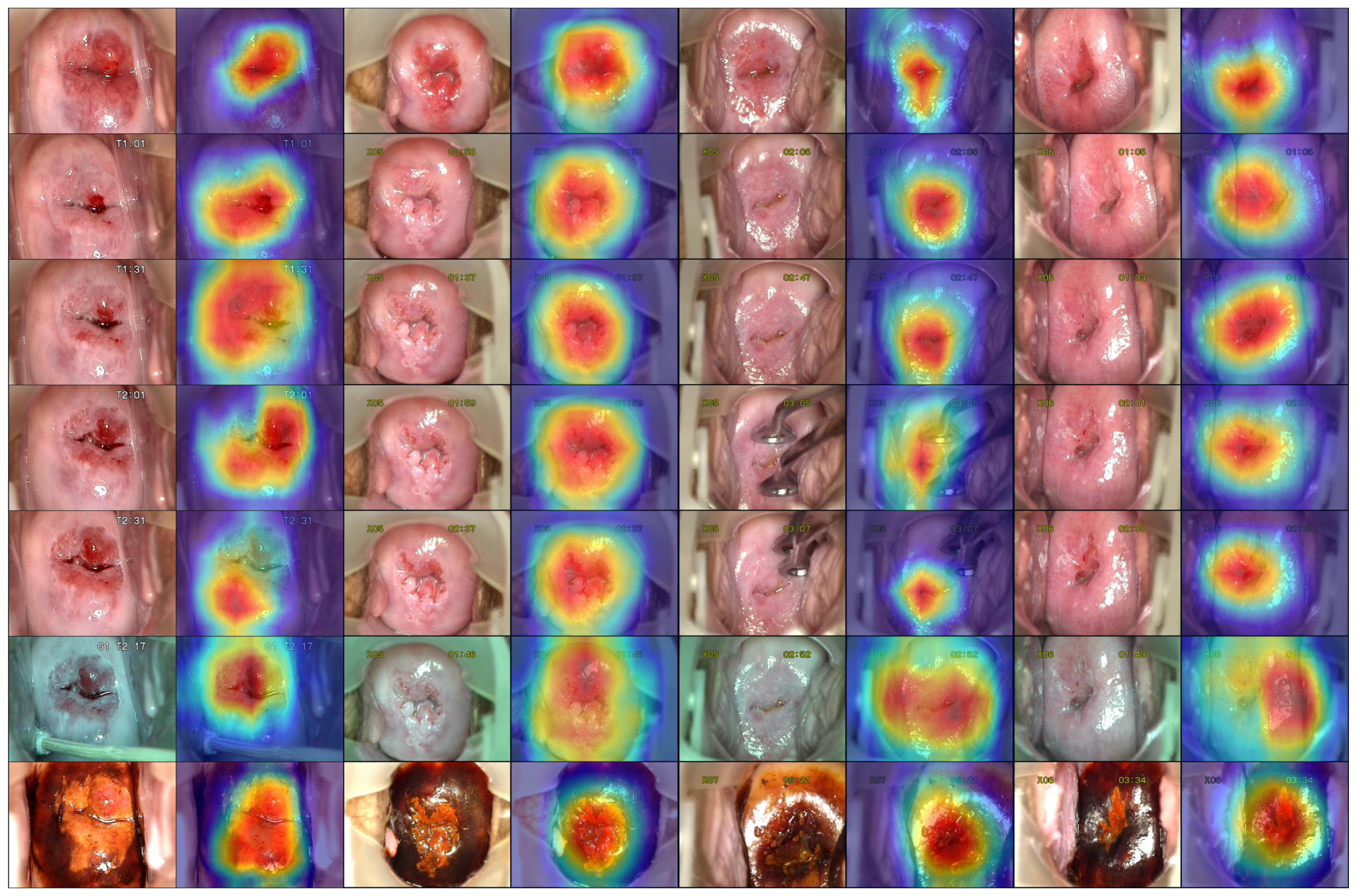
The last experiment that we did was for visualizing the network activations. Most often, the models are evaluated by several metrics that describe their performance from different perspectives. However, the network visualization, even though less popular, could be used as a complementary means of network evaluation and understanding. In this case, there are no numbers to quantify the performance. Still, by observing the activations and reasoning about them, we can draw conclusions about the network. One way is to hunt for overfitting factors, for example if we notice that the network is paying attention to image features that should not matter for the classification problem. In our case, the pictures contained certain noise, such as text displayed by the camera and medical devices. At first, we were worried especially about the text displayed by the camera. We considered hiding that text or resizing the pictures in such a way that it disappeared. Fortunately, the network automatically learned that it was not a relevant feature for our problem. We found out about this through the network activations’ visualization. On the other hand, we learned that the model was sometimes distracted by the vaginal walls and the strong light reflections, a phenomenon also described in the paper [
47].
6. Conclusions
We proposed a method for analyzing multiple colposcopy images at once and achieved 83% and 91% accuracies in the four-class and binary classification problems. In this paper, we did not design a network architecture that solved a generic problem; instead, we applied the recent technological advances in deep learning to a specific problem and dataset. For arriving at the solutions described, we performed more than 100 experiments, each consisting of training a network. To keep the article readable and avoid getting lost in less important details, we documented only the relevant findings. In our opinion, one of the hardest obstacles encountered was overfitting. It was overcome through very careful design and parameter tuning. While arriving at our solution, we brought the following innovations:
The comparison of multiple convolutional network models on the current dataset.
The use of the MobileNetV2 architecture for colposcopy image understanding and the tuning of this architecture for a dataset with a reduced size. We would like to emphasize that this architecture should not be used only in scenarios where execution speed and memory consumption are constrained; it should also be an important candidate whenever there is a need for a light-weight solution, when overfitting is a major concern, and when fast experimentation is required.
The design of a convolutional ensemble composed of multiple MobileNetV2 networks for taking advantage of different kinds of images available in a colposcopy procedure. The use of representational learning was an important aspect of the ensemble design and training.
The techniques used for handling the dataset imbalance cannot be considered original; however, their application to this specific dataset and model were.
Apart from the design of the network, the current paper stresses the importance of properly measuring the performance from the perspective of dataset imbalance, an aspect that many times is neglected by researchers. Furthermore, we believe that the use of the network activations’ visualization could complement the usual network evaluation. This could help in targeting the problems of the model.
We will end this paper by highlighting the advantages, disadvantage, and further research directions. The current solution presented the following advantages:
Resistant to overfitting and well adapted to reduced datasets.
Can take advantage of different types of images at the same time.
Fast execution speed and low memory consumption. If this might not be of interest to the end-user, it is important in the experimentation and design phase, allowing performing more experiments and better parameter tuning given that both computational power and time are not unlimited resources.
Even with the current regularizations in place, the model promises further performance gains if presented with more data, as noted in
Section 3.4. Furthermore, if enough data is available, the regularizations can be relaxed to allow learning of more complex features.
Adaptable, in the sense that additional MobileNetV2 networks could be added or removed from the ensemble to fit a different setting.
The limitations of the proposed solution were:
Suboptimal if applied to a very large dataset.
Would not be able to take advantage of a large sequence of images because we used a convolutional network instead of a recurrent one for modeling the ensemble.
Does not take advantage of multiple image scales.
Does not provide hints to the user on the location of the cervical lesions in the image, which would have been a very useful feature.
In our opinion, the reported results were encouraging to further research in this direction; however, there is still considerable room for improvement, and applying the system to the real world is not feasible at this moment.
There are many possibilities for modeling the task of colposcopy image classification, and as variations, one could try to replace the ensemble inverted residual unit with a recurrent network, try out visual attention mechanisms, images at higher resolutions or at multiple scales, and aligning the images when processing more of them. Using unsupervised learning could provide another research direction in which extracted features are compared and clustered. Moreover, segmentation would be very useful for this system, because it would indicate the problematic tissues to the physician. Such features can be used as the subject of further research.
Colposcopy plays a very important role in the prevention of cervical cancer and it has the potential to save human lives. We believe that this procedure would greatly benefit from computer image processing and the deep learning technology. We hope that such practices will have a positive impact on medicine and will be deployed in many production systems soon.