Abstract
The spike protein of the pandemic human corona virus is essential for its entry into human cells. In fact, most neutralizing antibodies against Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) are directed against the Virus-surface exposed spike protein, making it the antigen of choice for use in vaccines and diagnostic tests. In the current pandemic context, global demand for spike proteins has rapidly increased and could exceed hundreds of grams to kilograms annually. Coronavirus spikes are large heavily glycosylated homo-trimeric complexes, with inherent instability. The poor manufacturability now threatens the availability of these proteins for vaccines and diagnostic tests. Here, we outline scalable, Good Manufacturing Practice (GMP) compliant, and chemically defined processes for the production of two cell-secreted stabilized forms of the trimeric spike proteins (Wuhan and D614G variant). The processes are chemically defined and based on clonal suspension-CHO cell populations and on protein purification via a two-step scalable downstream process. The trimeric conformation was confirmed using electron microscopy and HPLC analysis. Binding to susceptible cells was shown using a virus-inhibition assay. The diagnostic sensitivity and specificity for detection of serum SARS-CoV-2-specific-immunoglobulin molecules was found to exceed that of spike fragments (Spike subunit-1, S1 and Receptor Binding Domain, RBD). The process described here will enable production of sufficient high-quality trimeric spike protein to meet the global demand for SARS-CoV-2 diagnostic tests and potentially vaccines.
1. Introduction
SARS-CoV-2 is the virus responsible for the 2019 coronavirus disease (COVID-19) pandemic [1,2], which presents an unprecedented challenge to societies globally. Effective vaccines and sensitive diagnostic tools for COVID-19 are urgently needed, and systems to produce and deliver these tools in sufficient quantities are required. It has become clear that protein synthesis technologies used thus far for spike proteins are insufficient to meet the unprecedented global demand for these critical ingredients. As the SARS-CoV-2 trimeric spike complex is a major target of the immune system, it could be a useful ingredient for vaccine and diagnostic applications. Stabilized trimeric spike proteins have been selected as the antigen of choice for RNA and virus-vector-based vaccine candidates that are currently under development (Moderna Inc., Novavax Inc., Pfizer Inc., J&J Inc. etc.). The efficacy of these vaccines was recently reported to be as high as 95% [3,4]. However, to maintain long-term protective immunity, follow-up vaccinations may be required. An adjuvated subunit vaccine based on the stabilized trimeric spike protein could be ideal for this purpose. Subunit vaccines can be produced in a cost-effective way and can be transported and stored lyophilized at ambient temperatures. Additionally, whether any vaccination approach has elicited a sufficient immune response and how long it can be maintained needs to be verified. To address this, the quantification of antibody levels against SARS-CoV-2 in serum is the most practical approach. For this reason, there will be a high demand for diagnostic tests for quantification of SARS-CoV-2-specific antibodies. The antigen that provides the best sensitivity and specificity for detection of SARS-CoV-2 antibodies is the trimeric form of the spike protein [5].
The viral spike (S) protein complex is a surface-exposed homo-trimeric structure that mediates entry into host cells. The spike engages the cellular Angiotensin Converting Enzyme 2 (ACE-2) receptor and allows virus–host membrane fusion. The spike complex is considered the primary target of neutralizing antibodies [6,7,8,9]. The ideal diagnostic test for SARS-CoV-2 antibodies would detect all antibodies directed against the trimeric spike protein complex. The production of such diagnostic tests requires the production of large quantities of highly purified S protein in its natural prefusion conformation [10,11]. The coronavirus spike protein is a heavily glycosylated complex, with inherent structural flexibility and instability. In addition, the S protein is processed by Furin and another membrane-bound cell host protease [8,12]. Poor yield and pre-fusion instability of the S protein have hampered its use for the development of vaccines and diagnostic tests. We and others [13] explored the expression and eventually scaled-up manufacture of trimeric spike protein in a secreted soluble form using Chinese Hamster Ovary (CHO) cells.
In this study, we developed a production process that yields high-quality trimeric spike protein with a minimal trimerization domain. This protein can be used for diagnostic tests as well as for vaccine applications. We used the fully characterized and chemistry, manufacturing and control (CMC) compliant CHOExpress™ cell host (ExcellGene SA), single use equipment, chemically defined media, and additives. Additionally, regulatory CMC-requirements from DNA construction to production in bioreactors were strictly followed. The clonally derived CHO cell line and the scalable production processes outlined here have already allowed us to produce dozens of grams of trimeric spike protein. This process can be readily scaled up to produce kilograms of trimeric spike protein, should the demand rise to such a level.
2. Material and Methods
2.1. Design of SARS CoV-2 Spike Proteins
SARS-CoV-2 spike protein encoding DNA was constructed and codon-optimized for CHO cells. A total of 11 DNA constructs were designed (Figure 1) and synthetized by ATUM (Menlo Park, CA, USA) and provided with CMC relevant documentation. A functional IgG leader sequence was added to mediate efficient signal peptide cleavage. Histidine (8-mer)-encoding DNA was added for carboxyterminal expression. Where mentioned, the furin cleavage site RRAR is mutated to be non-functional. The transmembrane domain and the C-terminal intracellular tail were removed and replaced by a T4-foldon sequence [14] in trimer designs. For the RBD fragment (aa319-541), IgG leader and His-tag sequences were added. Because of the widespread emergence of SARS-CoV-2 variants that carry the D614G amino acid change [15,16], this variant of the spike protein was also included in our study.
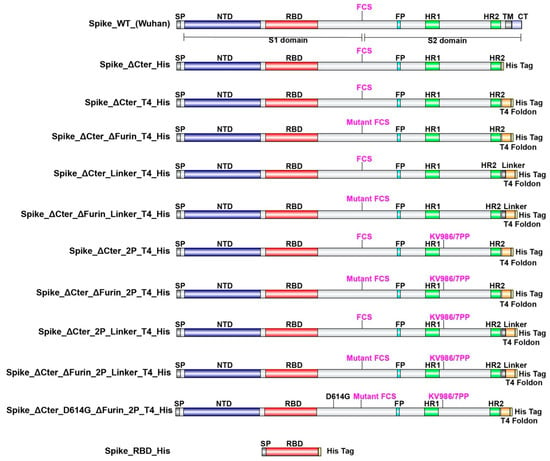
Figure 1.
Schematic representation of SARS-CoV-2 spike protein designs. SP: Signal Peptide, NTD: N Terminal Domain, RBD: Receptor Binding Domain, FCS: Furin Cleavage Site, FP: Fusion Peptide, HR1: Heptad Repeat 1, HR2: Heptad Repeat 2, TM: Transmembrane Domain, CT: C terminal Tail.
2.2. Expression and Purification of SARS-CoV-2 Proteins
Optimized protocols were followed for transfection and culturing of CHOExpress™ cells (ExcellGene SA, Monthey, Switzerland). A transfection reagent mix (CHO4Tx®, ExcellGene SA, Monthey, Switzerland) was used to transfect cells in animal-component-free media with the vector pXLG-6 (ExcellGene SA, Monthey, Switzerland) containing SARS-CoV-2 spike DNA sequences, driven by promoter/enhancer sequences and associated elements. The vector includes an expression cassette for a puromycin resistance maker. Supernatants of high-density transient transfections under suspension culture were harvested after 10 days, mimicking a fed-batch process with ExcellGene’s animal component-free and protein-free CHO4Tx® PM (production medium) in 50-mL TubeSpin® bioreactor 50 tubes (TPP, Trasadingen, Switzerland). Cell culture process conditions, such as medium formulations, feeds, feed, and temperature shift timing. were evaluated at a 10-mL scale in TPP® TubeSpin 50 bioreactor tubes as previously described [17,18]. Cultures were shaken in a humidified ISF1-X/ISF3-X shaker (Kuhner, Birsfelden, Switzerland).
Recombinant CHO cell lines: After transfections, suspension cells were stringently selected with puromycin. Resulting stable pools of recombinant CHO cells with satisfying yields were kept. Clonal cell lines were obtained by image-assisted cell distribution into single wells of 96-well plates (f-sight, Cytena GmbH, Freiburg, Germany) for the Wuhan variant. Expanded cell populations with high-level expression for expected trimers and RDB monomers were frozen in mini-banks. Both a recombinant pool cell line for the RBD spike fragment, and the lead clonal cell line for the trimeric spike were used for scale-up in an optimized fed-batch process at the 0.2-, 10-, and 50-L bioreactor scale of operation.
The production medium employed EX-CELL® Advanced™ CHO Fed-batch medium (Merck KGaA, Darmstadt, Germany). Bioreactors were seeded at 5 × 105 cells/mL at 37 °C. On day 4, the cell culture was shifted to 31 °C, and animal component-free 7a and 7b feeds (HyClone Cell Boost, Cytiva, Marlborogh, MA, USA) were used according to an ExcellGene optimized procedure. Production culture fluids were harvested, clarified, and subjected to purification by affinity chromatography after pumping them through layered (5, 0.6, and 0.2 µm) harvest filters to remove cells. Loading onto, washing of, and elution from a Ni-Sepharose column were optimized, following the resin producers’ suggestions. The eluted product stream was loaded on a Size-Exclusion column (SEC, Superdex 200 pg, for further purification, following a tangential flow filtration (TFF) with a cut-off of 100 kD for trimeric spike products. All materials for down-stream processing were purchased from Cytiva, Marlborogh, MA, USA or its’ European distributors. The step of TFF was omitted for the RBD fragment.
2.3. Electron Microscopy Analysis of Trimeric Spikes
Frozen aliquots of SARS-CoV-2 trimeric spike (Wuhan) and D614G-variant trimeric spike were thawed and purified by size exclusion chromatography (Superose 6 Increase 10/300, Cytiva, GE Healthcare, Marlborogh, MA, USA) using 2 mM Tris pH 8.0, 200 mM NaCl, and 0.02% (w/v) NaN3. Elution fractions containing trimeric spike were collected and diluted to 0.05 mg/mL. Samples were deposited onto plasma-cleaned carbon-coated copper grids (CF400 mesh, Electron Microscopy Sciences, Hatfield, PA, USA), and stained in 2% (w/v) uranyl acetate at pH 7.0. Grids were imaged at a resolution of 92,000× in a Talos F200C transmission electron microscope equipped with a Ceta 16M detector (ThermoFisher Scientific, Waltham, MA, USA). The pixel size was 1.63 Å. Contrast transfer function estimation and particle picking were performed in cisTEM [19]. Extracted particles were exported to cryoSPARC-v2 (Structura Biotechnology Inc., Toronto, ON, Canada) for 2-D classification, ab initio 3-D reconstruction, and homogeneous refinement. Three-fold symmetry (C3) was imposed during the final round of refinement.
2.4. Inhibition of SARS-CoV-2 Infection by SARS-CoV-2 Proteins
Briefly, SARS-CoV-2 virus (MOI of 0.01) was mixed with cell culture medium containing two-fold serial dilutions of RBD or trimeric spike, ranging in concentration from 240 to 0.244 µM. This mixture was inoculated onto Vero E6 cells and at 48 h post infection, SARS-CoV-2 infection was visualized using virus nucleocapsid antigen-specific staining (red) and by a cell nucleus-specific staining (blue), as described in [20]. Quantification of fluorescence was done as previously described [21].
2.5. Detection of SARS-CoV-2 Antibodies Using SARS-CoV-2 Proteins
A bead-based serological assay [22] was used. Patient sera were collected and reacted with commercially available RBD and monomeric S1 domain protein, as well as trimeric spike and RBD. Briefly, 11 µg of RBD (Sino Biologics, Bejing, China, Cat. No. 40592-V08H), RBD (ExcellGene), monomeric spike S1 (Sino Biologics, Bejing, China, Cat. No. 40591-V08H), or trimeric spike (ExcellGene), were loaded on 100 µL of Microplex fluorescent beads and reacted with IgG-containing sera. Relative optical readings were taken for each protein-specific assay using the control sera and the COVID-19 sera. The relative reading for individual samples ranged from 0.01 to about 1000 AU/mL. The sensitivity and specificity of the assay for each of the tested proteins was plotted in a receiver operating characteristic (ROC) plot.
3. Results and Discussion
3.1. Design and Selection of CHO Manufacturable SARS CoV-2 Trimeric Spikes
In order to define the optimal construct design to produce a soluble trimeric spike protein, we evaluated the expression with various S protein variants (Figure 1) by CHO transient expression. The transmembrane domain and the C terminal intracellular tail were removed and replaced by a T4 foldon DNA sequence [14] with or without (GGGS)n linker and an 8×His tag encoding DNA. The furin cleavage site RRAR is mutated to be non-functional. WT spike sequences of both the Wuhan strain and the D614G strain and amino-acid changes K986P/V987P (“2P”) for locking the protein into a prefusion conformation [11] were also used. In addition, a receptor-binding domain truncation of the trimeric spike was used (RBD-His). S proteins variants with a scrambled furin cleavage site were found to be better expressed. Mutation of the furin cleavage site seemed to increase trimer assembly and/or stability, suggesting that trimer assembly via T4 foldon occurs in the Golgi (trans-Golgi network where furin is active). The same construct modifications were also applied for the D614G mutation in the spike protein, which has recently replaced to a large extent the Wuhan version.
Based on the relative expression levels in transient expressions with CHO cells combined with the efficiency of trimerization (Table 1), the spike_ΔCter_ΔFurin_2P_T4_His design was selected for further evaluation. This design will be referred to in the following as trimeric spike.

Table 1.
Overview of the relative expression levels of the different SARS-CoV-2 Spike protein designs evaluated under fed-batch conditions, as well as their ability to form trimers in culture supernatants. ΔCter in the table refers to a construct that has the transmembrane and intracellular (TM, CT) section of the protein deleted.
3.2. CHO Expression and Purification of SARS-CoV-2 Proteins
Stable recombinant cell pools expressing trimeric spike and RBD were generated using puromycin selection. From a total of 300 clonally derived cell populations, those with the highest expression levels of trimeric spike and RBD were expanded and a fed-batch production process was developed. Optimal conditions were selected on the basis of product yield, product quality, viable cell density (VCD), and cell viability. The production process was scaled up to 200-mL shake flasks, to 10- and to 40-L stirred tank bioreactors (STR). Viable cell density (VCD) and viability remained high for at least 10 days (Figure 2). Final harvest titers of hundreds of milligram/L were observed for both trimer protein and for RDB monomers at all scales. In addition, viable cell density (VCD) and viability profiles were highly comparable between shake flasks, 10- and 40-L STRs, indicating process scalability. From the authors’ experience in developing high-yielding manufacturing processes for clinically relevant recombinant antibodies and other proteins for almost 20 years, there is high confidence to assure feasibility to any scale of operation, up and beyond the 2000-L scale.
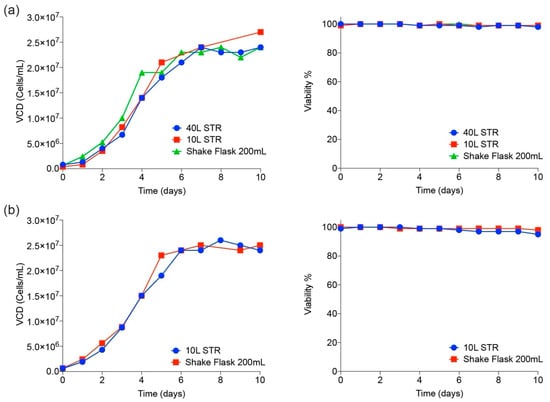
Figure 2.
Cell culture scale-up performance in shake flasks and bioreactors. Viable cell densities (cells/mL) and viability (%) are shown at various scales of operation for both trimeric spike (a) and RBD (b).
Krammer [23] rightly points to expression and scale-up difficulties for the production of large and complex proteins in global SARS-CoV-2 vaccines, such as the trimeric spike. However, since the mid 1980s, CHO cells have delivered hundreds of kilograms and tons of proteins from bioreactors at scales of up to 20,000 L [24]. One leading subunit vaccine candidate similar to the one presented here includes the transmembrane region of the trimeric spike protein and is produced and purified from Baculovirus-infected insect cells (SF-9). To the knowledge of the authors, this technology has not been scaled up to any scale larger than 100 L. This vaccine may face a serious manufacturability issue, when intended for global use. Additionally, insect cells present a different glycosylation pattern on proteins (pauci-manosidic, alpha 1-3 fucose) than mammalian cell systems [25]. One could argue that a manufacturing system with more human-like glycosylation patterns could be a superior vaccine, since it mimics more closely the authentic protein. This is particularly important since each spike monomer has over 20 predicted N- and O-glycosylation sites [26] that reveal the polypeptide backbone for potential antigenic epitopes only at a part of its surface [27].
3.3. Molecular Characterization and Electron Microscopy Analysis of Trimeric Spikes
Trimeric spike and RBD from cell-free culture supernatant were purified using an immobilized metal affinity chromatography (IMAC) capture step followed by preparative size exclusion. Eluates were concentrated and formulated at 1 mg/mL in phophate-buffered salt solution, H 7.4. Purity of the final products was estimated to be over 95% as analyzed by HPLC-SEC (Figure 3a,b).
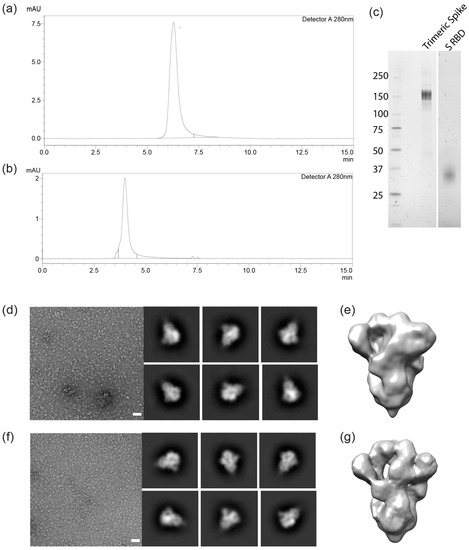
Figure 3.
Characterization of the recombinant trimeric spike and RBD. (a,b): Size-exclusion chromatography (SEC) analysis plot with a single peak at 4.0 and 6.4 min for trimeric spike and RBD, respectively. SDS-PAGE gel showing bands for trimeric spike (c) and for RBD monomer (d). Negative-stain electron microscopy of the SARS-CoV-2 trimeric spike of the Wuhan strain (d,e) and for the D614G variant of the virus (f,g). White bars in d and f: 50 nm. Six 2-D class averages are shown to the right of each representative micrograph. 3-D reconstructions are shown in (e,g).
HPLC-SEC (non-denaturing conditions) indicates that the S protein complex is trimeric (about 460 kDa) and the RBD is monomeric. The expected sizes of 150–160 and 29 kDa for the spike and RBD monomers, respectively, were confirmed by reducing SDS-PAGE (Figure 3c,d). The heavy glycosylation of trimeric spike contributes significantly to size heterogeneity, as has been seen with other CHO cell-produced virus-derived proteins, such as truncated surface proteins from Ebola and HIV. Presently ongoing work will characterize the glycosylation of the trimeric spike from the CHO cell process. Trimeric confirmation was also confirmed using negative-stain electron microscopy and 3-D modeling (Figure 3d,g).
3.4. Inhibition of SARS-CoV-2 Infection by SARS-CoV-2 Proteins
SARS-CoV-2 virus was mixed with serially diluted RBD or trimeric spike and the mixture was inoculated on Vero E6 cells. Then, 48 h later, SARS-CoV-2 infection was visualized and quantified (Figure 4a,b). The presence of both the RBD spike fragment and trimeric spike reduced infection of Vero E6 cells. For RBD, a reduction of infectivity was observed at a concentration of 20 µM and higher. For the trimeric spike (both for the Wuhan variant and the D614G variant), a near 100-fold lower concentration reduced infection (Figure 4b). Taken together, these results show that CHO-produced trimeric spike and RBD are able to compete with SARS-CoV-2 for binding to host cells. No differences were observed between the 614D (Wuhan) and the D614G variants of the trimeric spike.
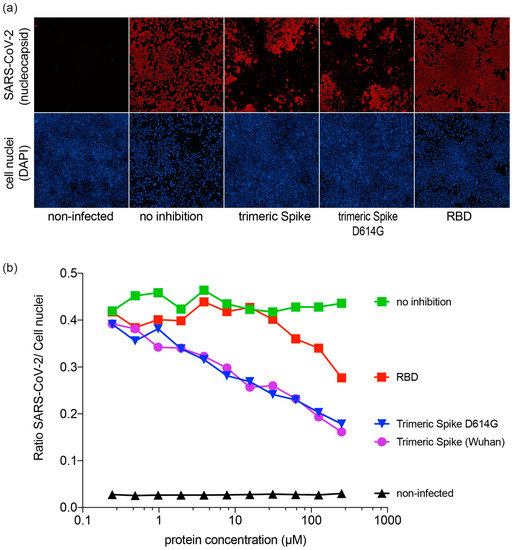
Figure 4.
Inhibition of SARS-CoV-2 infection in Vero E6 cells; (a) Inhibitory effect of trimeric spike and RBD on infection of Vero E6 cells. SARS-CoV-2 antigen-positive cells were visualized by immunofluorescent staining. Virus nucleocapsid antigen staining (red), cell nucleus staining (blue); (b) Inhibitory effect of trimeric spike and RBD on infection of Vero E6 cells.
3.5. Sensitivity and Specificity of SARS CoV-2 Spike Proteins for Detection of Antibodies
The trimeric spike and RBD proteins were used to detect SARS-CoV-2-specific IgG in a semi-quantitative bead-based serological assay [22]. We specifically selected a set of COVID-19 serum samples that cover a large range of SARS-CoV-2-specific antibody concentrations. Sera (n = 72) were collected from PCR-confirmed non-hospitalized (n = 63) and hospitalized (n = 9) COVID-19 patients between days 4 and 40 after disease onset. Control sera (n = 79) were collected before the emergence of SARS-CoV-2 from patients with seasonal corona-induced influenza-like illness (n = 44), non-corona influenza-like illness (n = 29), and from healthy individuals (n = 6).
For all proteins, the median signal intensity for the COVID samples was over 400-fold higher than for the control samples (Figure 5a). Receiver operator characteristic (ROC) analysis (Figure 5b) showed that trimeric spike (both Wuhan and D614G) provided higher test sensitivity and higher specificity characteristics compared to both S1 monomer and commercial RBD produced in Human Embryo Kidney 293 cells (p = 0.03; 0.04), and RBD produced in CHO (p = 0.05). Statistical analysis [22,28] showed no differences between the ROC of S1 monomer and both RBDs, neither between Wuhan nor D614G trimeric spikes. Additionally, taken together, our results indicate that CHO-produced trimeric spike can deliver higher diagnostic specificity and sensitivity than S1 monomer and RBD. These observations are in line with previous reports [5].
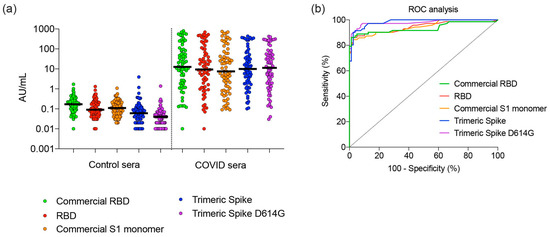
Figure 5.
Performance of RBD, monomeric S1, and trimeric spike in a diagnostic assay to identify COVID-19 patients. (a) Control sera (n = 151) and COVID-19 sera (n = 72) collected at day 4–40 of symptoms were tested and compared for concentrations of immunoglobulin G. Median concentration and 95% confidence intervals are shown. (b) The sera tested in (a) were analyzed by ROC. Abbreviations: AU, arbitrary unit; RBD, receptor-binding domain; ROC, receiver operator characteristic; S1, Spike protein subunit 1.
4. Conclusions
Using an optimized CHO expression system, and a scalable chemically defined production process, trimeric SARS CoV-2 spike proteins with mammalian-type glycosylation could be provided in sufficient quantities. Analysis of the resulting protein shows that it is of high purity and trimeric. Functional analysis showed it to be efficient in blocking virus infectivity in an in vitro model. In addition, the diagnostic performance of the trimeric spike for SARS-CoV-2-specific IgG was shown to exceed that of RDB and monomeric S1 protein.
Author Contributions
Conceptualization, P.P., D.K., M.J.W., J.K. and F.M.W.; methodology, P.P., J.K., R.D., V.A., G.C., J.S.M., D.K., M.J.W.; validation, P.P., J.K., D.K., M.J.W.; molecular analysis and EM work, D.K., J.S.M., P.O.B.; reactivity toward sera, P.R., R.D., G.d.H.; formal analysis, F.M.W., P.P., J.K., J.S.M., R.D.; writing—first draft preparation, F.M.W.; writing—review and editing, P.P., J.K., F.M.W.; supervision, P.P., J.K., D.K.; project administration, M.J.W., D.K.; strategy and planning, A.V.K. All authors have read and agreed to the published version of the manuscript.
Funding
ExcellGene funded internal research and provided materials cost free to academic partners. The work of V.A. was partially funded by a post-doctoral grant from the Swiss Secretary of State for Research, Education and Innovation.
Acknowledgments
Frederico Pratesi and Paola Migliorini (Clinical Immunology Laboratory, University of Pisa, Italy, are thanked for rapid and early provisioning of Covid-19 sera providing encouraging data to the team towards further and more profound work on this program.
Conflicts of Interest
The authors declare no conflict of interest, but disclose that ExcellGene is providing some materials as the result of this work commercially to interested parties. ExcellGene authors declare that the interpretation of the results is done with the highest standards of objectivity. Conclusions and interpretation of results of non-ExcellGene authors (blocking virus infectivity, reactivity against patient sera, etc.) were based on the judgement of the co-authoring expert scientists outside of the ExcellGene community.
References
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- New Pfizer Results: Coronaviurs Vaccine Is Safe and 95% protective. Available online: https://www.nytimes.com/2020/11/18/health/pfizer-covid-vaccine.html (accessed on 18 November 2020).
- Early Data Show Moderna’s Coronavirus Vaccine Is 94.5% Effective. Available online: https://www.nytimes.com/2020/11/16/health/Covid-moderna-vaccine.html (accessed on 22 November 2020).
- Fenwick, C.; Croxatto, A.; Coste, A.T.; Pojer, F.; André, C.; Pellaton, C.; Farina, A.; Campos, J.; Hacker, D.; Lau, K.; et al. Changes in SARS-CoV-2 Spike versus nucleoprotein antibody responses impact the estimates of infections in population-based seroprevalence studies. J. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Su, B.; Guo, X.; Sun, W.; Deng, Y.; Bao, L.; Zhu, Q.; Zhang, X.; Zheng, Y.; Geng, C.; et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent Patients’ B cells. Cell 2020, 182, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, N.C.; Yuan, M.; Bangaru, S.; Torres, J.L.; Caniels, T.G.; van Schooten, J.; Zhu, X.; Lee, C.D.; Brouwer, P.J.M.; et al. Cross-neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. bioRxiv 2020. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Seydoux, E.; Homad, L.J.; MacCamy, A.J.; Parks, K.R.; Hurlburt, N.K.; Jennewein, M.F.; Akins, N.R.; Stuart, A.B.; Wan, Y.-H.; Feng, J.; et al. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity 2020, 53, 98–105. [Google Scholar] [CrossRef]
- Hsieh, C.-L.; Goldsmith, J.A.; Schaub, J.M.; DiVenere, A.M.; Kuo, H.-C.; Javanmardi, K.; Le, K.C.; Wrapp, D.; Lee, A.G.; Liu, Y.; et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020, 369, 1501–1505. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Benton, D.J.; Wrobel, A.G.; Xu, P.; Roustan, C.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nat. Cell Biol. 2020, 1–8. [Google Scholar] [CrossRef]
- Johari, Y.B.; Jaffé, S.R.P.; Scarrott, J.M.; Johnson, A.O.; Mozzanino, T.; Pohle, T.H.; Maisuria, S.; Bhayat-Cammack, A.; Lambiase, G.; Brown, A.J.; et al. Production of trimeric SARS-CoV-2 spike protein by CHO cells for serological COVID-19 testing. Biotechnol. Bioeng. 2020. [Google Scholar] [CrossRef] [PubMed]
- Güthe, S.; Kapinos, L.; Möglich, A.; Meier, S.; Grzesiek, S.; Kiefhaber, T. Very fast folding and association of a trimerization domain from bacteriophage T4 fibritin. J. Mol. Biol. 2004, 337, 905–915. [Google Scholar] [CrossRef]
- Zhao, Z.; Sokhansanj, B.A.; Malhotra, C.; Zheng, K.; Rosen, G.L. Genetic grouping of SARS-CoV-2 coronavirus sequences using informative subtype markers for pandemic spread visualization. PLoS Comput. Biol. 2020, 16, e1008269. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 812–827. [Google Scholar] [CrossRef] [PubMed]
- De Jesus, M.J.; Girard, P.; Bourgeois, M.; Baumgartner, G.; Jacko, B.; Amstutz, H.; Wurm, F.M. TubeSpin satellites: A fast track approach for process development with animal cells using shaking technology. Biochem. Eng. J. 2004, 17, 217–223. [Google Scholar] [CrossRef]
- Gomez, N.; Ambhaikar, M.; Zhang, L.; Huang, C.-J.; Barkhordarian, H.; Lull, J.; Gutierrez, C. Analysis of Tubespins as a suitable scale-down model of bioreactors for high cell density CHO cell culture. Biotechnol. Prog. 2017, 33, 490–499. [Google Scholar] [CrossRef]
- Grant, T.; Rohou, A.; Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. eLife 2018, 7, e35383. [Google Scholar] [CrossRef]
- Thao, T.T.N.; Labroussaa, F.; Ebert, N.; V’Kovski, P.; Stalder, H.; Portmann, J.; Kelly, J.; Steiner, S.; Holwerda, M.; Kratzel, A.; et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nat. Cell Biol. 2020, 582, 561–565. [Google Scholar]
- Holwerda, M.; V’kovski, P.; Wider, M.; Thiel, V.; Dijkman, R. Identification of Five Antiviral Compounds from the Pandemic Response Box Targeting SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hartog, G.D.; Schepp, R.M.; Kuijer, M.; GeurtsvanKessel, C.; Van Beek, J.; Rots, N.; Koopmans, M.P.G.; Van Der Klis, F.R.M.; Van Binnendijk, R.S. SARS-CoV-2–specific antibody detection for seroepidemiology: A multiplex analysis approach accounting for accurate seroprevalence. J. Infect. Dis. 2020, 222, 1452–1461. [Google Scholar] [CrossRef]
- Krammer, F. SARS-CoV-2 vaccines in development. Nat. Cell Biol. 2020, 586, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Wurm, F.M. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 2004, 22, 1393–1398. [Google Scholar] [CrossRef]
- Shi, X. Protein N-glycosylation in the baculovirus-insect cell system. Curr. Drug Targets 2007, 8, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Grant, O.C.; Montgomery, D.; Ito, K.; Woods, R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Hanley, J.A.; McNeil, B.J. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 1983, 148, 839–843. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).