Adjusting Organic Load as a Strategy to Direct Single-Stage Food Waste Fermentation from Anaerobic Digestion to Chain Elongation
Abstract
1. Introduction
2. Materials and Methods
3. Results
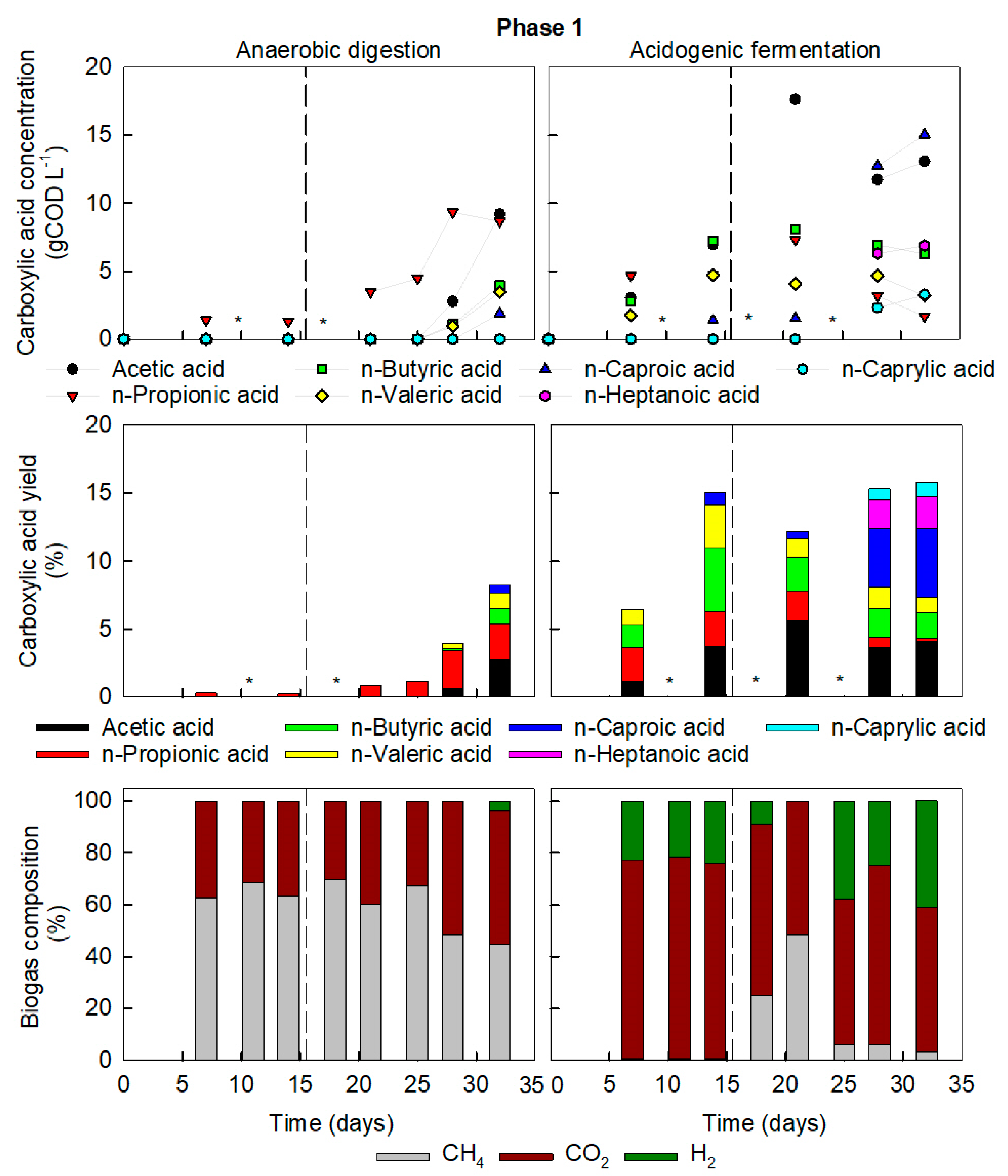
3.1. Elevated Organic Load Directed Anaerobic Digester Sludge towards Acidogenic Fermentation
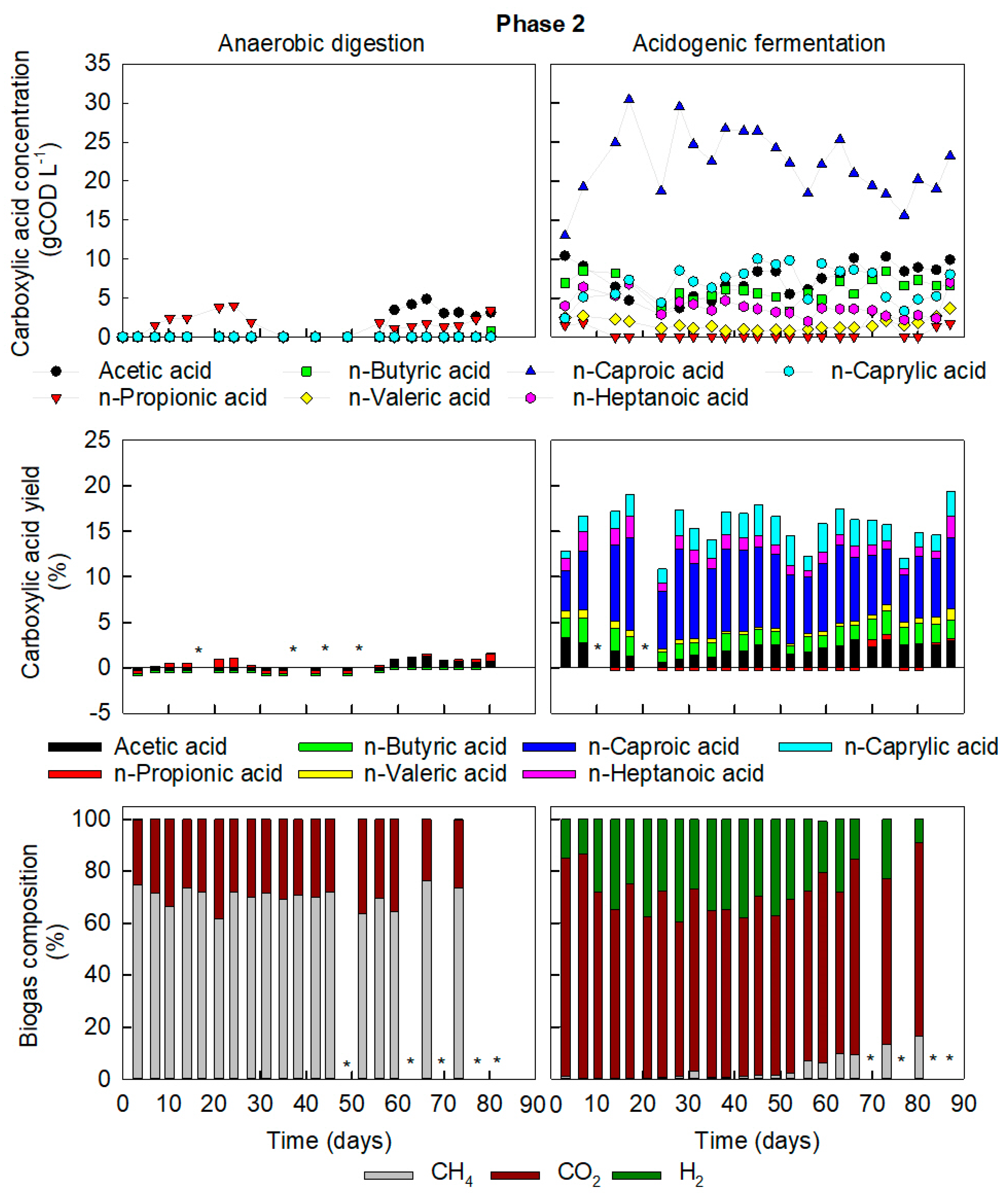
3.2. High-COD Food Waste Required Increased Retention Times for AD but Promoted Chain Elongation in AF
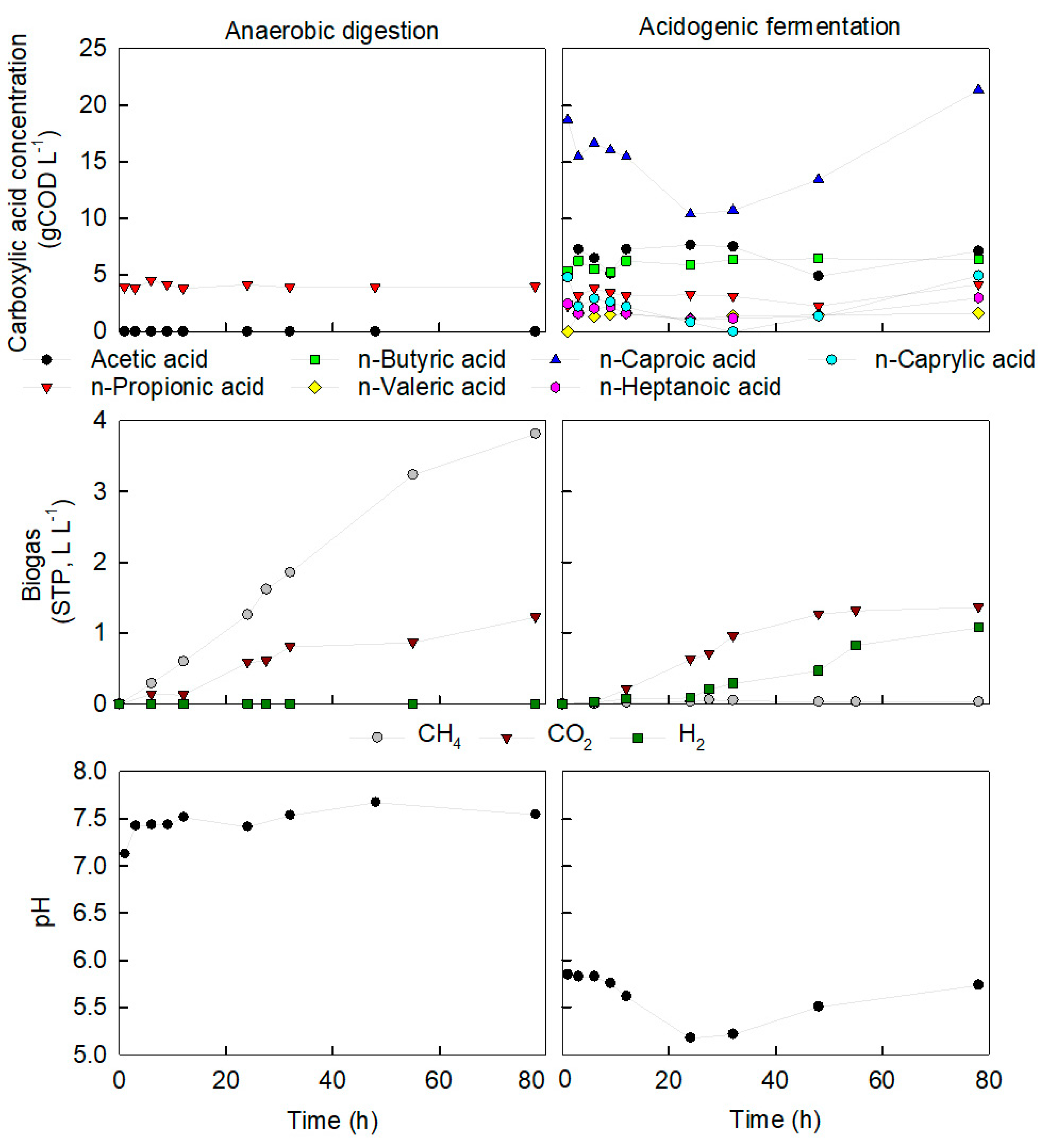
3.3. Presence of Hydrogen and pH Stabilisation Indicate Chain Elongation
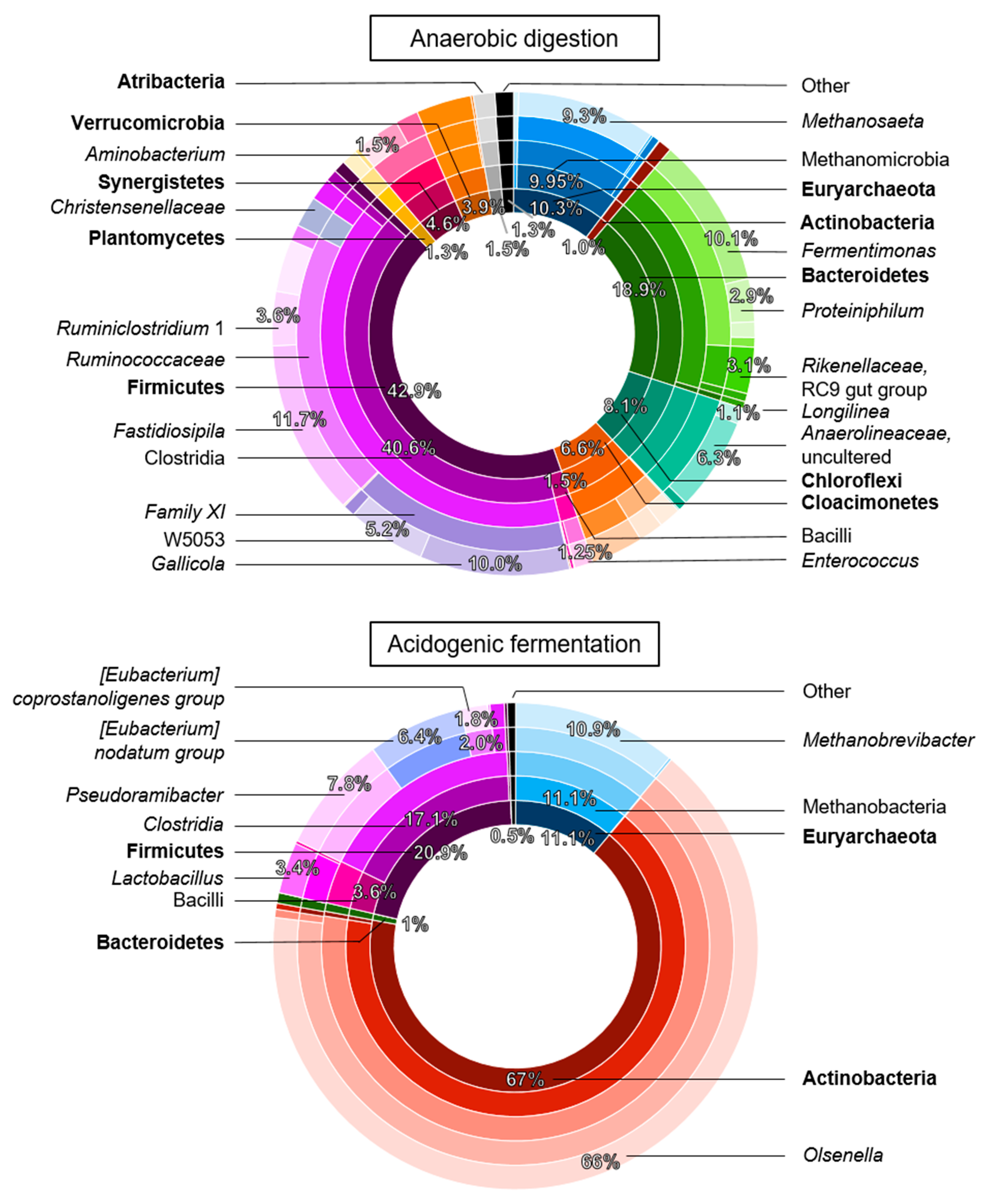
3.4. A Distinct Enriched Microbiome for Chain Elongation
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Teigiserova, D.A.; Hamelin, L.; Thomsen, M. Review of high-value food waste and food residues biorefineries with focus on unavoidable wastes from processing. Resour. Conserv. Recycl. 2019, 149, 413–426. [Google Scholar] [CrossRef]
- Stenmarck, Å.; Jensen, C.; Quested, T.; Moates, G. Estimates of European food Waste Levels. 2016. Available online: https://www.eu-fusions.org/phocadownload/Publications/Estimates%20of%20European%20food%20waste%20levels.pdf (accessed on 17 November 2020).
- Xu, F.; Li, Y.; Ge, X.; Yang, L.; Li, Y. Anaerobic digestion of food waste—Challenges and opportunities. Bioresour. Technol. 2018, 247, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Agler, M.T.; Wrenn, B.A.; Zinder, S.H.; Angenent, L.T. Waste to bioproduct conversion with undefined mixed cultures: The carboxylate platform. Trends Biotechnol. 2011, 29, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Urban, C.; Xu, J.; Sträuber, H.; dos Santos Dantas, T.R.; Mühlenberg, J.; Härtig, C.; Angenent, L.T.; Harnisch, F. Production of drop-in fuels from biomass at high selectivity by combined microbial and electrochemical conversion. Energy Environ. Sci. 2017, 10, 2231–2244. [Google Scholar] [CrossRef]
- De Groof, V.; Coma, M.; Arnot, T.; Leak, D.J.; Lanham, A.B. Medium chain carboxylic acids from complex organic feedstocks by mixed culture fermentation. Molecules 2019, 24, 398. [Google Scholar] [CrossRef]
- Han, W.; He, P.; Shao, L.; Lü, F. Road to full bioconversion of biowaste to biochemicals centering on chain elongation: A mini review. J. Environ. Sci. 2019, 86, 50–64. [Google Scholar] [CrossRef]
- Vanwonterghem, I.; Jensen, P.D.; Rabaey, K.; Tyson, G.W. Temperature and solids retention time control microbial population dynamics and volatile fatty acid production in replicated anaerobic digesters. Sci. Rep. 2015, 5, 8496. [Google Scholar] [CrossRef] [PubMed]
- Greses, S.; Tomás-Pejó, E.; González-Fernández, C. Short-chain fatty acids and hydrogen production in one single anaerobic fermentation stage using carbohydrate-rich food waste. J. Clean. Prod. 2020. [Google Scholar] [CrossRef]
- Kim, M.S.; Na, J.G.; Lee, M.K.; Ryu, H.; Chang, Y.K.; Triolo, J.M.; Yun, Y.M.; Kim, D.H. More value from food waste: Lactic acid and biogas recovery. Water Res. 2016, 96, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, W.; Gu, X.; Guo, Z.; Song, J.; Zhu, D.; Liu, Y.; Liu, Y.; Xue, G.; Li, X.; et al. Stabilizing lactate production through repeated batch fermentation of food waste and waste activated sludge. Bioresour. Technol. 2020, 300, 122709. [Google Scholar] [CrossRef]
- Cavinato, C.; Giuliano, A.; Bolzonella, D.; Pavan, P.; Cecchi, F. Bio-hythane production from food waste by dark fermentation coupled with anaerobic digestion process: A long-term pilot scale experience. Int. J. Hydrog. Energy 2012, 37, 11549–11555. [Google Scholar] [CrossRef]
- Im, S.; Lee, M.-K.; Yun, Y.-M.; Cho, S.-K.; Kim, D.-H. Effect of storage time and temperature on hydrogen fermentation of food waste. Int. J. Hydrog. Energy 2020, 45, 3769–3775. [Google Scholar] [CrossRef]
- Spirito, C.M.; Richter, H.; Rabaey, K.; Stams, A.J.; Angenent, L.T. Chain elongation in anaerobic reactor microbiomes to recover resources from waste. Curr. Opin. Biotechnol. 2014, 27, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Weimer, P.J.; Kohn, R.A. Impacts of ruminal microorganisms on the production of fuels: How can we intercede from the outside? Appl. Microbiol. Biotechnol. 2016, 100, 3389–3398. [Google Scholar] [CrossRef]
- Chen, W.S.; Strik, D.; Buisman, C.J.N.; Kroeze, C. Production of caproic acid from mixed organic waste: An Environmental life cycle perspective. Environ. Sci. Technol. 2017, 51, 7159–7168. [Google Scholar] [CrossRef]
- Arslan, D.; Steinbusch, K.J.; Diels, L.; De Wever, H.; Hamelers, H.V.; Buisman, C.J. Selective carboxylate production by controlling hydrogen, carbon dioxide and substrate concentrations in mixed culture fermentation. Bioresour. Technol. 2013, 136, 452–460. [Google Scholar] [CrossRef]
- Steinbusch, K.J.; Arvaniti, E.; Hamelers, H.V.; Buisman, C.J. Selective inhibition of methanogenesis to enhance ethanol and n-butyrate production through acetate reduction in mixed culture fermentation. Bioresour. Technol. 2009, 100, 3261–3267. [Google Scholar] [CrossRef]
- Zhang, W.; Xing, W.; Li, R. Real-time recovery strategies for volatile fatty acid-inhibited anaerobic digestion of food waste for methane production. Bioresour. Technol. 2018, 265, 82–92. [Google Scholar] [CrossRef]
- Coma, M.; Martinez-Hernandez, E.; Abeln, F.; Raikova, S.; Donnelly, J.; Arnot, T.C.; Allen, M.J.; Hong, D.D.; Chuck, C.J. Organic waste as a sustainable feedstock for platform chemicals. Faraday Discuss 2017, 202, 175–195. [Google Scholar] [CrossRef]
- Jarboe, L.R.; Royce, L.A.; Liu, P. Understanding biocatalyst inhibition by carboxylic acids. Front. Microbiol. 2013, 4, 272. [Google Scholar] [CrossRef]
- Zhu, X.; Tao, Y.; Liang, C.; Li, X.; Wei, N.; Zhang, W.; Zhou, Y.; Yang, Y.; Bo, T. The synthesis of n-caproate from lactate: A new efficient process for medium-chain carboxylates production. Sci. Rep. 2015, 5, 14360. [Google Scholar] [CrossRef] [PubMed]
- Kucek, L.A.; Xu, J.; Nguyen, M.; Angenent, L.T. Waste conversion into n-Caprylate and n-Caproate: Resource recovery from wine lees using anaerobic reactor microbiomes and in-line extraction. Front. Microbiol. 2016, 7, 1892. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.J.; Candry, P.; Basadre, T.; Khor, W.C.; Roume, H.; Hernandez-Sanabria, E.; Coma, M.; Rabaey, K. Electrolytic extraction drives volatile fatty acid chain elongation through lactic acid and replaces chemical pH control in thin stillage fermentation. Biotechnol. Biofuels 2015, 8, 221. [Google Scholar] [CrossRef]
- Duber, A.; Jaroszynski, L.; Zagrodnik, R.; Chwialkowska, J.; Juzwa, W.; Ciesielski, S.; Oleskowicz-Popiel, P. Exploiting the real wastewater potential for resource recovery—n-Caproate production from acid whey. Green Chem. 2018, 20, 3790–3803. [Google Scholar] [CrossRef]
- Carvajal-Arroyo, J.M.; Candry, P.; Andersen, S.J.; Props, R.; Seviour, T.; Ganigué, R.; Rabaey, K. Granular fermentation enables high rate caproic acid production from solid-free thin stillage. Green Chem. 2019, 21, 1330–1339. [Google Scholar] [CrossRef]
- Wu, Q.; Feng, X.; Guo, W.; Bao, X.; Ren, N. Long-term medium chain carboxylic acids production from liquor-making wastewater: Parameters optimization and toxicity mitigation. Chem. Eng. J. 2020, 388. [Google Scholar] [CrossRef]
- Grootscholten, T.I.M.; Strik, D.P.B.T.B.; Steinbusch, K.J.J.; Buisman, C.J.N.; Hamelers, H.V.M. Two-stage medium chain fatty acid (MCFA) production from municipal solid waste and ethanol. Appl. Energy 2014, 116, 223–229. [Google Scholar] [CrossRef]
- Xu, J.; Hao, J.; Guzman, J.J.L.; Spirito, C.M.; Harroff, L.A.; Angenent, L.T. Temperature-phased conversion of acid whey waste into medium-chain carboxylic acids via lactic acid: No external e-donor. Joule 2018, 2, 280–295. [Google Scholar] [CrossRef]
- Khor, W.C.; Andersen, S.; Vervaeren, H.; Rabaey, K. Electricity-assisted production of caproic acid from grass. Biotechnol. Biofuels 2017, 10, 180. [Google Scholar] [CrossRef]
- Nzeteu, C.O.; Trego, A.C.; Abram, F.; O’Flaherty, V. Reproducible, high-yielding, biological caproate production from food waste using a single-phase anaerobic reactor system. Biotechnol. Biofuels 2018, 11, 108. [Google Scholar] [CrossRef]
- Ma, H.; Lin, Y.; Jin, Y.; Gao, M.; Li, H.; Wang, Q.; Ge, S.; Cai, L.; Huang, Z.; van Le, Q.; et al. Effect of ultrasonic pretreatment on chain elongation of saccharified residue from food waste by anaerobic fermentation. Environ. Pollut. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, N. Design and engineering of biogas plants. In Biogas Handbook: Science, Production and Applications; Wellinger, A., Murphy, J.D., Baxter, D., Braun, R., Eds.; Elsevier Science & Technology: Amsterdam, The Netherlands, 2013; Volume 52, pp. 191–211. [Google Scholar]
- Murphy, J.D.; Thamsiriroj, T. Fundametal science and engineering of the anaerobic digestion process for biogas production. In Biogas Handbook: Science, Production and Applications; Wellinger, A., Murphy, J.D., Baxter, D., Braun, R., Eds.; Elsevier Science & Technology: Amsterdam, The Netherlands, 2013; Volume 52. [Google Scholar]
- Lambrecht, J.; Cichocki, N.; Schattenberg, F.; Kleinsteuber, S.; Harms, H.; Muller, S.; Strauber, H. Key sub-community dynamics of medium-chain carboxylate production. Microb. Cell Fact. 2019, 18, 92. [Google Scholar] [CrossRef]
- Scarborough, M.J.; Lynch, G.; Dickson, M.; McGee, M.; Donohue, T.J.; Noguera, D.R. Increasing the economic value of lignocellulosic stillage through medium-chain fatty acid production. Biotechnol. Biofuels 2018, 11, 200. [Google Scholar] [CrossRef]
- Contreras-Davila, C.A.; Carrion, V.J.; Vonk, V.R.; Buisman, C.N.J.; Strik, D. Consecutive lactate formation and chain elongation to reduce exogenous chemicals input in repeated-batch food waste fermentation. Water Res. 2020, 169, 115215. [Google Scholar] [CrossRef] [PubMed]
- APHA. Standard methods for the examination of water and wastewater. Am. Public Health Assoc. 2017, 23. [Google Scholar] [CrossRef]
- Manni, G.; Caron, F. Calibration and determination of volatile fatty acids in waste leachates by gas chromatography. J. Chromatogr. A 1995, 690, 237–242. [Google Scholar] [CrossRef]
- Albertsen, M.; Karst, S.M.; Ziegler, A.S.; Kirkegaard, R.H.; Nielsen, P.H. Back to basics—The influence of DNA extraction and primer choice on phylogenetic analysis of activated sludge communities. PLoS ONE 2015, 10, e0132783. [Google Scholar] [CrossRef]
- Illumina Inc, 16S Metagenomic Sequencing Library Preparation. 2015. Available online: https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html (accessed on 17 November 2020).
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic. Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Envionment for Statistical Computing; Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.R-project.org (accessed on 16 November 2020).
- de Groof, V.; Coma Bech, M.; Leak, D.J.; Arnot, T.; Lanham, A. Dataset for “Adjusting organic load as a strategy to direct single-stage food waste fermentation from anaerobic digestion to chain elongation”. Bath Univ. Bath Res. Data Arch. 2020. [Google Scholar] [CrossRef]
- Siegert, I.; Banks, C. The effect of volatile fatty acid additions on the anaerobic digestion of cellulose and glucose in batch reactors. Process Biochem. 2005, 40, 3412–3418. [Google Scholar] [CrossRef]
- Voelklein, M.A.; O’Shea, R.; Jacob, A.; Murphy, J.D. Role of trace elements in single and two-stage digestion of food waste at high organic loading rates. Energy 2017, 121, 185–192. [Google Scholar] [CrossRef]
- Angenent, L.T.; Richter, H.; Buckel, W.; Spirito, C.M.; Steinbusch, K.J.; Plugge, C.M.; Strik, D.P.; Grootscholten, T.I.; Buisman, C.J.; Hamelers, H.V. Chain Elongation with Reactor Microbiomes: Open-culture biotechnology to produce biochemicals. Environ. Sci. Technol. 2016, 50, 2796–2810. [Google Scholar] [CrossRef]
- Prabhu, R.; Altman, E.; Eiteman, M.A. Lactate and acrylate metabolism by Megasphaera elsdenii under batch and steady-state conditions. Appl. Environ. Microbiol. 2012, 78, 8564–8570. [Google Scholar] [CrossRef]
- Kucek, L.A.; Nguyen, M.; Angenent, L.T. Conversion of L-lactate into n-caproate by a continuously fed reactor microbiome. Water Res. 2016, 93, 163–171. [Google Scholar] [CrossRef]
- Candry, P.; Radic, L.; Favere, J.; Carvajal-Arroyo, J.M.; Rabaey, K.; Ganigue, R. Mildly acidic pH selects for chain elongation to caproic acid over alternative pathways during lactic acid fermentation. Water Res. 2020, 186, 116396. [Google Scholar] [CrossRef] [PubMed]
- Sträuber, H.; Schröder, M.; Sabine Kleinsteuber, S. Metabolic and microbial community dynamics during the hydrolytic and acidogenic fermentation in a leach-bed process. Energy Sustain. Soc. 2012, 2, 1–10. [Google Scholar] [CrossRef]
- Yasin, N.H.; Mumtaz, T.; Hassan, M.A.; Abd Rahman, N. Food waste and food processing waste for biohydrogen production: A review. J. Environ. Manag. 2013, 130, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Cardinali-Rezende, J.; Rojas-Ojeda, P.; Nascimento, A.M.; Sanz, J.L. Proteolytic bacterial dominance in a full-scale municipal solid waste anaerobic reactor assessed by 454 pyrosequencing technology. Chemosphere 2016, 146, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhang, S.; Li, L.; Zhao, X.; Ma, Y.; Shi, D. Long-term high-solids anaerobic digestion of food waste: Effects of ammonia on process performance and microbial community. Bioresour. Technol. 2018, 262, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Hahnke, S.; Langer, T.; Koeck, D.E.; Klocke, M. Description of Proteiniphilum saccharofermentans sp. nov., Petrimonas mucosa sp. nov. and Fermentimonas caenicola gen. nov., sp. nov., isolated from mesophilic laboratory-scale biogas reactors, and emended description of the genus Proteiniphilum. Int. J. Syst. Evol. Microbiol. 2016, 66, 1466–1475. [Google Scholar] [CrossRef] [PubMed]
- Krieg, N.R.; Ludwig, W.; Euzéby, J.; Whitman, W.B. Phylum XIV. Bacteroidetes phyl. nov. In Bergey’s Manual® of Systematic Bacteriology; Krieg, N.R., Staley, J.T., Brown, D.R., Hedlund, B.P., Paster, B.J., Ward, N.L., Ludwig, W., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2010. [Google Scholar] [CrossRef]
- Baena, S.; Fardeau, M.-L.; Labat, M.; Ollivier, B.; Thomas, P.; Garcia, J.-L.; Patel, B.K.C. Aminobacterium colobiense gen. nov. sp. nov., an amino acid-degrading anaerobe isolated from anaerobic sludge. Anaerobe 1998, 4, 241–250. [Google Scholar] [CrossRef]
- Nelson, M.C.; Morrison, M.; Yu, Z. A meta-analysis of the microbial diversity observed in anaerobic digesters. Bioresour. Technol. 2011, 102, 3730–3739. [Google Scholar] [CrossRef]
- Braguglia, C.M.; Gallipoli, A.; Gianico, A.; Pagliaccia, P. Anaerobic bioconversion of food waste into energy: A critical review. Bioresour. Technol. 2018, 248, 37–56. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Paster, B.J.; Tzellas, N.; Coleman, B.; Downes, J.; Spratt, D.A.; Wade, W.G. Characterization of novel human oral isolates and closed 16S rDNA sequences that fall in the family coriobacteriaceae: Description of olsenella gen. nov., reclassification of lactobacillus ulo as olsenella uli comb. nov. and description of olsenella profusa sp. nov. Int. J. Syst. Evol. Microbiol. 2001, 51, 1797–1804. [Google Scholar]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kraatz, M.; Wallace, R.J.; Svensson, L. Olsenella umbonata sp. nov., a microaerotolerant anaerobic lactic acid bacterium from the sheep rumen and pig jejunum, and emended descriptions of Olsenella, Olsenella uli and Olsenella profusa. Int. J. Syst. Evol. Microbiol. 2011, 61, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Kleinsteuber, S.; Centler, F.; Harms, H.; Strauber, H. Competition between butyrate fermenters and chain-elongating bacteria limits the efficiency of medium-chain carboxylate production. Front. Microbiol. 2020, 11, 336. [Google Scholar] [CrossRef] [PubMed]
- Scarborough, M.J.; Myers, K.S.; Donohue, T.J.; Noguera, D.R. Medium-chain fatty acid synthesis by “Candidatus Weimeria bifida” gen. nov., sp. nov., and "Candidatus Pseudoramibacter fermentans" sp. nov. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef] [PubMed]
- Willems, A.; Collins, M.D. Genus VI. Pseudoramibacter. In Bergey’s manual® of Systematic Bacteriology: The Firmicutes, 2th ed.; de Vos, P., Garrity, G.M., Jones, D., Krieg, N.R., ludwig, W., Rainey, F.A., Schleifer, K., Whitman, W.B., Eds.; Springer: Berlin/Heidelberg, Germany, 1996; Volume 3, pp. 902–909. [Google Scholar]
- Freier, T.A.; Beitz, D.C.; Li, L.; Hartman, P.A. Characterization of Eubacterium coprostanoligenes sp. nov., a cholesterol-reducing anaerobe. Int. J. Syst. Bacteriol. 1994, 44, 137–142. [Google Scholar] [CrossRef]
- Niu, Q.; Kobayashi, T.; Takemura, Y.; Kubota, K.; Li, Y.Y. Evaluation of functional microbial community’s difference in full-scale and lab-scale anaerobic digesters feeding with different organic solid waste: Effects of substrate and operation factors. Bioresour. Technol. 2015, 193, 110–118. [Google Scholar] [CrossRef]
- Regueiro, L.; Veiga, P.; Figueroa, M.; Alonso-Gutierrez, J.; Stams, A.J.; Lema, J.M.; Carballa, M. Relationship between microbial activity and microbial community structure in six full-scale anaerobic digesters. Microbiol. Res. 2012, 167, 581–589. [Google Scholar] [CrossRef]
- Qiao, J.T.; Qiu, Y.L.; Yuan, X.Z.; Shi, X.S.; Xu, X.H.; Guo, R.B. Molecular characterization of bacterial and archaeal communities in a full-scale anaerobic reactor treating corn straw. Bioresour. Technol. 2013, 143, 512–518. [Google Scholar] [CrossRef]
- Jang, H.M.; Kim, J.H.; Ha, J.H.; Park, J.M. Bacterial and methanogenic archaeal communities during the single-stage anaerobic digestion of high-strength food wastewater. Bioresour. Technol. 2014, 165, 174–182. [Google Scholar] [CrossRef]
- Li, L.; He, Q.; Wei, Y.; He, Q.; Peng, X. Early warning indicators for monitoring the process failure of anaerobic digestion system of food waste. Bioresour. Technol. 2014, 171, 491–494. [Google Scholar] [CrossRef]
- Cavalcante, W.d.A.; Leitão, R.C.; Gehring, T.A.; Angenent, L.T.; Santaella, S.T. Anaerobic fermentation for n-caproic acid production: A review. Process Biochem. 2017, 54, 106–119. [Google Scholar] [CrossRef]
- Arslan, D.; Steinbusch, K.J.J.; Diels, L.; Hamelers, H.V.M.; Strik, D.P.B.T.B.; Buisman, C.J.N.; de Wever, H. Selective short-chain carboxylates production: A review of control mechanisms to direct mixed culture fermentations. Crit. Rev. Environ. Sci. Technol. 2016, 46, 592–634. [Google Scholar] [CrossRef]
- Roghair, M.; Liu, Y.; Strik, D.; Weusthuis, R.A.; Bruins, M.E.; Buisman, C.J.N. Development of an effective chain elongation process from acidified food waste and ethanol into n-caproate. Front. Bioeng. Biotechnol. 2018, 6, 50. [Google Scholar] [CrossRef]
- Duber, A.; Zagrodnik, R.; Chwialkowska, J.; Juzwa, W.; Oleskowicz-Popiel, P. Evaluation of the feed composition for an effective medium chain carboxylic acid production in an open culture fermentation. Sci. Total Environ. 2020, 728, 138814. [Google Scholar] [CrossRef]
- Daly, S.E.; Usack, J.G.; Harroff, L.A.; Booth, J.G.; Keleman, M.P.; Angenent, L.T. Systematic analysis of factors that affect food-waste storage: Toward maximizing lactate accumulation for resource recovery. ACS Sustain. Chem. Eng. 2020, 8, 13934–13944. [Google Scholar] [CrossRef]
- Fisgativa, H.; Tremier, A.; Dabert, P. Characterizing the variability of food waste quality: A need for efficient valorisation through anaerobic digestion. Waste Manag. 2016, 50, 264–274. [Google Scholar] [CrossRef]
- Andersen, S.J.; De Groof, V.; Khor, W.C.; Roume, H.; Props, R.; Coma, M.; Rabaey, K. A clostridium group iv species dominates and suppresses a mixed culture fermentation by tolerance to medium chain fatty acids products. Front. Bioeng. Biotechnol. 2017, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Domingos, J.M.B.; Puccio, S.; Martinez, G.A.; Amaral, N.; Reis, M.A.M.; Bandini, S.; Fava, F.; Bertin, L. Cheese whey integrated valorisation: Production, concentration and exploitation of carboxylic acids for the production of polyhydroxyalkanoates by a fed-batch culture. Chem. Eng. J. 2018, 336, 47–53. [Google Scholar] [CrossRef]
- Li, L.; Peng, X.; Wang, X.; Wu, D. Anaerobic digestion of food waste: A review focusing on process stability. Bioresour. Technol. 2018, 248, 20–28. [Google Scholar] [CrossRef]
- Louca, S.; Polz, M.F.; Mazel, F.; Albright, M.B.N.; Huber, J.A.; O’Connor, M.I.; Ackermann, M.; Hahn, A.S.; Srivastava, D.S.; Crowe, S.A.; et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018, 2, 936–943. [Google Scholar] [CrossRef]
- Marshall, C.W.; LaBelle, E.V.; May, H.D. Production of fuels and chemicals from waste by microbiomes. Curr. Opin. Biotechnol. 2013, 24, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Agler, M.T.; Spirito, C.M.; Usack, J.G.; Werner, J.J.; Angenent, L.T. Chain elongation with reactor microbiomes: Upgrading dilute ethanol to medium-chain carboxylates. Energy Environ. Sci. 2012, 5. [Google Scholar] [CrossRef]
- Gupta, H.; Barua, M.K. A framework to overcome barriers to green innovation in SMEs using BWM and Fuzzy TOPSIS. Sci. Total Environ. 2018, 633, 122–139. [Google Scholar] [CrossRef] [PubMed]
- Garrone, P.; Grilli, L.; Groppi, A.; Marzano, R. Barriers and drivers in the adoption of advanced wastewater treatment technologies: A comparative analysis of Italian utilities. J. Clean. Prod. 2018, 171, S69–S78. [Google Scholar] [CrossRef]
| Parameter | FW 1 | FW 2 |
|---|---|---|
| pH | 5.0 ± 0.1 | 5.4 ± 0.1 |
| Conductivity (mS cm−1) | 6.20 | 6.16 |
| Solid Content (% w/w) | ||
| Total Solids | 9.94 ± 0.07 | 17.7 ± 1.6 |
| Volatile Solids | 8.832 ± 0.002 | 16.3 ± 1.1 |
| Chemical Oxygen Demand (gCOD L−1) | ||
| Total COD | 150 ± 1 | 297 ± 9 |
| Soluble COD | 37.4 ± 1.1 | 38.2 ± 0.1 |
| Soluble Compounds (g L−1) | ||
| Acetic acid | 1.27 ± 0.18 | 0.89 ± 0.16 |
| n-Propionic acid | 0.63 ± 0.02 | 0.63 ± 0.19 |
| n-Butyric acid | <0.31 | 0.35 ± 0.24 |
| MCCA (C5–C8) | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Glucose | 3.61 ± 0.14 | 5.03 ± 0.87 |
| Sugar compounds * | 0.55 ± 0.07 | 5.34 ± 2.13 |
| Lactic acid | 7.27 ± 0.34 | 2.48 ± 0.02 |
| Ethanol | 1.39 ± 0.71 | 0.80 ± 0.16 |
| STR | Feedstock | Days | OLR (gCOD L−1 d−1) | HRT (d) |
|---|---|---|---|---|
| Phase 1—Shift functionality with increased organic load | ||||
| AD | FW 1 | 0–14 | 4.2 ± 0.4 | 35 ± 3 |
| FW 2 | 14–32 | 8.5 ± 0.8 | 35 ± 3 | |
| AF | FW 1 | 0–14 | 8.5 ± 0.7 | 18 ± 2 |
| FW 2 | 14–32 | 17.1 ± 1.5 | 18 ± 2 | |
| Phase 2—Establish longer term operation | ||||
| AD | FW 2 | 0–80 | 4.4 ± 0.5 | 69 ± 6 |
| AF | FW 2 | 0–10 | NA * | NA * |
| FW 2 | 10–87 | 21.3± 1.6 | 14 ± 1 | |
| Index | AD | AF |
|---|---|---|
| Observed OTUs | 159 ± 4 | 102 ± 7 |
| Shannon | 3.49 ± 0.04 | 1.70 ± 0.08 |
| Simpson | 0.945 ± 0.007 | 0.66 ± 0.04 |
| InvSimpson | 18.6 ± 0.8 | 2.9 ± 0.3 |
| Chao1 (richness) | 182 ± 16 | 129 ± 5 |
| ACE (richness) | 184 ± 18 | 135 ± 14 |
| Pielou evenness | 0.625 ± 0.007 | 0.31 ± 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Groof, V.; Coma, M.; Arnot, T.C.; Leak, D.J.; Lanham, A.B. Adjusting Organic Load as a Strategy to Direct Single-Stage Food Waste Fermentation from Anaerobic Digestion to Chain Elongation. Processes 2020, 8, 1487. https://doi.org/10.3390/pr8111487
De Groof V, Coma M, Arnot TC, Leak DJ, Lanham AB. Adjusting Organic Load as a Strategy to Direct Single-Stage Food Waste Fermentation from Anaerobic Digestion to Chain Elongation. Processes. 2020; 8(11):1487. https://doi.org/10.3390/pr8111487
Chicago/Turabian StyleDe Groof, Vicky, Marta Coma, Tom C. Arnot, David J. Leak, and Ana B. Lanham. 2020. "Adjusting Organic Load as a Strategy to Direct Single-Stage Food Waste Fermentation from Anaerobic Digestion to Chain Elongation" Processes 8, no. 11: 1487. https://doi.org/10.3390/pr8111487
APA StyleDe Groof, V., Coma, M., Arnot, T. C., Leak, D. J., & Lanham, A. B. (2020). Adjusting Organic Load as a Strategy to Direct Single-Stage Food Waste Fermentation from Anaerobic Digestion to Chain Elongation. Processes, 8(11), 1487. https://doi.org/10.3390/pr8111487






