Applying Differential Neural Networks to Characterize Microbial Interactions in an Ex Vivo Gastrointestinal Gut Simulator
Abstract
:1. Introduction
1.1. Microbial Interactions
1.2. Microbial Interactions in the Human Colon
1.3. Mathematical Models Describing Microbial Interactions
1.4. Differential Neural Networks for Model Approximation
1.5. The Objective of the Study
2. Results and Discussion
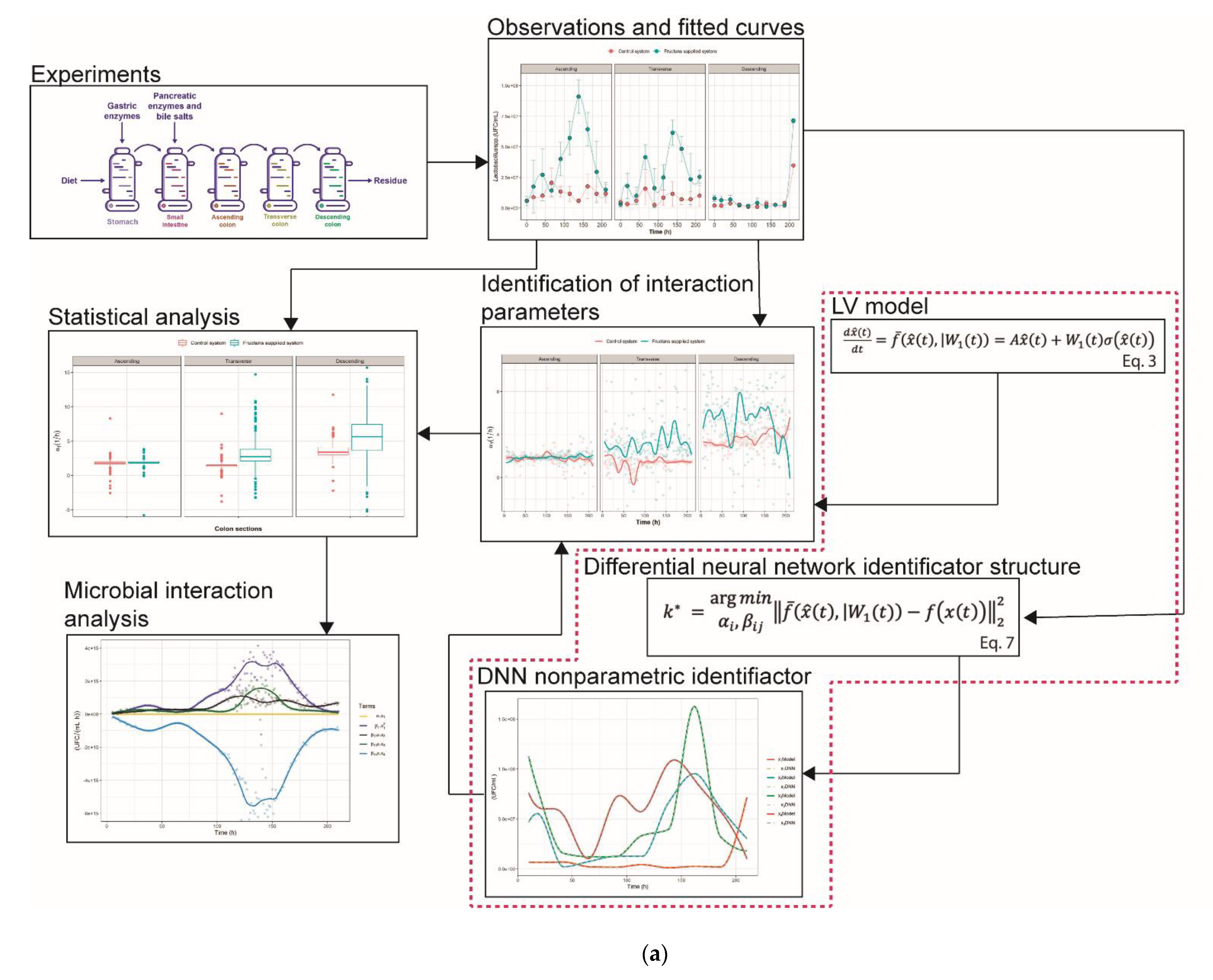
2.1. The Growth Kinetics of Four Microbial Populations
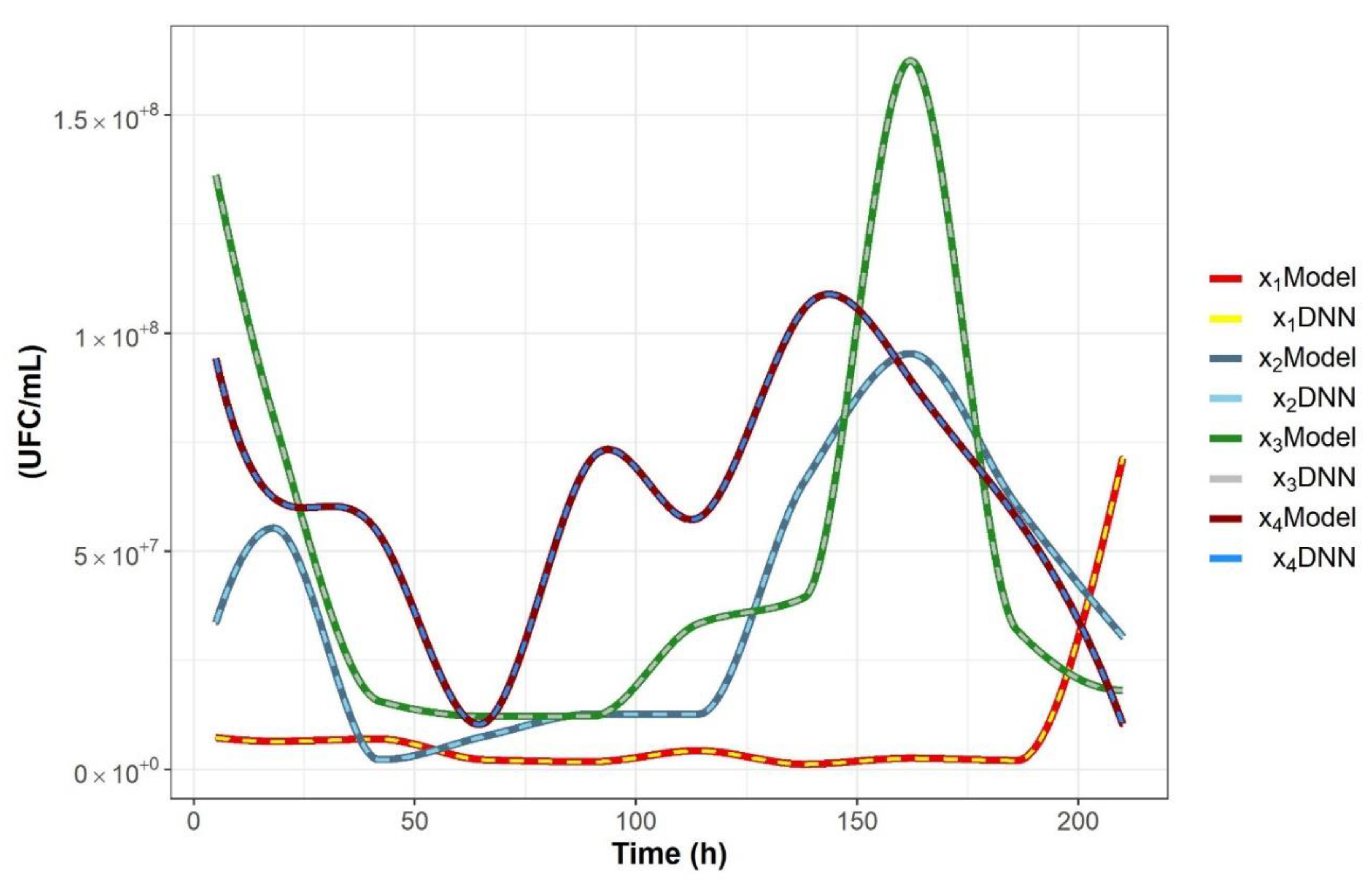
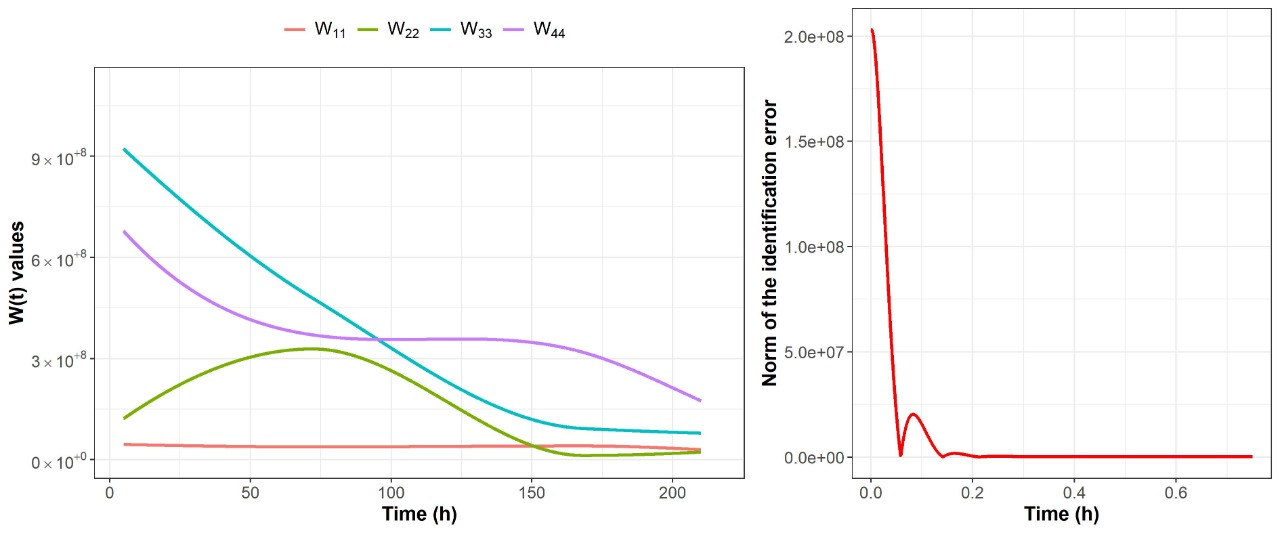
2.2. Nonparametric Identification of the Experimental Data by the DNN Identifier
2.3. Analysis of Microbial Interaction Parameters
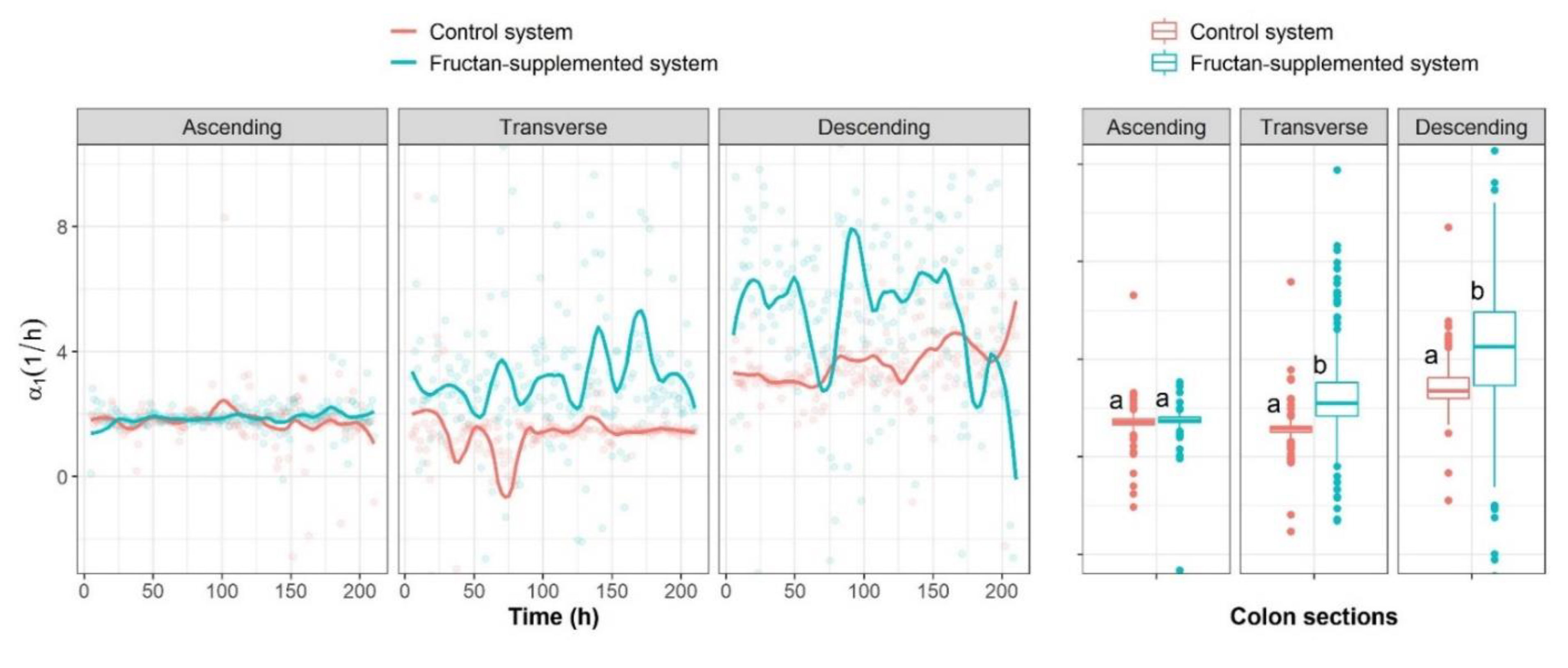
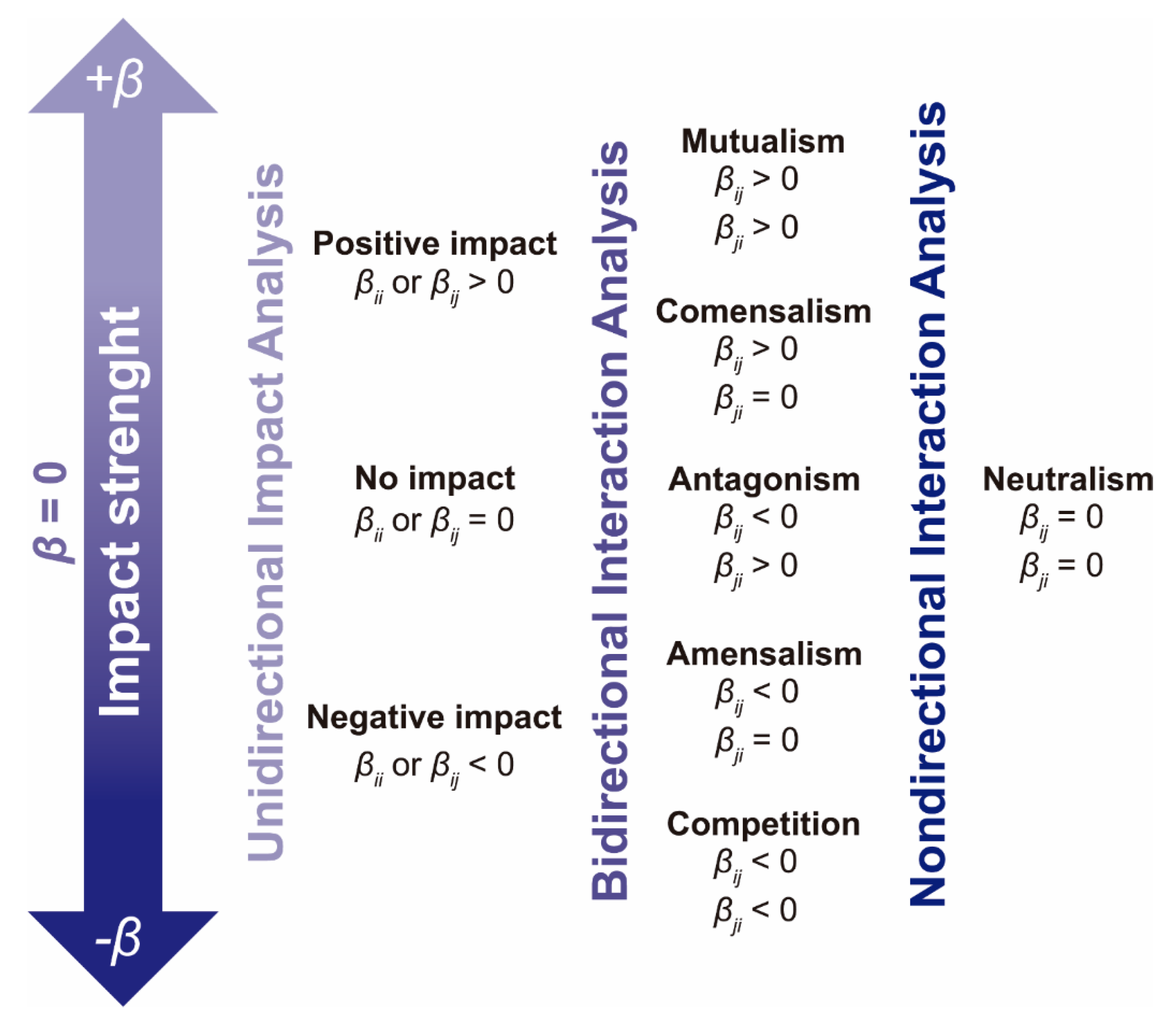
2.3.1. Unidirectional Impact Analysis
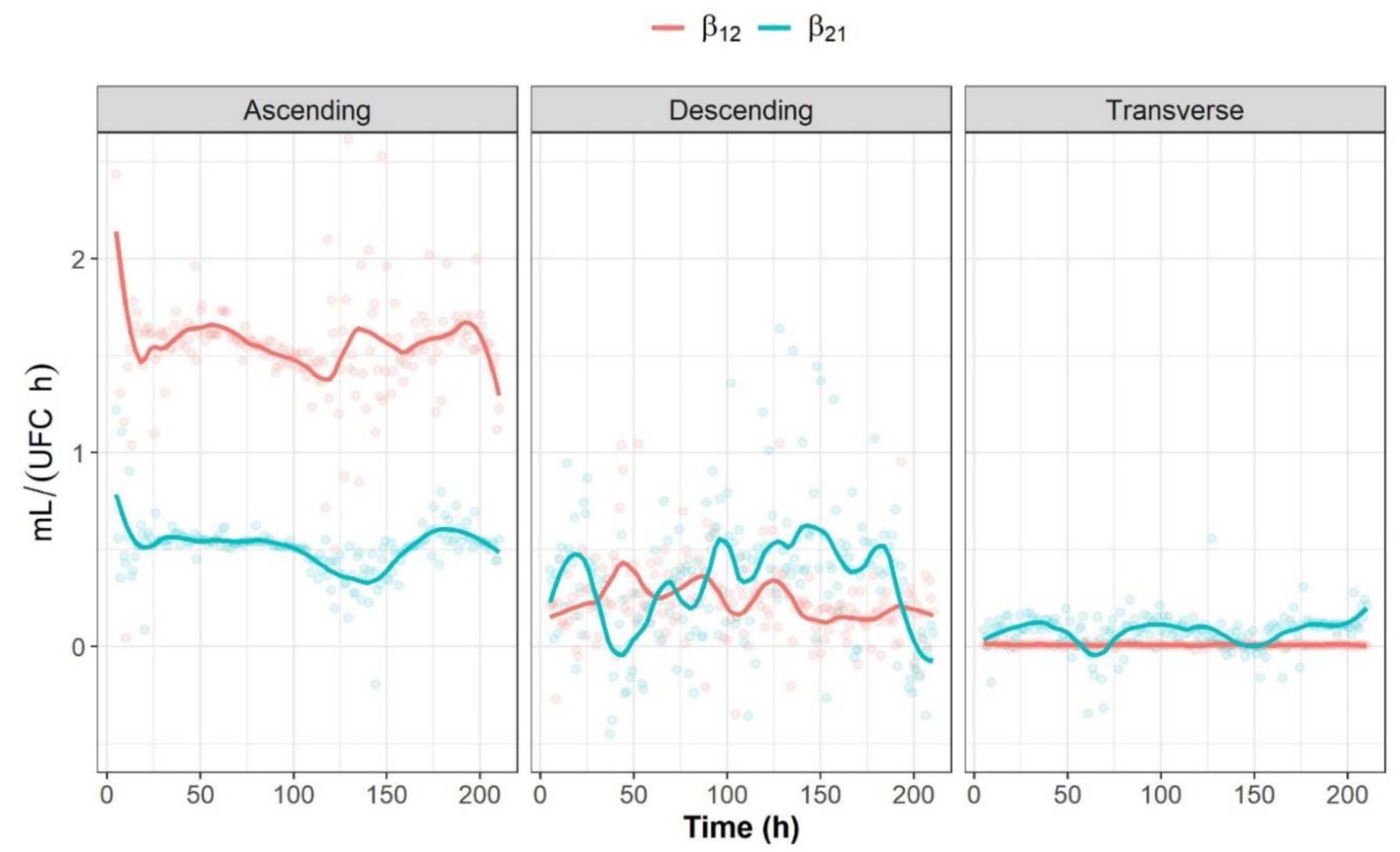
2.3.2. Analysis of Bidirectional Microbial Interactions for the Fructan-Supplemented System
2.3.3. Graphical Representation of Bidirectional Interactions
3. Materials and Methods
3.1. Agave Fructans
3.2. Conditioning the Experimental System
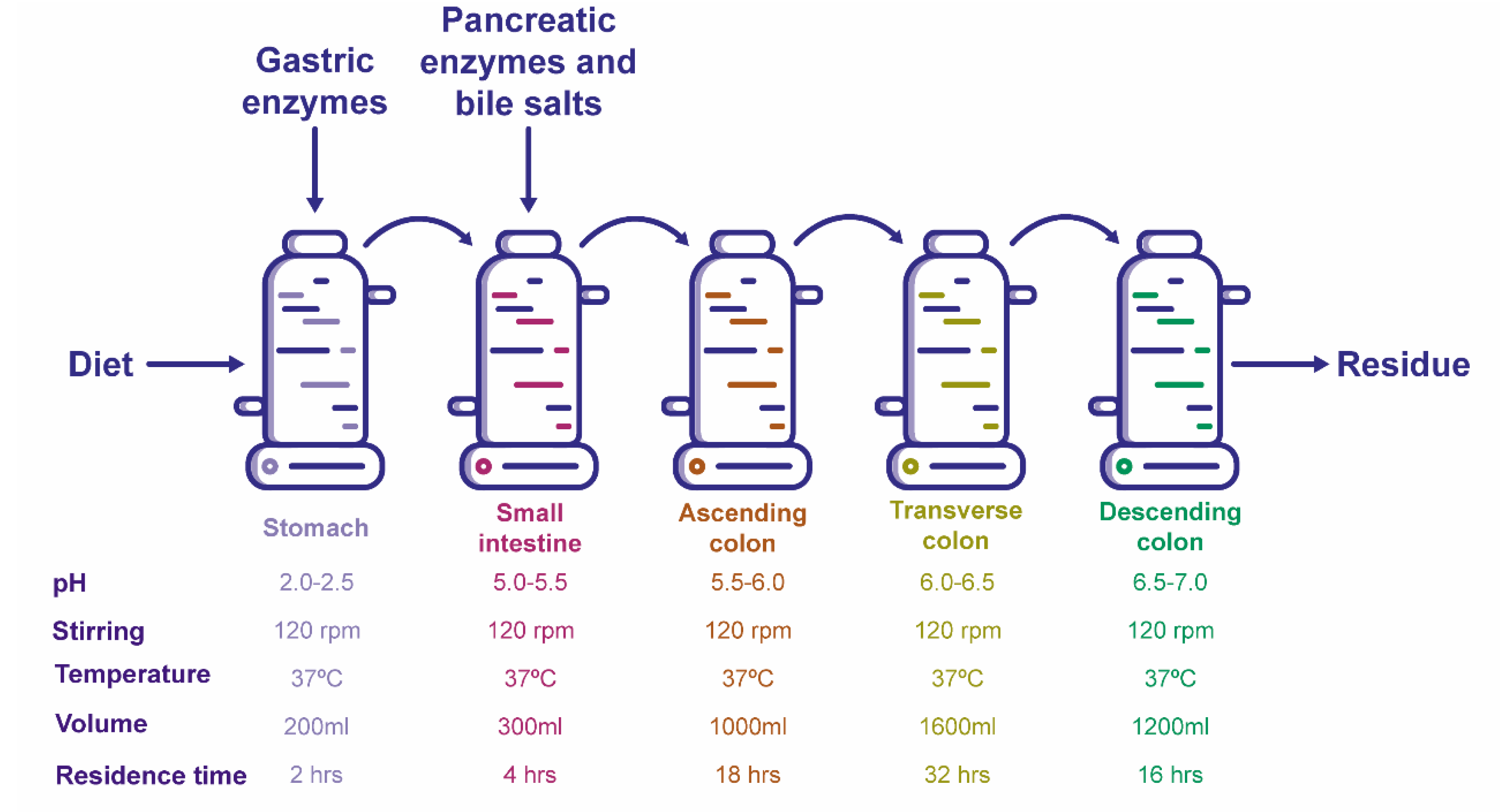
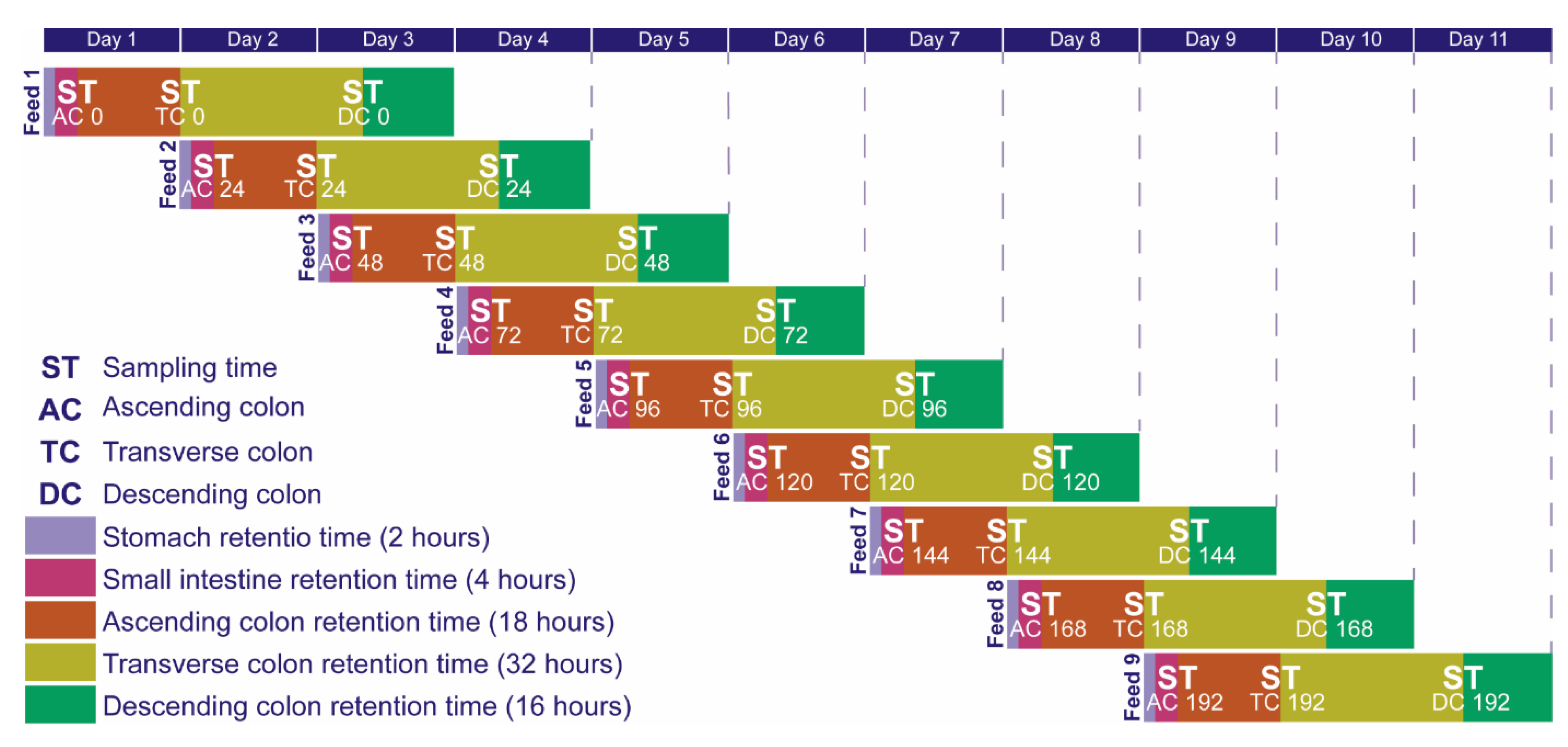
3.3. Ex-Vivo Simulations
3.4. Analysis of Microbial Interactions
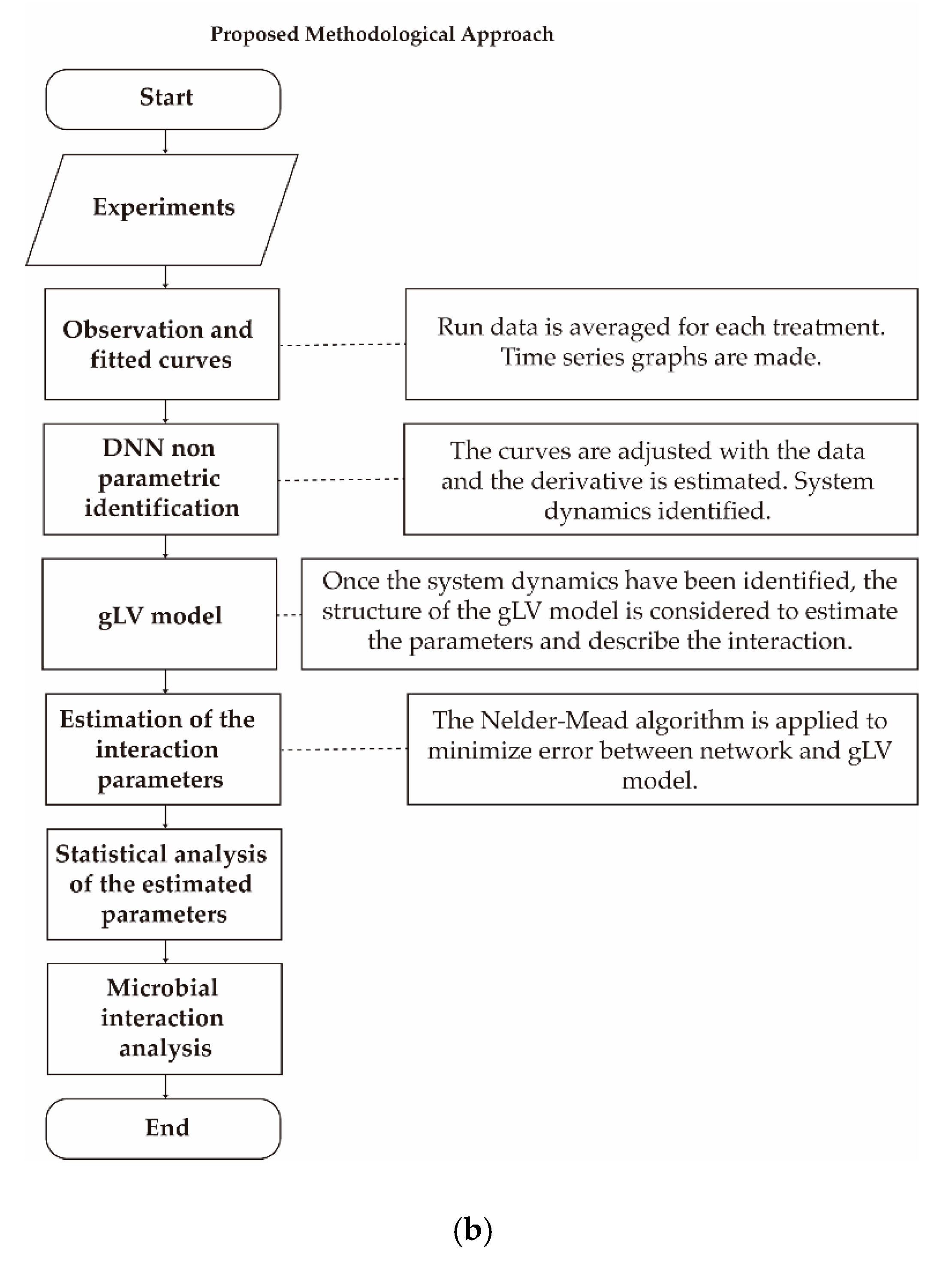
3.4.1. General Dynamic System Structure for the Dynamic Microbiota
3.4.2. Nonparametric System Identification of the Dynamic Microbial Behavior by a Differential Neural Network
3.4.3. Study of Microbial Interactions with Generalized Lotka-Volterrra Model
3.4.4. Identification of the gLV Model Parameters Applying the DNN Identifier and the Nelder-Mead Optimization Algorithm
3.4.5. Unidirectional Impact Analysis
3.4.6. Bidirectional Interaction Analysis
3.4.7. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Garcia, S.L. Mixed cultures as model communities: Hunting for ubiquitous microorganisms, their partners, and interactions. Aquat. Microb. Ecol. 2016, 77, 79–85. [Google Scholar] [CrossRef]
- Brenner, K.; You, L.; Arnold, F.H. Engineering microbial consortia: A new frontier in synthetic biology. Trends Biotechnol. 2008, 26, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, R.; Van Loosdrecht, M.C. Mixed culture biotechnology for bioenergy production. Curr. Opin. Biotechnol. 2007, 18, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Smid, E.J.; Lacroix, C. Microbe–microbe interactions in mixed culture food fermentations. Curr. Opin. Biotechnol. 2013, 24, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Kho, Z.Y.; Lal, S.K. The Human Gut Microbiome—A Potential Controller of Wellness and Disease. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordero, O.X.; Datta, M.S. Microbial interactions and community assembly at microscales. Curr. Opin. Microbiol. 2016, 31, 227–234. [Google Scholar] [CrossRef] [Green Version]
- Song, H.-S.; Cannon, W.R.; Beliaev, A.S.; Konopka, A. Mathematical Modeling of Microbial Community Dynamics: A Methodological Review. Processes 2014, 2, 711–752. [Google Scholar] [CrossRef] [Green Version]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Ivey, M.; Massel, M.; Phister, T.G. Microbial Interactions in Food Fermentations. Annu. Rev. Food Sci. Technol. 2013, 4, 141–162. [Google Scholar] [CrossRef]
- Kedia, G.; Wang, R.; Patel, H.; Pandiella, S.S. Use of mixed cultures for the fermentation of cereal-based substrates with potential probiotic properties. Process Biochem. 2007, 42, 65–70. [Google Scholar] [CrossRef]
- Sieuwerts, S.; Bok, F.A.M.; De Hugenholtz, J.; Vlieg, J.E.T.; Van Hylckama Vlieg, J.E. Unraveling Microbial Interactions in Food Fermentations: From Classical to Genomics Approaches. Appl. Environ. Microbiol. 2008, 74, 4997–5007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faust, K.; Lahti, L.; Gonze, D.; de Vos, W.M.; Raes, J. Metagenomics meets time series analysis: Unraveling microbial community dynamics. Curr. Opin. Microbiol. 2015, 25, 56–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falony, G.; Vandeputte, D.; Caenepeel, C.; Vieira-Silva, S.; Daryoush, T.; Vermeire, S.; Raes, J. The human microbiome in health and disease: Hype or hope. Acta Clin. Belg. 2019, 74, 53–64. [Google Scholar] [CrossRef] [Green Version]
- Gentile, C.L.; Weir, T.L. The gut microbiota at the intersection of diet and human health. Science 2018, 362, 776–780. [Google Scholar] [CrossRef] [Green Version]
- Jakobsson, H.E.; Rodríguez-Piñeiro, A.M.; Schütte, A.; Ermund, A.; Boysen, P.; Bemark, M.; Sommer, F.; Bäckhed, F.; Hansson, G.C.; Johansson, M.E. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015, 16, 164–177. [Google Scholar] [CrossRef]
- Dieterich, W.; Schink, M.; Zopf, Y. Microbiota in the Gastrointestinal Tract. Med. Sci. 2018, 6. [Google Scholar] [CrossRef] [Green Version]
- Sung, J.; Kim, S.; Cabatbat, J.J.T.; Jang, S.; Jin, Y.-S.; Jung, G.Y.; Chia, N.; Kim, P.-J. Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Proal, A.D.; Lindseth, I.A.; Marshall, T.G. Microbe-Microbe and Host-Microbe Interactions Drive Microbiome Dysbiosis and Inflammatory Processes. Discov. Med. 2017, 23, 51–60. [Google Scholar]
- Quigley, E.M.M. Prebiotics and probiotics; Modifying and mining the microbiota. Pharmacol. Res. 2010, 61, 213–218. [Google Scholar] [CrossRef]
- Gibson, G.R. Dietary Modulation of the Human Gut Microflora Using the Prebiotics Oligofructose and Inulin. J. Nutr. 1999, 129, 1438S–1441S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, G.; Hutkins, R.; Sanders, M.; Prescott, S.; Reimer, R.; Salminen, S.; Scott, K.; Stanton, C.; Swanson, K.; Cani, P.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Prebiotics. Fac. Publ. Food Sci. Technol. 2017. [Google Scholar]
- Krumbeck, J.A.; Maldonado-Gomez, M.X.; Ramer-Tait, A.E.; Hutkins, R.W. Prebiotics and synbiotics: Dietary strategies for improving gut health. Curr. Opin. Gastroenterol. 2016, 32, 110–119. [Google Scholar] [CrossRef] [PubMed]
- García Gamboa, R.; Ortiz Basurto, R.I.; Calderón Santoyo, M.; Bravo Madrigal, J.; Ruiz Álvarez, B.E.; González Avila, M. In vitro evaluation of prebiotic activity, pathogen inhibition and enzymatic metabolism of intestinal bacteria in the presence of fructans extracted from agave: A comparison based on polymerization degree. LWT 2018, 92, 380–387. [Google Scholar] [CrossRef]
- Kolida, S.; Gibson, G.R. Prebiotic Capacity of Inulin-Type Fructans. J. Nutr. 2007, 137, 2503S–2506S. [Google Scholar] [CrossRef] [Green Version]
- De Vries, J.; Le Bourgot, C.; Calame, W.; Respondek, F. Effects of β-fructans fiber on bowel function: A systematic review and meta-analysis. Nutrients 2019, 11, 91. [Google Scholar] [CrossRef] [Green Version]
- Wiele, T.V.D.; Boon, N.; Possemiers, S.; Jacobs, H.; Verstraete, W. Inulin-type fructans of longer degree of polymerization exert more pronounced in vitro prebiotic effects. J. Appl. Microbiol. 2007, 102, 452–460. [Google Scholar] [CrossRef]
- Andrade, A.I.C.; Bautista, C.R.; Cabrera, M.A.R.; Guerra, R.E.S.; Chávez, E.G.; Ahumada, C.F.; Lagunes, A.G. Agave salmiana fructans as gut health promoters: Prebiotic activity and inflammatory response in Wistar healthy rats. Int. J. Biol. Macromol. 2019, 136, 785–795. [Google Scholar] [CrossRef]
- Biedrzycka, E.; Bielecka, M. Prebiotic effectiveness of fructans of different degrees of polymerization. Trends Food Sci. Technol. 2004, 15, 170–175. [Google Scholar] [CrossRef]
- Makras, L.; Acker, G.V.; Vuyst, L.D. Lactobacillus paracasei subsp. paracasei 8700:2 Degrades Inulin-Type Fructans Exhibiting Different Degrees of Polymerization. Appl. Environ. Microbiol. 2005, 71, 6531–6537. [Google Scholar] [CrossRef] [Green Version]
- Márquez-Aguirre, A.L.; Camacho-Ruiz, R.M.; Arriaga-Alba, M.; Padilla-Camberos, E.; Kirchmayr, M.R.; Blasco, J.L.; González-Avila, M. Effects of Agave tequilana fructans with different degree of polymerization profiles on the body weight, blood lipids and count of fecal Lactobacilli/Bifidobacteria in obese mice. Food Funct. 2013, 4, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Fuller, R. Aspects of in vitro and in vivo research approaches directed toward identifying probiotics and prebiotics for human use. J. Nutr. 2000, 130, 391S–395S. [Google Scholar] [CrossRef] [PubMed]
- D’hoe, K.; Vet, S.; Faust, K.; Moens, F.; Falony, G.; Gonze, D.; Lloréns-Rico, V.; Gelens, L.; Danckaert, J.; De Vuyst, L.; et al. Integrated culturing, modeling and transcriptomics uncovers complex interactions and emergent behavior in a three-species synthetic gut community. eLife 2018, 7, e37090. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Bauchinger, F.; Laroche, B.; De Buyl, S.; Lahti, L.; Washburne, A.D.; Gonze, D.; Widder, S. Signatures of ecological processes in microbial community time series. Microbiome 2018, 6, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zomorrodi, A.R.; Segrè, D. Synthetic Ecology of Microbes: Mathematical Models and Applications. J. Mol. Biol. 2016, 428, 837–861. [Google Scholar] [CrossRef] [Green Version]
- Vet, S.; Buyl, S.; De Faust, K.; Danckaert, J.; Gonze, D.; Gelens, L. Bistability in a system of two species interacting through mutualism as well as competition: Chemostat vs. Lotka-Volterra equations. PLoS ONE 2018, 13, e0197462. [Google Scholar] [CrossRef]
- Stein, R.R.; Bucci, V.; Toussaint, N.C.; Buffie, C.G.; Rätsch, G.; Pamer, E.G.; Sander, C.; Xavier, J.B. Ecological Modeling from Time-Series Inference: Insight into Dynamics and Stability of Intestinal Microbiota. PLOS Comput. Biol. 2013, 9, e1003388. [Google Scholar] [CrossRef]
- Gonze, D.; Lahti, L.; Raes, J.; Faust, K. Multi-stability and the origin of microbial community types. ISME J. 2017, 11, 2159–2166. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Tamayo, R.; Laroche, B.; Walter, É.; Doré, J.; Leclerc, M. Mathematical modelling of carbohydrate degradation by human colonic microbiota. J. Theor. Biol. 2010, 266, 189–201. [Google Scholar] [CrossRef]
- Jeyamkondan, S.; Jayas, D.S.; Holley, R.A. Microbial growth modelling with artificial neural networks. Int. J. Food Microbiol. 2001, 64, 343–354. [Google Scholar] [CrossRef]
- Becker, T.; Enders, T.; Delgado, A. Dynamic neural networks as a tool for the online optimization of industrial fermentation. Bioprocess Biosyst. Eng. 2002, 24, 347–354. [Google Scholar] [CrossRef]
- Zhang, C.; Vinyals, O.; Munos, R.; Bengio, S. A Study on Overfitting in Deep Reinforcement Learning. arXiv 2018, arXiv:180406893. [Google Scholar]
- Vega, R.; Hernandez-Reynoso, A.G.; Linn, E.K.; Fuentes-Aguilar, R.Q.; Sanchez-Ante, G.; Santos-Garcia, A.; Garcia-Gonzalez, A. Hemodynamic Pattern Recognition During Deception Process Using Functional Near-infrared Spectroscopy. J. Med. Biol. Eng. 2016, 36, 22–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopkins, M.; Cummings, J.; Macfarlane, G. Inter-species differences in maximum specific growth rates and cell yields of bifidobacteria cultured on oligosaccharides and other simple carbohydrate sources. J. Appl. Microbiol. 1998, 85, 381–386. [Google Scholar] [CrossRef]
- Poznyak, A.S.; Sanchez, E.N.; Yu, W. Differential Neural Networks for Robust Nonlinear Control: Identification, State Estimation and Trajectory Tracking; World Scientific: Singapore, 2001; ISBN 978-981-02-4624-2. [Google Scholar]
- Gupta, M.; Jin, L.; Homma, N. Static and Dynamic Neural Networks: From Fundamentals to Advanced Theory; John Wiley & Sons: Hoboken, NJ, USA, 2004; ISBN 978-0-471-46092-3. [Google Scholar]
- Gueguim Kana, E.B.; Oloke, J.K.; Lateef, A.; Adesiyan, M.O. Modeling and optimization of biogas production on saw dust and other co-substrates using Artificial Neural network and Genetic Algorithm. Renew. Energy 2012, 46, 276–281. [Google Scholar] [CrossRef]
- Jorjani, E.; Chehreh Chelgani, S.; Mesroghli, S. Prediction of microbial desulfurization of coal using artificial neural networks. Miner. Eng. 2007, 20, 1285–1292. [Google Scholar] [CrossRef]
- Gradilla-Hernández, S.J.; Herrera-López, E.; Gschaedler, A.; González-Avila, M.; Fuentes-Aguilar, R.; Garcia-Gonzalez, A. Differential neural network identifier for parameter determination of a mixed microbial culture model. IFAC-Pap. 2018, 51, 479–484. [Google Scholar] [CrossRef]
- Heeney, D.D.; Gareau, M.G.; Marco, M.L. Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr. Opin. Biotechnol. 2018, 49, 140–147. [Google Scholar] [CrossRef] [Green Version]
- Tojo, R.; Suárez, A.; Clemente, M.G.; De los Reyes-Gavilán, C.G.; Margolles, A.; Gueimonde, M.; Ruas-Madiedo, P. Intestinal microbiota in health and disease: Role of bifidobacteria in gut homeostasis. World J. Gastroenterol. 2014, 20, 15163–15176. [Google Scholar] [CrossRef]
- Ferreira, R.B.R.; Gill, N.; Willing, B.P.; Antunes, L.C.M.; Russell, S.L.; Croxen, M.A.; Finlay, B.B. The Intestinal Microbiota Plays a Role in Salmonella-Induced Colitis Independent of Pathogen Colonization. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Ji, B.; Nielsen, J. From next-generation sequencing to systematic modeling of the gut microbiome. Front. Genet. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Gamboa, R.; Gradilla-Hernández, M.S.; Ortiz-Basurto, R.I.; García-Reyes, R.A.; González-Avila, M. Assessment of intermediate and long chains agave fructan fermentation on the growth of intestinal bacteria cultured in a gastrointestinal tract simulator. Rev. Mex. Ing. Quím. 2020, 19, 827–838. [Google Scholar] [CrossRef] [Green Version]
- Cardarelli, H.R.; Martinez, R.C.; Albrecht, S.; Schols, H.; Franco, B.D.G.M.; Saad, S.M.I.; Smidt, H. In vitro fermentation of prebiotic carbohydrates by intestinal microbiota in the presence of Lactobacillus amylovorus DSM 16998. Benef Microbes 2016, 7, 119–133. [Google Scholar] [CrossRef]
- Salazar, N.; Dewulf, E.M.; Neyrinck, A.M.; Bindels, L.B.; Cani, P.D.; Mahillon, J.; De Vos, W.M.; Thissen, J.-P.; Gueimonde, M.; De los Reyes-Gavilán, C.G.; et al. Inulin-type fructans modulate intestinal Bifidobacterium species populations and decrease fecal short-chain fatty acids in obese women. Clin. Nutr. 2015, 34, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Roberfroid, M.B.; Van Loo, J.A.E.; Gibson, G.R. The Bifidogenic Nature of Chicory Inulin and Its Hydrolysis Products. J. Nutr. 1998, 128, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loo, J.V.; Cummings, J.; Delzenne, N.; Englyst, H.; Franck, A.; Hopkins, M.; Kok, N.; Macfarlane, G.; Newton, D.; Quigley, M.; et al. Functional food properties of non-digestible oligosaccharides: A consensus report from the ENDO project (DGXII AIRII-CT94-1095). Br. J. Nutr. 1999, 81, 121–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rastall, R.A.; Gibson, G.R. Recent developments in prebiotics to selectively impact beneficial microbes and promote intestinal health. Curr. Opin. Biotechnol. 2015, 32, 42–46. [Google Scholar] [CrossRef]
- Barroso, E.; Sánchez-Patán, F.; Martín-Alvarez, P.J.; Bartolomé, B.; Moreno-Arribas, M.V.; Peláez, C.; Requena, T.; Van de Wiele, T.; Martínez-Cuesta, M.C. Lactobacillus plantarum IFPL935 favors the initial metabolism of red wine polyphenols when added to a colonic microbiota. J. Agric. Food Chem. 2013, 61, 10163–10172. [Google Scholar] [CrossRef] [Green Version]
- Medina, D.A.; Pinto, F.; Ovalle, A.; Thomson, P.; Garrido, D. Prebiotics Mediate Microbial Interactions in a Consortium of the Infant Gut Microbiome. Int. J. Mol. Sci. 2017, 18, 2095. [Google Scholar] [CrossRef] [Green Version]
- Moens, F.; De Vuyst, L. Inulin-type fructan degradation capacity of Clostridium cluster IV and XIVa butyrate-producing colon bacteria and their associated metabolic outcomes. Benef. Microbes 2017. [Google Scholar] [CrossRef]
- Perrin, S.; Warchol, M.; Grill, J.P.; Schneider, F. Fermentations of fructo-oligosaccharides and their components by Bifidobacterium infantis ATCC 15697 on batch culture in semi-synthetic medium. J. Appl. Microbiol. 2001, 90, 859–865. [Google Scholar] [CrossRef]
- Meulen, R.V.; Der Avonts, L.; Vuyst, L.D. Short Fractions of Oligofructose Are Preferentially Metabolized by Bifidobacterium animalis DN-173 010. Appl. Environ. Microbiol. 2004, 70, 1923–1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuyst, L.D.; Makras, L.; Avonts, L.; Holo, H.; Yi, Q.; Servin, A.; Fayol-Messaoudi, D.; Berger, C.; Zoumpopoulou, G.; Tsakalidou, E.; et al. Antimicrobial potential of probiotic or potentially probiotic lactic acid bacteria, the first results of the international European research project PROPATH of the PROEUHEALTH cluster. Microb. Ecol. Health Dis. 2004, 16, 125–130. [Google Scholar] [CrossRef] [Green Version]
- De Keersmaecker, S.C.J.; Verhoeven, T.L.A.; Desair, J.; Marchal, K.; Vanderleyden, J.; Nagy, I. Strong antimicrobial activity of Lactobacillus rhamnosus GG against Salmonella typhimurium is due to accumulation of lactic acid. FEMS Microbiol. Lett. 2006, 259, 89–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bielecka, M.; Biedrzycka, E.; Biedrzycka, E.; Smoragiewicz, W.; Smieszek, M. Interaction of Bifidobacterium and Salmonella during associated growth. Int. J. Food Microbiol. 1998, 45, 151–155. [Google Scholar] [CrossRef]
- Fayol-Messaoudi, D.; Berger, C.N.; Coconnier-Polter, M.-H.; Moal, V.L.-L.; Servin, A.L. pH-, Lactic Acid-, and Non-Lactic Acid-Dependent Activities of Probiotic Lactobacilli against Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 2005, 71, 6008–6013. [Google Scholar] [CrossRef] [Green Version]
- Servin, A.L. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 2004, 28, 405–440. [Google Scholar] [CrossRef] [Green Version]
- Gibson, G.R. Dietary modulation of the human gut microflora using prebiotics. Br. J. Nutr. 1998, 80, S209–S212. [Google Scholar] [CrossRef] [Green Version]
- Molly, K.; Vande Woestyne, M.; Verstraete, W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 1993, 39, 254–258. [Google Scholar] [CrossRef]
- Santiaguín-Padilla, A.J.; Peña-Ramos, E.A.; Pérez-Gallardo, A.; Rascón-Chu, A.; González-Ávila, M.; González-Ríos, H.; González-Noriega, J.A.; Islava-Lagarda, T. In vitro Digestibility and Quality of an Emulsified Meat Product Formulated With Animal Fat Encapsulated With Pectin. J. Food Sci. 2019, 84, 1331–1339. [Google Scholar] [CrossRef]
- Gutiérrez-Zamorano, C.; González-Ávila, M.; Díaz-Blas, G.; Smith, C.T.; González-Correa, C.; García-Cancino, A. Increased anti-Helicobacter pylori effect of the probiotic Lactobacillus fermentum UCO-979C strain encapsulated in carrageenan evaluated in gastric simulations under fasting conditions. Food Res. Int. Ott. Ont 2019, 121, 812–816. [Google Scholar] [CrossRef]
- García, A.; Luviano-Juárez, A.; Chairez, I.; Poznyak, A.; Poznyak, T. Projectional dynamic neural network identifier for chaotic systems: Application to Chua’s circuit. Int. J. Artif. Intell. 2011, 6, 1–18. [Google Scholar]
- Bucci, V.; Xavier, J.B. Towards Predictive Models of the Human Gut Microbiome. J. Mol. Biol. 2014, 426, 3907–3916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barati Reza Parameter Estimation of Nonlinear Muskingum Models Using Nelder-Mead Simplex Algorithm. J. Hydrol. Eng. 2011, 16, 946–954. [CrossRef]
- Dochain, D. Automatic Control of Bioprocesses; John Wiley & Sons: Hoboken, NJ, USA, 2013; ISBN 978-1-118-62391-6. [Google Scholar]
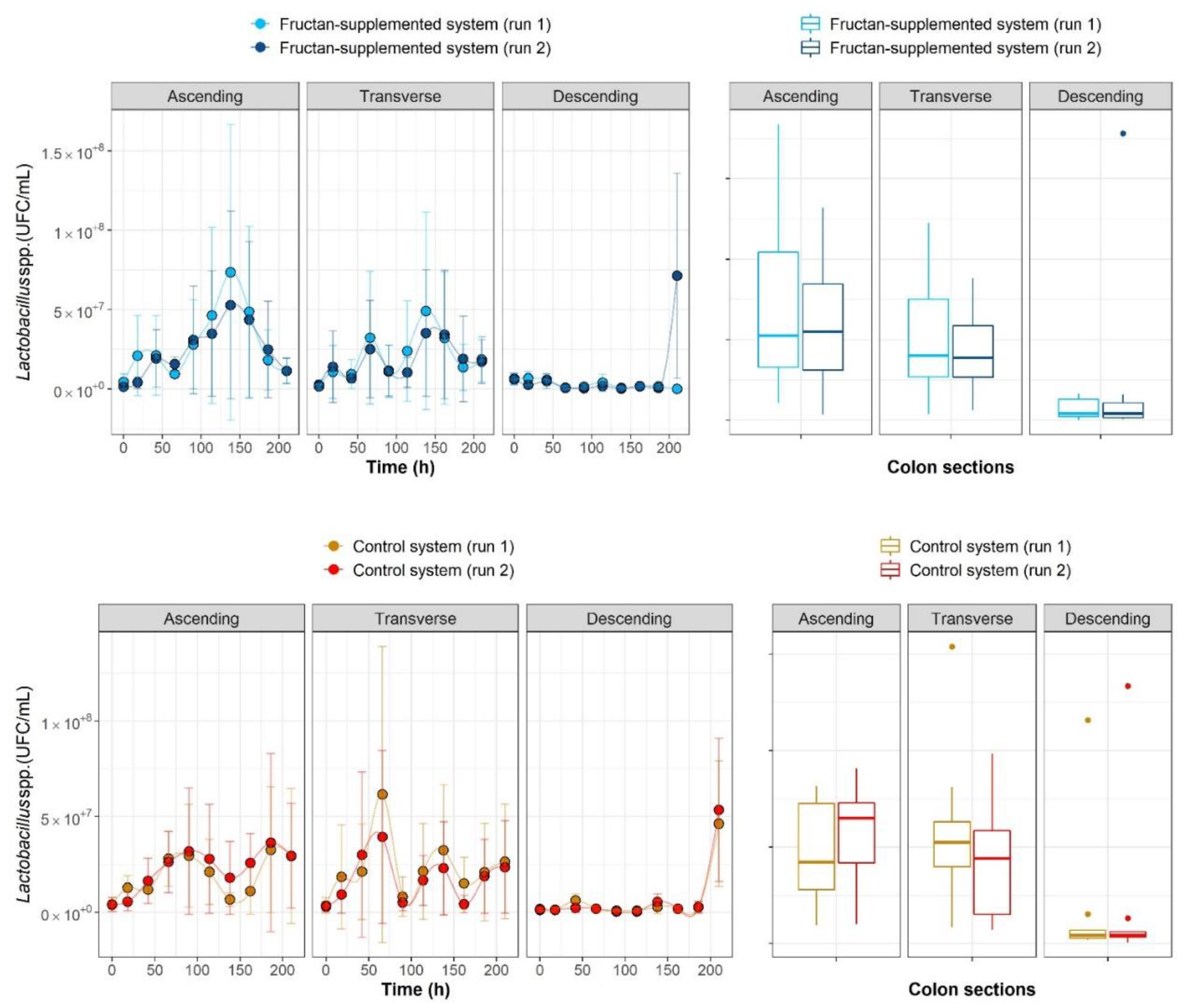
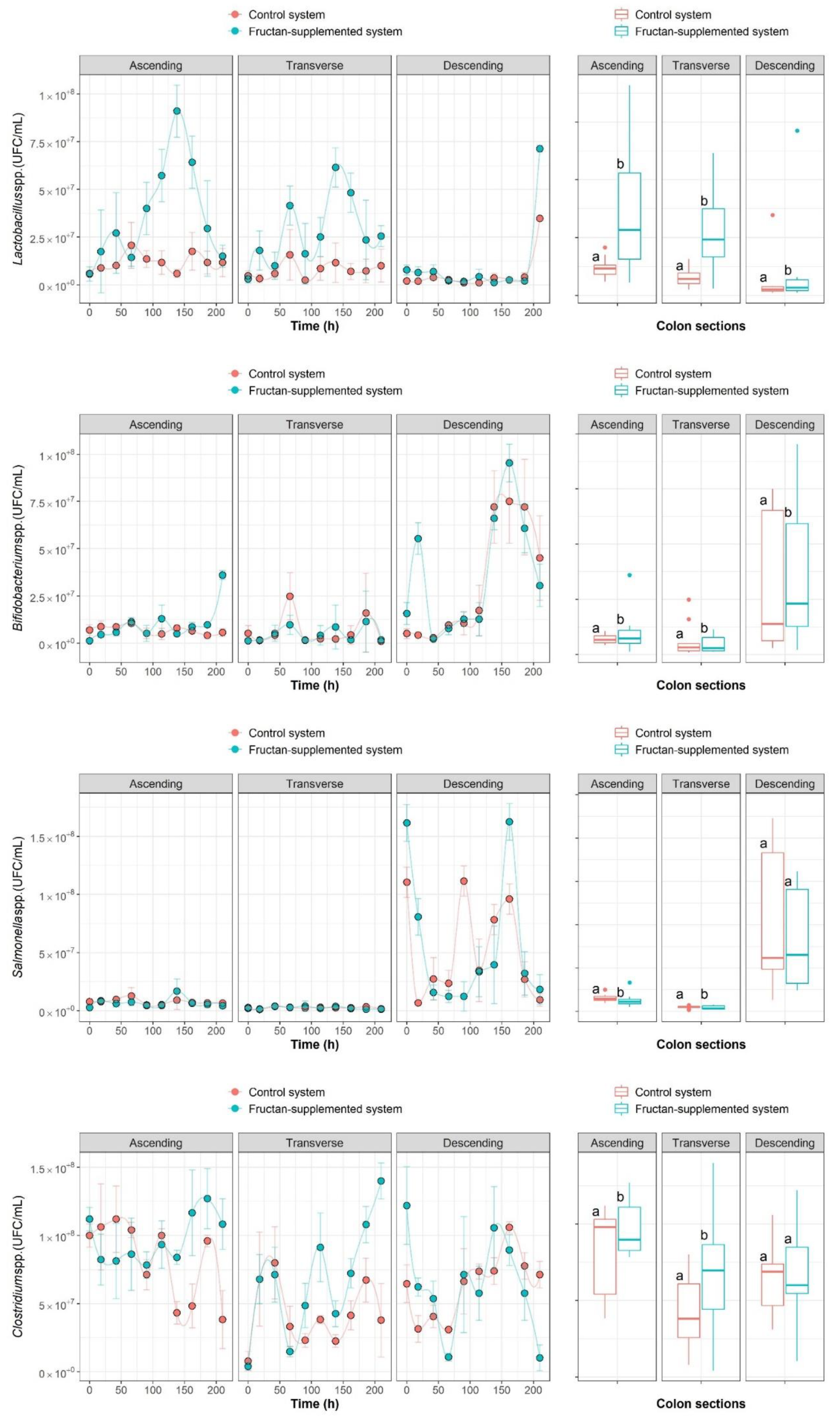

| Colon Section | Ascending Colon | Transverse Colon | Descending Colon | ||||
|---|---|---|---|---|---|---|---|
| Treatment | Control | Fructans | Control | Fructans | Control | Fructans | |
| Lactobacillus spp. | α1 | 1.788 ± 0.797 | 1.864 ± 0.667 | 1.331 ± 1.304 | 3.142 ± 2.962 * | 3.642 ± 1.265 | 5.201 ± 4.747 * |
| β11 | 0.929 ± 0.281 * | 0.620 ± 0.141 | 0.553 ± 0.280 * | 0.008 ± 0.015 | 1.459 ± 0.439 | 0.039 ± 1.928 * | |
| β12 | 1.318 ± 0.913 | 1.565 ± 0.325 * | 1.576 ± 1.645 * | 0.010 ± 0.008 | 0.406 ± 0.132 * | 0.237 ± 0.202 | |
| β13 | 1.163 ± 0.516 | 1.328 ± 0.290 * | 2.362 ± 2.296 * | 0.029 ± 0.337 | 0.188 ± 0.051 | 0.170 ± 0.190 | |
| β14 | −0.374 ± 0.125 * | −0.493 ± 0.136 | −0.452 ± 0.182 | −0.001 ± 0.018 * | −0.379 ± 0.130 | −0.079 ± 0.151 * | |
| Bifidobacterium spp. | α2 | 1.152 ± 0.380 | 2.068 ± 0.612 | 2.158 ± 1.102 | 3.317 ± 4.295 | 2.743 ± 0.770 | 3.295 ± 3.542 |
| β21 | 1.064 ± 0.313 | 0.511 ± 0.129 | 0.681 ± 0.392 | 0.077 ± 0.092 | 1.555 ± 0.437 | 0.336 ± 0.368 | |
| β22 | 1.218 ± 0.379 | 1.461 ± 0.336 | 1.085 ± 0.523 | 0.279 ± 0.316 | 0.252 ± 0.068 | 0.066 ± 0.118 | |
| β23 | 1.109 ± 0.364 | 1.209 ± 0.315 | 1.453 ± 0.533 | 0.861 ± 1.193 | 0.167 ± 0.062 | 0.097 ± 0.060 | |
| β24 | −0.378 ± 0.143 | −0.430 ± 0.125 | −0.350 ± 0.144 | −0.083 ± 0.069 | −0.312 ± 0.075 | −0.023 ± 0.050 | |
| Salmonella spp. | α3 | 1.869 ± 0.697 | 2.313 ± 0.738 | 1.051 ± 0.635 | 2.800 ± 9.005 | 2.052 ± 0.753 | 2.678 ± 4.782 |
| β31 | 0.936 ± 0.294 | 0.227 ± 0.089 | 0.567 ± 0.277 | 0.094 ± 0.084 | 1.709 ± 0.497 | 0.121 ± 0.708 | |
| β32 | 1.226 ± 0.383 | 1.382 ± 0.310 | 0.452 ± 0.174 | 0.198 ± 0.822 | 0.369 ± 0.101 | 0.051 ± 0.045 | |
| β33 | 1.203 ± 0.353 | 1.382 ± 0.429 | 1.203 ± 0.553 | 1.452 ± 1.557 | 0.193 ± 0.057 | 0.043 ± 0.082 | |
| β34 | −0.372 ± 0.119 | −0.322 ± 0.104 | −0.246 ± 0.140 | −0.176 ± 0.137 | −0.400 ± 0.106 | 0.013 ± 0.018 | |
| Clostridium spp. | α4 | 1.607 ± 1.607 | 2.302 ± 0.592 | 2.657 ± 0.966 | 2.665 ± 3.124 | 2.802 ± 0.782 | 2.088 ± 4.116 |
| β41 | 1.326 ± 0.0.554 | 0.865 ± 0.366 | 1.063 ± 0.458 | 0.360 ± 0.679 | 1.827 ± 0.561 | 0.341 ± 0.810 | |
| β42 | 1.894 ± 0.622 | 1.399 ± 0.449 | 0.831 ± 0.523 | 1.674 ± 1.986 | 0.776 ± 0.247 | 0.510 ± 0.921 | |
| β43 | 0.944 ± 0.469 | 1.417 ± 0.396 | 1.208 ± 0.544 | 1.465 ± 3.279 | 0.518 ± 0.154 | 0.489 ± 0.583 | |
| β44 | −0.481 ± 0.0.194 | −0.336 ± 0.158 | −0.370 ± 0.160 | −0.451 ± 0.459 | −0.015 ± 0.259 | −0.685 ± 0.517 | |
| Colon Section | Ascending Colon | Transverse Colon | Descending Colon | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Interaction Parameters Difference | Average Difference | Interaction | Observation | Average Difference | Interaction | Observation | Average Difference | Interaction | Observation |
| β12-β21 | 1.054 * | Mutualism (+/+) | Lactobacillus spp. (most benefited) | −0.067 * | Neutralism (0/0) | Both effects are negligible <0.1 | −0.099 * | Mutualism (+/+) | Bifidobacterium spp. (most benefited) |
| β13-β31 | 1.101 * | Mutualism (+/+) | Lactobacillus spp. (most benefited) | −0.084 * | Neutralism (0/0) | Both effects are negligible <0.1 | −0.049 | Neutralism (+/+) | Equally benefitted |
| |β14|-β41 | −0.372 * | Antagonism (−/+) | Clostridium spp. (is benefited) | −0.359 * | Commensalism (0/+) | Clostridium spp. (is benefited) | −0.262 * | Commensalism (0/+) | Clostridium spp. (is benefited) |
| β23-β32 | −0.173 * | Mutualism (+/+) | Salmonella spp. (most benefited) | 0.663 * | Mutualism (+/+) | Bifidobacterium spp. (most benefited) | 0.046 * | Neutralism (0/0) | Both effects are negligible <0.1 |
| |β24|-β42 | −0.969 * | Antagonism (−/+) | Clostridium spp. (is benefited) | −1.591 * | Commensalism (0/+) | Clostridium spp. (is benefited) | −0.487 * | Commensalism (0/+) | Clostridium spp. (is benefited) |
| |β34|-β43 | −1.095 * | Antagonism (−/+) | Clostridium spp. (is benefited) | −1.289 * | Antagonism (−/+) | Clostridium spp. (is benefited) | −0.476 * | Commensalism (0/+) | Clostridium spp. (is benefited) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gradilla-Hernández, M.S.; García-González, A.; Gschaedler, A.; Herrera-López, E.J.; González-Avila, M.; García-Gamboa, R.; Yebra Montes, C.; Fuentes-Aguilar, R.Q. Applying Differential Neural Networks to Characterize Microbial Interactions in an Ex Vivo Gastrointestinal Gut Simulator. Processes 2020, 8, 593. https://doi.org/10.3390/pr8050593
Gradilla-Hernández MS, García-González A, Gschaedler A, Herrera-López EJ, González-Avila M, García-Gamboa R, Yebra Montes C, Fuentes-Aguilar RQ. Applying Differential Neural Networks to Characterize Microbial Interactions in an Ex Vivo Gastrointestinal Gut Simulator. Processes. 2020; 8(5):593. https://doi.org/10.3390/pr8050593
Chicago/Turabian StyleGradilla-Hernández, Misael Sebastián, Alejandro García-González, Anne Gschaedler, Enrique J. Herrera-López, Marisela González-Avila, Ricardo García-Gamboa, Carlos Yebra Montes, and Rita Q. Fuentes-Aguilar. 2020. "Applying Differential Neural Networks to Characterize Microbial Interactions in an Ex Vivo Gastrointestinal Gut Simulator" Processes 8, no. 5: 593. https://doi.org/10.3390/pr8050593
APA StyleGradilla-Hernández, M. S., García-González, A., Gschaedler, A., Herrera-López, E. J., González-Avila, M., García-Gamboa, R., Yebra Montes, C., & Fuentes-Aguilar, R. Q. (2020). Applying Differential Neural Networks to Characterize Microbial Interactions in an Ex Vivo Gastrointestinal Gut Simulator. Processes, 8(5), 593. https://doi.org/10.3390/pr8050593







