Abstract
Presented herein is a model-driven strategy for characterizing the production capability of expression host and subsequently identifying targets for strain improvement by resorting to network structural comparison with reference strain and in silico analysis of genome-scale metabolic model. The applicability of the strategy was demonstrated by exploring the capability of Zymomonas mobilis, as a succinic acid producer. Initially, the central metabolism of Z. mobilis was compared with reference producer, Mannheimia succiniciproducens, in order to identify gene deletion targets. It was followed by combinatorial gene deletion analysis. Remarkably, resultant in silico strains suggested that knocking out pdc, ldh, and pfl genes encoding pyruvate-consuming reactions as well as the cl gene leads to fifteen-fold increase in succinic acid molar yield. The current exploratory work could be a promising support to wet experiments by providing guidance for metabolic engineering strategies and lowering the number of trials and errors.
1. Introduction
Succinic acid (SA) is a valuable specialty chemical with a wide range of applications in food, pharmaceutical, and chemical industries, currently being synthesized through petrochemical processes by the catalytic hydrogenation of petroleum-derived maleic anhydride [1]. However, the increase in oil prices along with raising environmental awareness has made bio-based SA production an attractive option. Thus, a multitude of studies have attempted to develop SA-producing microbial fermentation systems coupled with the use of renewable biomasses [2,3]. Several well-known SA producers include Anaerobiospirillum succiniciproducens, Actinobacillus succinogenes, Mannheimia succiniciproducens, and recombinant Escherichia coli. Generally, SA is excreted during anaerobic fermentation, which has some operational disadvantages such as low cell growth and slow carbon uptake, leading to its low productivity. In addition, these producers secrete other competing fermentative byproducts, e.g., acetic acid, lactic acid, and formic acid, thus reducing SA yield and making its purification difficult.
To overcome the above limitations, researchers have tried to develop various metabolic engineering strategies to enhance SA production mostly in E. coli [4,5,6,7,8]. For example, yield was significantly increased by amplifying the enzymatic reactions involved in SA pathway [9,10,11] and by introducing non-indigenous enzymes to E. coli [12,13]. Other interesting options are the inactivation of pathways that compete with SA [14,15] and the construction of glyoxylate cycle to enable aerobic succinate production [16,17,18]. For other producers, several studies were also carried out to reduce byproduct formations in A. succiniciproducens [19,20], A. succinogenes [21,22,23], and M. succiniciproducens [24,25].
In summary, various strategies described above have successfully enhanced SA yields and reduced byproduct formations. Nonetheless, there still remain issues regarding slow carbon uptake under anaerobic condition leading to low productivity. Therefore, we can consider other microorganisms that can grow well under anaerobic condition. One potential candidate is Zymomonas mobilis that is known to have relatively high catabolic and high glucose uptake rates, producing ethanol as the major fermentative product [26]. Moreover, Swings and DeLey [27] observed that SA was secreted in Z. mobilis as a minor byproduct at the yield of 0.0025–0.014 (g/g), when yeast extract was added into the medium. This evidence in conjunction with the operational advantages of anaerobic fermentation motivated us to explore metabolic engineering strategies for substantially enhancing SA production.
One strategy for improving strain performance is to identify potential targets for genetic modification. To this end, gene (or reaction) targets can be conventionally identified by random mutagenesis, followed by intelligent screening. Recently, various omics data generated by high-throughput experimental techniques have been exploited for gene identification through comparative genomic, transcriptomic, proteomic, and metabolomic studies within the context of systems biotechnology [28,29,30,31,32]. Apart from this high-throughput omics profiling, in silico analysis of genome-scale metabolic models of various microbes have been successfully employed to investigate their cellular behavior, pose new engineering hypotheses, and identify and evaluate potential genetic targets for strain improvements [33,34,35,36,37,38,39]. Several in silico analysis methods for strain optimization have been designed to select such candidates for genetic manipulations (addition and/or deletion), thereby giving rise to enhanced productivity and cellular properties. They include OptKnock [40], OptStrain [41], OptGene [42], OptForce [43], Flux-sum analysis [44], and cofactor modification analysis [45,46]. These studies have shown that genome-scale models can assist experimental studies for strain improvement [47]. In this work, we employ our previously developed genome-scale model for Z. mobilis and perform in silico analyses to explore and evaluate various strain engineering strategies to enhance SA production in Z. mobilis. In particular, we use constraint-based flux analyses for gene knockout simulations to develop optimization strategies for overproducing SA.
2. Materials and Methods
2.1. Genome-Scale Metabolic Model of Z. mobilis
The genome-scale metabolic model for Z. mobilis ZM4 (ATCC31821), iZM411, has been developed on the basis of its genome annotations [48]. This previous model was further improved using the updated information from KEGG and BioCyc, resulting in 738 reactions and 705 metabolites in 49 specific pathways or subsystems based on their functional roles. It includes major metabolic pathways, i.e., central metabolism, amino acid biosynthesis, lipid metabolism, cell wall metabolism, and vitamin and cofactor metabolism along with the necessary transport reactions for extracellular metabolites. Biomass equation is also derived from the drain of various biosynthetic precursors and relevant cofactors into Z. mobilis biomass at their appropriate ratios to quantify the cell specific growth rate. The complete updated model and biomass equation could be found in Supplementary 1 and 2, respectively.
2.2. Constraints-Based Flux Analysis
Mass balance on various metabolites under the steady state assumption gives rise to a set of linear equations with reactions fluxes as unknown quantities. It is mathematically constructed as an underdetermined system since the number of metabolites constraints is less than the number of unknown fluxes to be determined. Thus, unknown fluxes can then be evaluated by applying linear programming to find optimum value for a given cellular objective, subjected to known uptake rates as the constraints in the model. In this study, the objective function considered was maximization of biomass growth to describe the physiological behavior under the growing condition. Various in silico analyses were carried out by using MetaFluxNet [49] and CPLEX 12.1.0 solver under the general algebraic modeling system (GAMS) [50].
2.3. Combinatorial Knockout Simulation
Combinatorial knockout analysis was performed by applying additional constraints to the current linear flux model. We removed individual reaction(s) associated with the gene(s) being knocked out from in silico models, and evaluated the alteration in flux distribution as the consequence of the deletion of reaction(s). For this analysis, anaerobic growth on glucose was assumed by setting glucose and oxygen uptake rates at 10 mmol/gDCW/h and zero, respectively. Biomass maximization for a steady-state metabolic network, comprised of a set N = {1, …, n} of metabolites and a set M = {1, …, m} of reactions, can be expressed in the following linear programming (LP) problem:
where Sij is the stoichiometric coefficient of metabolite i in reaction j, vj is flux value reaction j, and yj is binary variable, with 1 for deleted reaction and 0 for active reaction. Gene deletion conducted in this study was for single (k = 1), double (k = 2), triple (k = 3), and quadruple (k = 4).
maximize vbiomass
2.4. Flux-Sum Quantification
In order to quantify turnover rate of intermediate metabolites, flux-sum (Φ) can be defined as the summation of incoming or outgoing fluxes of given metabolite i as follows [44]:
where Pi and Ci denote the sets of reactions producing and consuming metabolite i, respectively. Under the pseudo-steady state assumption, i is the mass flow contributed by all of the fluxes producing (consuming) metabolite i.
3. Results and Discussion
The current study aimed at exploring Z. mobilis as potential SA producer or expression host organism and developing the in silico model-driven systematic strategy for strain improvement. To this end, we first evaluated the potential of Z. mobilis to produce SA by computing its theoretical yield using our genome-scale model. Then, we compared and contrasted the central metabolic networks of Z. mobilis and a known SA producer, M. succiniciproducens, to identify engineering targets for enhancing SA production. This network comparison allowed us to find a few target genes/reactions to be added/deleted, which were then evaluated via in silico analysis of the model as successfully demonstrated by Lee et al. [51]. Additionally, having narrowed down the pool of possible targets from the central metabolic reactions, combinatorial knockout analyses were carried out to obtain a list of possible candidates for genetic modification and to investigate their KO effects on SA production. We now describe and discuss each of the above steps in detail.
3.1. Exploring Metabolic Capabilities for Succinic Acid Production in Z. mobilis
As mentioned earlier, Swings and Deley [27] experimentally observed SA yield of 0.0038–0.024 mol per mol glucose in wild-type Z. mobilis. Recently, Seo et al. [52] have shown the existence of frdABCD genes associated with succinic acid dehydrogenase which is responsible for converting fumarate to SA, from the genome sequence of Z. mobilis. This clearly suggests the fumarate reduction to SA is a plausible biosynthetic pathway in Z. mobilis. This route includes malic enzyme that converts pyruvate (PYR) to malate (MAL), which is then followed by fumarate hydratase and SA dehydrogenase. This is indeed the pathway present in M. succiniciproducens which is well-known succinic acid producer. Thus, our genome-scale model includes both pathways for SA production, and interestingly subsequent simulations of wild-type Z. mobilis on glucose confirmed that it naturally secreted SA at 0.06 molar yield during its exponential growth phase, as the byproducts in lysine and methionine biosynthesis. This yield is comparable to the yield observed by Swings and Deley [27].
Next, we maximized SA production in our in silico model satisfying the required cellular growth under growing condition in the wild-type Z. mobilis. Simulation results showed that Z. mobilis was capable of theoretically producing as much as 1.6 mol SA per mol glucose. Furthermore, an increase in SA production led to a decrease in ethanol production, which clearly suggests that ethanol is a competing metabolite for SA production. To understand this SA production further, we explored how the fluxes are diverted from ethanol to SA in this scenario of maximum SA yield. Comparing the flux distribution in this scenario with the growing condition of the wild-type showed the increase in fluxes through tricarboxylic acid (TCA) cycle and carbon dioxide (CO2) uptake, while the decrease in fluxes through ethanol production pathway was observed. The attenuation of these latter fluxes is obvious again, as ethanol is a competing metabolite for SA. Interestingly, the increased uptake of CO2 points to its consumption in SA production. This can be inferred from the increased flux of CO2-consuming malic enzyme that functions as the bridging reaction between glycolysis and TCA cycle in Z. mobilis.
3.2. Central Metabolism Comparison with SA Producer to Identify Gene Candidates in Z. mobilis
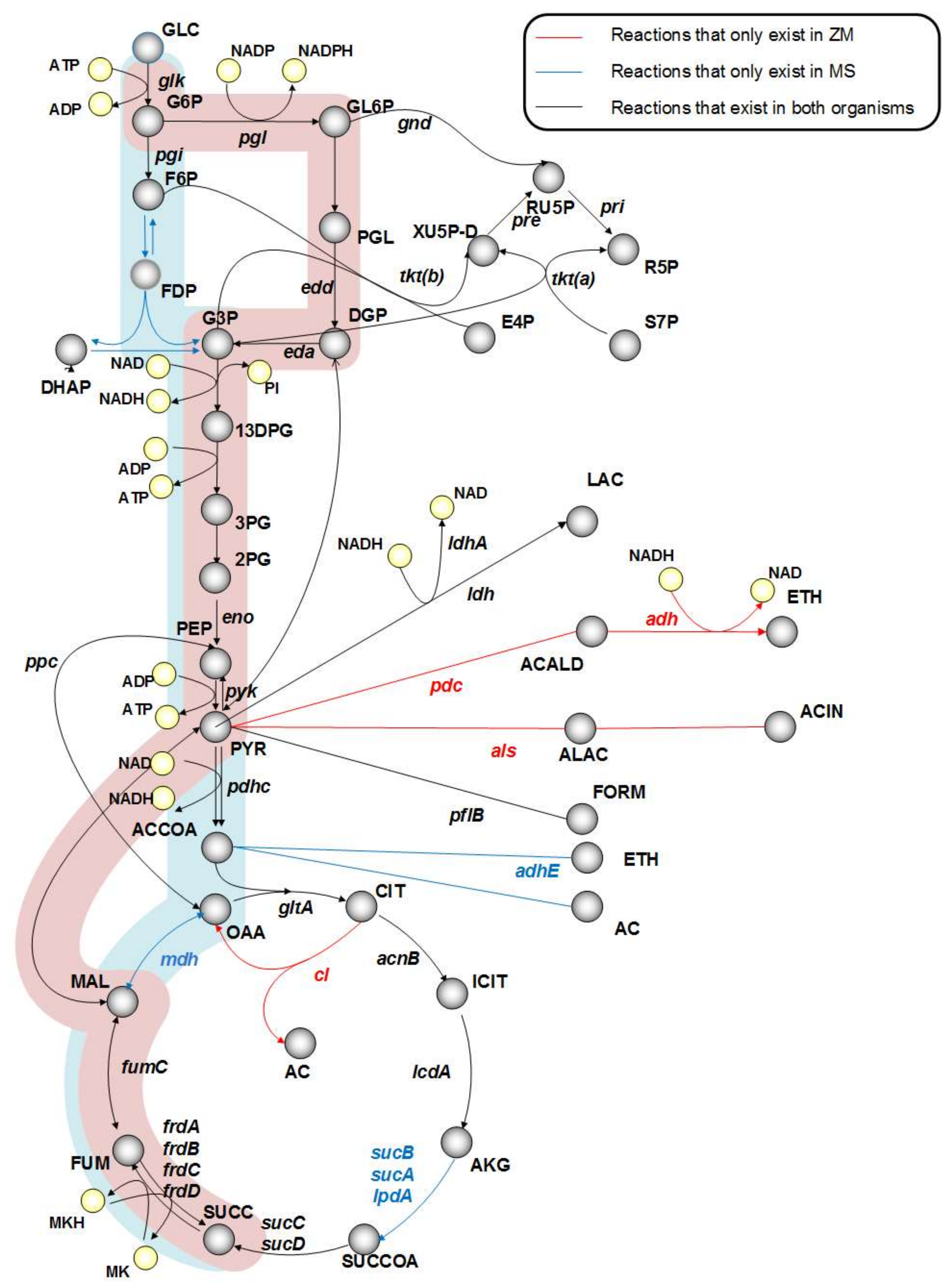
Of the several known SA producers, M. succiniciproducens has the highest SA yield of 1.2 mol per mol glucose, which is 60% of its theoretical SA yield [24]. Both Z. mobilis and M. succiniciproducens are anaerobic fermentative organisms without the phospho-transferring systems (PTS). Instead, they both utilize transportation protein for exchanging substrates between the cytoplasm and the surrounding environment via a process known as facilitated diffusion [14,53]. These common characteristics suggest that SA production in Z. mobilis can be enhanced by imitating M. succiniciproducens. Therefore, we compared the central metabolic pathways of Z. mobilis and M. succiniciproducens to identify gene candidates that may possibly impact fermentation profiles. One significant difference in their metabolisms is that Z. mobilis utilizes the Entner–Doudoroff (ED) pathway, while M. succiniciproducens has the Embden–Meyerhof–Parnas (EMP) pathway. Since the ED pathway in Z. mobilis is one major factor for its high catabolic rate, we focused more on the reactions that are uniquely present in Z. mobilis, but absent in M. succiniciproducens, which constitute potential gene deletion targets.
As illustrated in Figure 1, pyruvate decarboxylase (pdc, PYR → ACALD + CO2), acetolactate synthase (als, PYR → ALAC + CO2), and citrate lyase (cl, CIT → AC + OAA) are three genes that are unique to Z. mobilis. Therefore, we created in silico mutants without pdc, als, and cl to mimic M. succiniciproducens metabolism. The in silico simulation results of the wild-type strain and the knockout mutants are shown in Table 1. The deletion of pdc increased SA production to 0.5 mol SA per mol glucose. Not surprisingly, pdc is crucial for ethanol fermentation in Z. mobilis. Our simulations showed that its deletion diverts the flux from ethanol to acetic acid and SA in the TCA cycle on one hand, and the production of lactic acid and formic acid on the other. However, the deletion of acetolactate synthase (als) resulted in zero growth, because this gene is essential for valine synthesis in Z. mobilis. Clearly, this is not a viable candidate for strain improvement.
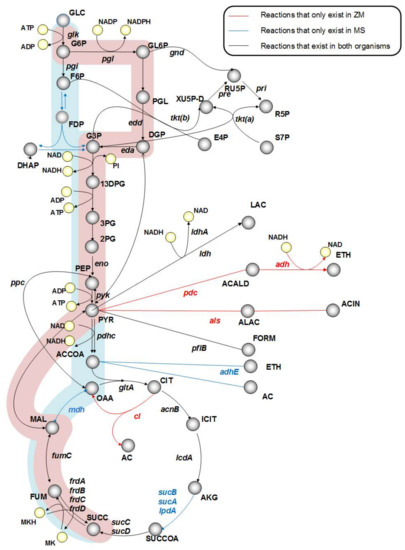
Figure 1.
Comparison of central metabolism in Zymomonas mobilis and Mannheimia succiniciproducens. Blue and red shadows highlight the succinic acid production pathway in M. succinicproducens and Z. mobilis, respectively.

Table 1.
Knockout simulations for target genes identified from central metabolism comparison.
The deletion of citrate lyase (cl) did not affect SA production. But, interestingly, the deletion of cl in addition to that of pdc increased SA production further from 0.5 to 0.6 mol SA per mol glucose. While this yield is 1.1 mol lower than the maximum possible yield predicted by our in silico model for Z. mobilis, it is much higher than 0.06 mol observed for the wild-type Z. mobilis. This clearly shows the success of a simple approach of mimicking M. succiniciproducens for enhancing SA production in Z. mobilis as similarly reported in the previous study [9]. However, the question is whether it is possible to increase this yield even more. Therefore, we now use a combinatorial approach in the following section to identify better knockout combinations.
3.3. Combinatorial Knockout Analysis
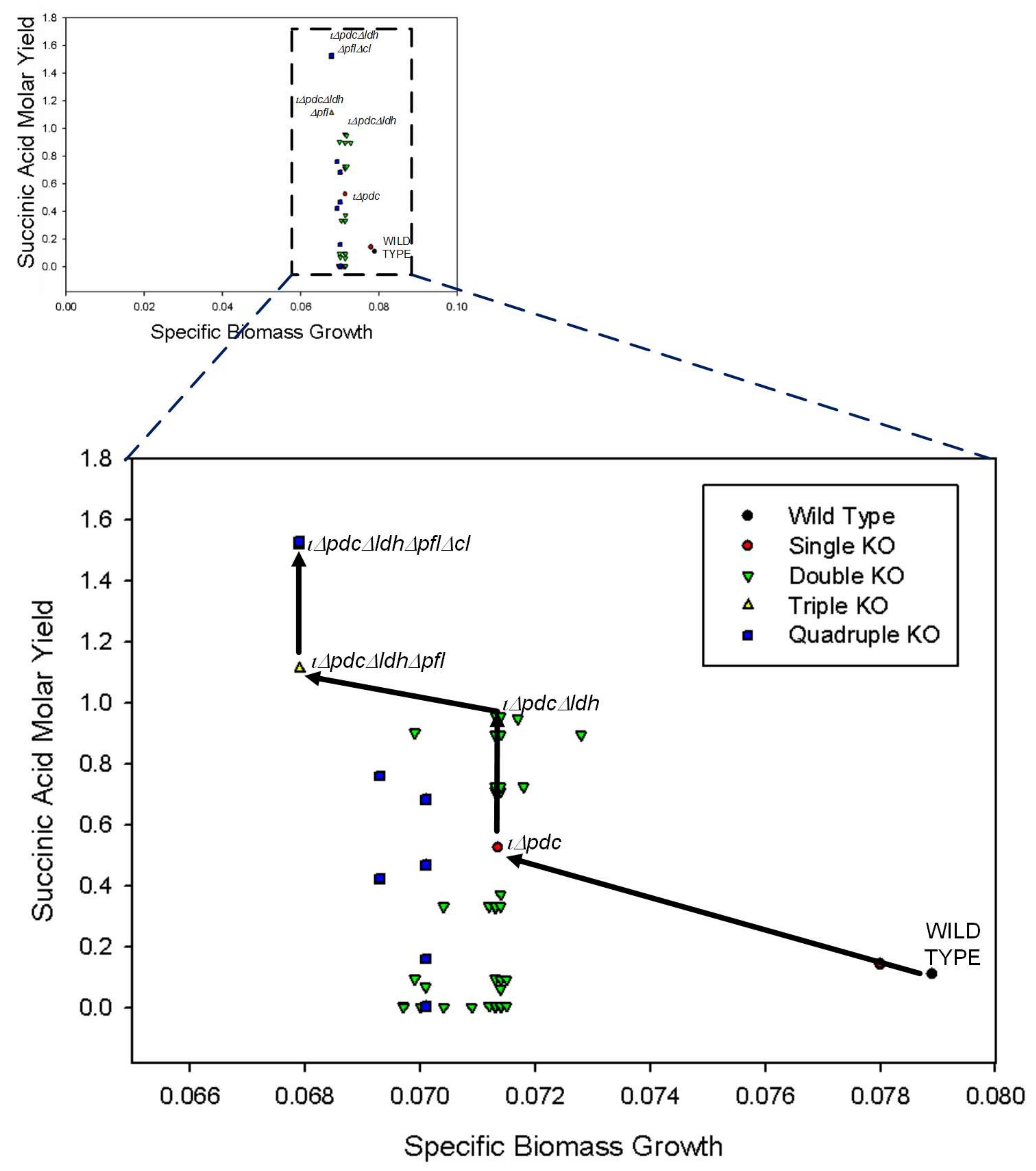
It should be noted that SA is a primary metabolite of Z. mobilis. Therefore, our possible knockout candidates are limited to the 44 genes for the 57 reactions within the central metabolism. Out of the 44 genes, 19 are lethal genes; hence only 25 genes were considered for strain improvement. Since this number is small, we studied all possible combinations of single, double, triple, and quadruple gene knockouts. The resulting predictions are presented in Figure 2.
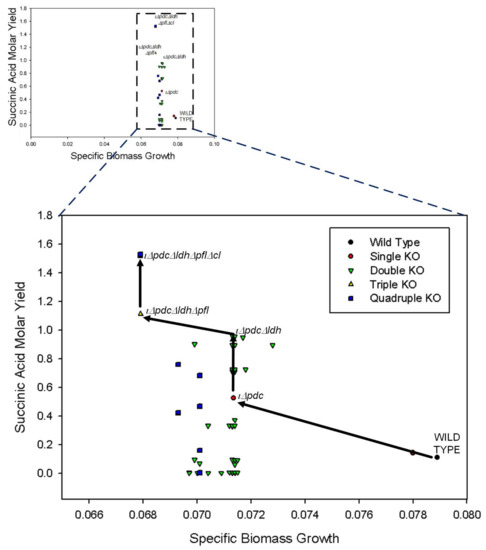
Figure 2.
Simulation results for gene knockout analysis on central metabolic pathways. The shift in biomass growth and succinic acid molar yield from wild type to quadruple knockout (KO) is shown. The best strain for SA production is also shown for single (iΔpdc), double (iΔpdcΔldh), triple (iΔpdcΔldhΔpfl), and quadruple (iΔpdcΔldhΔpflΔcl) knockouts.
From single gene knockouts, we find that pdc is the only gene whose knockout leads to the increase in SA production at the expense of growth. Without knocking out pdc, it is not possible to increase SA production. However, after pdc is deleted, several other knockout strategies can be considered to increase SA production further. Not expectedly, cell growth is not affected at all by these additional knockouts, which is a very useful characteristic of Z. mobilis. From Figure 2, we see that the additional deletions of ldh (lactate dehydrogenase), pfl (pyruvate formate lyase), and cl all increase SA production. Of these three 2-gene knockouts, pdc-ldh seems to be the best for SA production with a molar yield of 0.9. Note that Lee et al. [54] had also reported pdc and ldh as KO candidates for SA production and Seo et al. [55] improved SA production by removing the genes in Z. mobilis. We clarified that this is only the best 2-gene knockout with a yield of 0.9 mol per mol of glucose. Further improvement is possibly achievable through 3-gene knockouts. As seen from Figure 2, pdc–ldh–pfl seems to be the best 3-gene knockout with an SA molar yield of 1.1 mol per mol glucose. It should be noted that disrupting ldh and pfl genes led to the enhanced SA production in M. succiniciproducens and E. coli [24,56]. Finally, the knockout of all four genes (pdc–ldh–pfl–cl) enables us to obtain an even higher yield of 1.5 mol SA per mol glucose, which is very close to the maximum theoretical yield of 1.6 mol SA per mol glucose. Interestingly, the yield of SA was significantly increased by knockout of additional target, i.e., cl gene in Aspergillus niger [57].
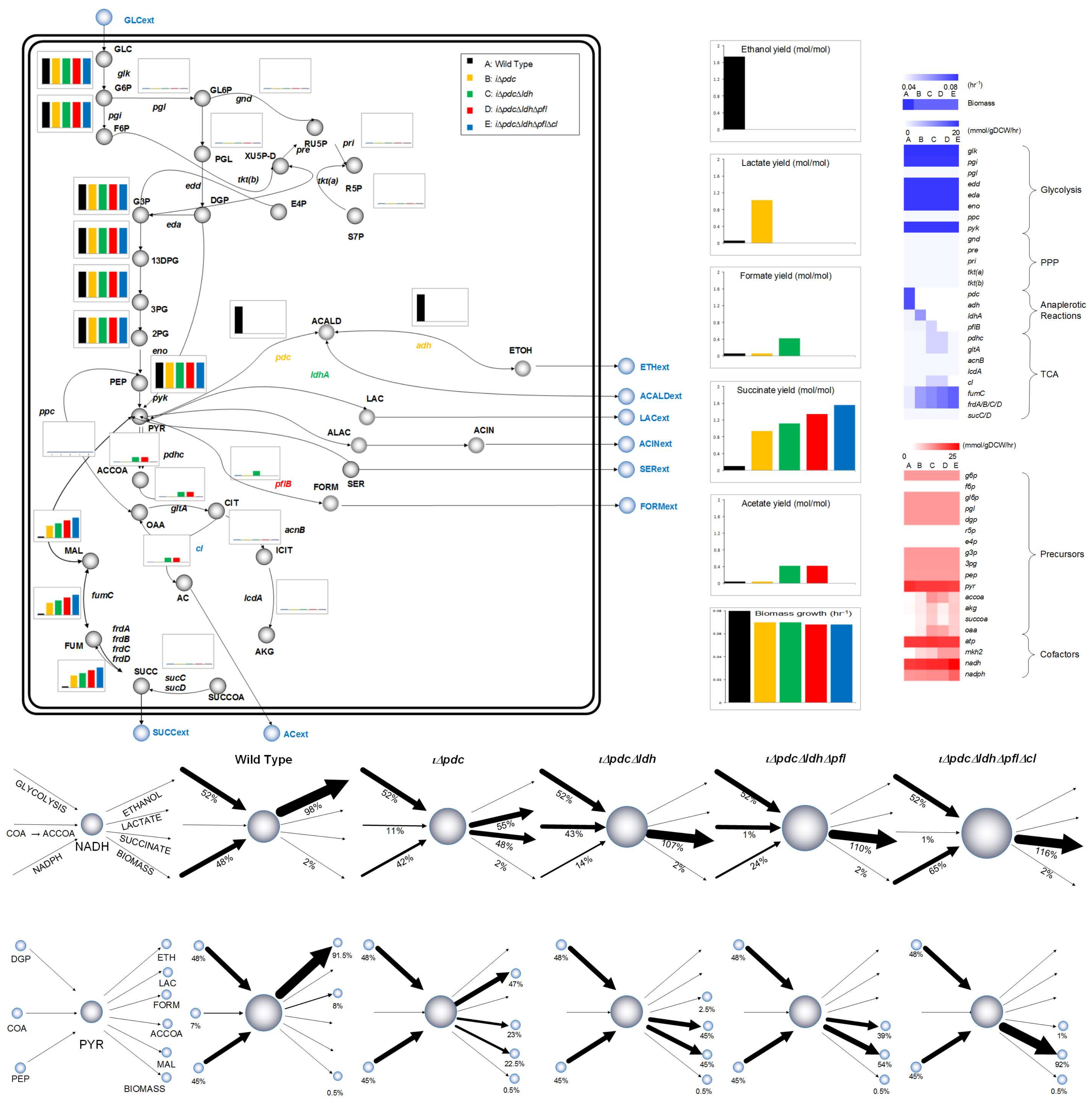
Figure 3 illustrates the redistribution of central metabolism fluxes with sequential knockouts of pdc, ldh, pfl, and cl, explaining how the strain can be genetically engineered to increase SA production. As we can see, this 4-gene knockout diverts carbon flux entirely from ethanol to SA sequentially through lactic acid (pdc), formic acid (ldh), acetic acid (pfl), and finally SA (cl). SA production is more expensive energetically, as it requires one extra mol of NADH compared to ethanol. While ethanol production needs one mol of NADH for pyruvate conversion, SA needs one for malate formation, and the other for menaquinone/menaquinol cycle. Figure 3 also shows how the NADH pool quantified by the flux-sum or turnover rate (see Materials and Methods) increases gradually, as ethanol is diverted to SA via lactic, formic, and acetic acids. Hong and Lee [56] have also observed in E. coli that higher SA production required more NADH. In contrast, ADP/ATP pool remains constant. It is also interesting to see that, while pyruvate is common to pdc, ldh, and pfl, only the knockout of pdc reduces its pool. This has a direct effect on growth, thus, the growth of Z. mobilis depends on the size of pyruvate pool. However, the additional knockouts of ldh, pfl, and cl do not affect the pyruvate pool, and hence have no effect on growth.
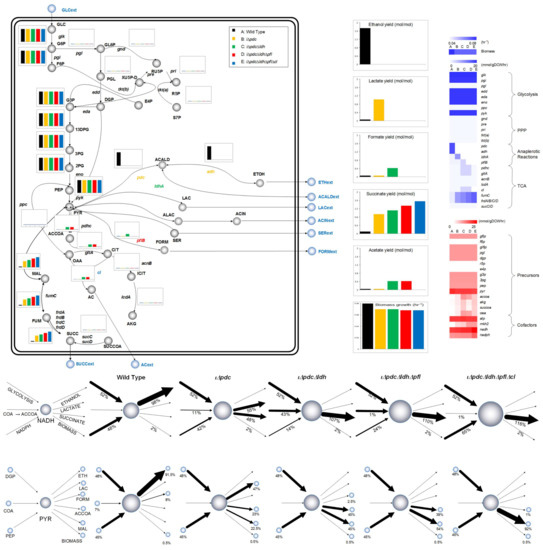
Figure 3.
Metabolic flux distribution across the central metabolic pathways during the exponential growth phase of the microbial culture and flux-sum across the metabolite pyruvate and cofactor NADH in Z. mobilis. Consumption and production of the cofactor NADH and the metabolite pyruvate (PYR) is shown using the flux-sum values across each of the strains (wild type, iΔpdc, iΔpdcΔldh, iΔpdcΔldhΔpfl, iΔpdcΔldhΔpflΔcl). Percentage contribution is also shown. NADH: reduced nicotinamide adenine dinucleotide; NADPH: reduced nicotinamide adenine dinucleotide phosphate; COA: coenzyme A; ACCOA: acetyl coenzyme A; PYR: pyruvate; PEP: phosphoenolpyruvate; DGP: 2-dehydro-3-deoxy-d-gluconate 6-phosphate; MAL: malate.
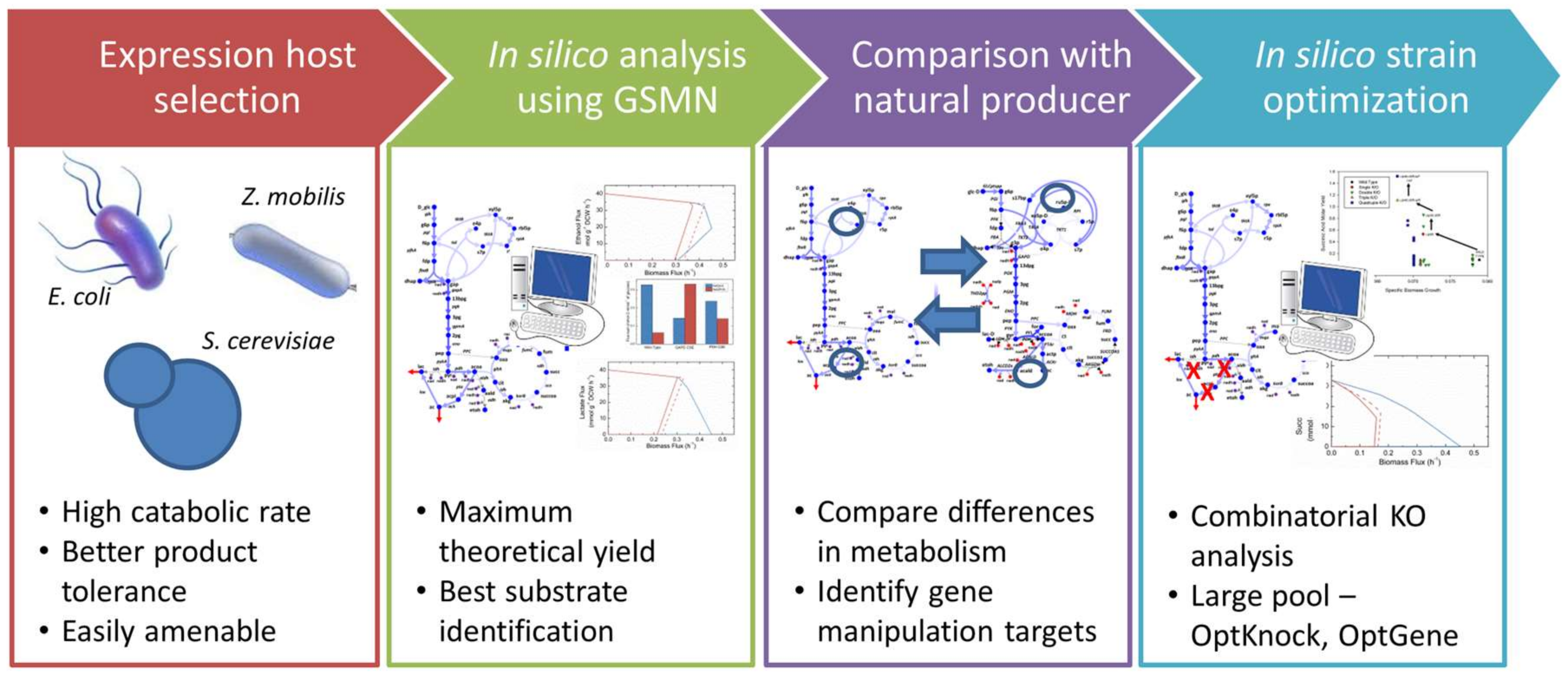
3.4. Model-Driven Systematic Framework for Strain Optimization
Based on our analyses in the previous sections, we now propose a systematic model-driven framework for strain engineering to produce a desired target metabolite using a microbial host (Figure 4). Given a target metabolite, the first step of this framework is to select an expression host from the microbes that could potentially produce it. This expression host should have some special characteristics that would address some deficiencies such as low productivity in other expression hosts. In case of Z. mobilis, the high catabolic rate in anaerobic fermentation was the key feature that may offer a faster metabolite production rate. The second step is to develop and use a genome-scale model for the expression host to predict its maximum theoretical yield for the target metabolite. If the theoretical yield is comparable to those for other known producers, then the selected microbe has the potential to become a host for strain improvement.
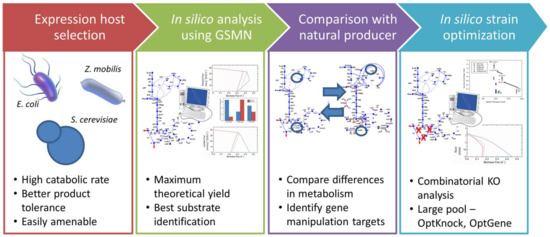
Figure 4.
Proposed framework for strain improvement to overproduce a desirable target metabolite in a potential expression host (succinic acid in Z. mobilis in the current study).
The third step is to explore various combinations of gene modifications (deletions and/or additions) in the expression host to overproduce the target metabolite. This can be done in several ways. One way is to compare the metabolic network of the selected expression host with that of a known reference strain to identify the key differences in genes and pathways. Such a rational approach may give unique gene candidates in the reference strain and expression host for additions and deletions, respectively. The hypothesis is that the reference has optimally evolved and developed relevant synthetic pathways towards the desired metabolite [9]. In cases where only a few genes can be manipulated, as was the case in this work, this approach can be used to narrow down the pool of possible gene targets. For a small pool, it may be feasible to exhaustively consider all possible knockout combinations as done in the current study. These targets can then be evaluated using the genome-scale metabolic model to predict yields. However, for a large pool, such an exhaustive search may not be effective due to the combinatorial explosion. Therefore, another approach would be to exploit optimization techniques for strain improvement such as OptKnock [40], OptStrain [41], OptGene [42], OptForce [43], etc. While these methods may require some computational effort, some of them may guarantee the best solutions, if they converge. Both OptKnock and OptGene suggest the same strain modification as the one we have obtained in this work (Table 1). The multiple solutions of metabolites production yield observed are the results of different tools used. Note that, the SA yields obtained from different tools are almost consistent with the yields obtained from combinatorial knockout.
4. Conclusions
In this study, we presented a systematic approach for overproducing SA in Z. mobilis by using an in silico constraints-based flux analysis. Our genome-scale model shows that Z. mobilis has the capability to produce SA. Our comparison of the central metabolisms of Z. mobilis and the SA-producer, M. succiniciproducens, suggested the removal of three genes (pdc, als, and cl) to mimic the metabolism of the latter. The inactivation of pdc and cl increased the SA yield, but the inactivation of als led to zero biomass growth. However, our combinatorial gene knockout analyses pointed to the inactivation of four genes pdc, ldh, pfl, and cl, which increased yield up to 1.5 mol SA per mol glucose. This is close to the maximum possible theoretical yield of 1.7 mol SA from glucose in Z. mobilis. Interestingly, pdc, ldh, and pfl are pyruvate-consuming reactions in Z. mobilis, whose sequential disruption seems to redistribute pyruvate flux from ethanol to lactic acid to formic acid to acetic acid, and then finally to succinic acid homofermentatively. Based on our work, we proposed a systematic framework to overproduce a desired metabolite from an organism by using the genome-scale metabolic model. The strain engineering strategies proposed in this work can be verified in the future through wet experiments.
Supplementary Materials
The supplements are available online at http://www.mdpi.com/2227-9717/6/4/30/s1, The supplements 1 and 2 contain complete updated model and biomass equations, respectively.
Acknowledgments
This work was supported by by the Academic Research Fund (R-279-000-476-112) of the National University of Singapore, Biomedical Research Council of A*STAR (Agency for Science, Technology and Research), Singapore and a grant from the Next-Generation BioGreen 21 Program (SSAC, No. PJ01334605), Rural Development Administration, Korea.
Author Contributions
H.W., I.A.K., and D.-Y.L. conceived and designed the in silico study; H.W. performed the computational simulations and drafted the manuscript; N.-R.L., and D.-Y.L. revised the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cukalovic, A.; Stevens, C.V. Feasibility of production methods for succinic acid derivatives: A marriage of renewable resources and chemical technology. Biofuels Bioprod. Biorefining 2008, 2, 505–529. [Google Scholar] [CrossRef]
- Beauprez, J.J.; De Mey, M.; Soetaert, W.K. Microbial succinic acid production: Natural versus metabolic engineered producers. Process Biochem. 2010, 45, 1103–1114. [Google Scholar] [CrossRef]
- Tan, T.; Liu, C.; Liu, L.; Zhang, K.; Zou, S.; Hong, J.; Zhang, M. Hydrogen sulfide formation as well as ethanol production in different media by cysND- and/or cysIJ-Inactivated mutant strains of Zymomonas mobilis ZM4. Bioprocess Biosyst. Eng. 2013, 36, 1363–1373. [Google Scholar] [CrossRef] [PubMed]
- Jantama, K.; Haupt, M.J.; Svoronos, S.A.; Zhang, X.; Moore, J.C.; Shanmugam, K.T.; Ingram, L.O. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng. 2008, 99, 1140–1153. [Google Scholar] [CrossRef] [PubMed]
- Thakker, C.; Martínez, I.; San, K.-Y.; Bennett, G.N. Succinate production in Escherichia coli. Biotechnol. J. 2012, 7, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.W.; Xia, S.T.; Wei, L.N.; Li, H.M.; Yuan, Z.P.; Tang, Y.J. Enhancing succinic acid biosynthesis in Escherichia coli by engineering its global transcription factor, catabolite repressor/activator (Cra). Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Guan, Z.; Wang, Y.; Ma, J.; Wu, H.; Jiang, M. Efficient succinic acid production by engineered Escherichia coli using ammonia as neutralizer. J. Chem. Technol. Biotechnol. 2016, 91, 2412–2418. [Google Scholar] [CrossRef]
- Mienda Faezah Mohd, B.S. Bio-succinic acid production: Escherichia coli strains design from genome-scale perspectives. AIMS Bioeng. 2017, 4, 418–430. [Google Scholar] [CrossRef]
- Lee, S.; Lee, D.; Kim, T.; Kim, B. Metabolic engineering of Escherichia coli for enhanced production of succinic acid, based on genome comparison and in silico gene knockout simulation. Appl. Environ. Microbiol. 2005, 71, 7880–7887. [Google Scholar] [CrossRef] [PubMed]
- Millard, C.S.; Chao, Y.P.; Liao, J.C.; Donnelly, M.I. Enhanced production of succinic acid by overexpression of phosphoenolpyruvate carboxylase in Escherichia coli. Appl. Environ. Microbiol. 1996, 62, 1808–1810. [Google Scholar] [PubMed]
- Wang, X.; Gong, C.S.; Tsao, G.T. Bioconversion of fumaric acid to succinic acid by recombinant E. coli. Appl. Biochem. Biotechnol. 1998, 70–72, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Vemuri, G.N.; Eiteman, M.A.; Altman, E. Effects of growth mode and pyruvate carboxylase on succinic acid production by metabolically engineered strains of Escherichia coli. Appl. Environ. Microbiol. 2002, 68, 1715–1727. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, Q.; Mao, Y.; Xing, J.; Su, Z. High-level succinic acid production and yield by lactose-induced expression of phosphoenolpyruvate carboxylase in ptsG mutant Escherichia coli. Appl. Microbiol. Biotechnol. 2010, 87, 2025–2035. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Kim, J.S.; Lee, S.Y.; In, Y.H.; Choi, S.S.; Rih, J.K.; Kim, C.H.; Jeong, H.; Hur, C.G.; Kim, J.J. The genome sequence of the capnophilic rumen bacterium Mannheimia succiniciproducens. Nat. Biotechnol. 2004, 22, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Stols, L.; Donnelly, M.I. Production of succinic acid through overexpression of NAD+-dependent malic enzyme in an Escherichia coli mutant. Appl. Environ. Microbiol. 1997, 63, 2695–2701. [Google Scholar] [PubMed]
- Lin, H.; Bennett, G.N.; San, K.Y. Fed-batch culture of a metabolically engineered Escherichia coli strain designed for high-level succinate production and yield under aerobic conditions. Biotechnol. Bioeng. 2005, 90, 775–779. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Bennett, G.N.; San, K.Y. Genetic reconstruction of the aerobic central metabolism in Escherichia coli for the absolute aerobic production of succinate. Biotechnol. Bioeng. 2005, 89, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Bennett, G.N.; San, K.Y. Metabolic engineering of aerobic succinate production systems in Escherichia coli to improve process productivity and achieve the maximum theoretical succinate yield. Metab. Eng. 2005, 7, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Chayabutra, C.; Wu, J.; Ju, L.K. Succinic acid production with reduced by-product formation in the fermentation of Anaerobiospirillum succiniciproducens using glycerol as a carbon source. Biotechnol. Bioeng. 2001, 72, 41–48. [Google Scholar] [CrossRef]
- Lee, P.C.; Lee, W.G.; Kwon, S.; Lee, S.Y.; Chang, H.N. Succinic acid production by Anaerobiospirillum succiniciproducens: Effects of the H2/CO2 supply and glucose concentration. Enzyme Microb. Technol. 1999, 24, 549–554. [Google Scholar] [CrossRef]
- McKinlay, J.B.; Laivenieks, M.; Schindler, B.D.; McKinlay, A.A.; Siddaramappa, S.; Challacombe, J.F.; Lowry, S.R.; Clum, A.; Lapidus, A.L.; Burkhart, K.B.; et al. A genomic perspective on the potential of Actinobacillus succinogenes for industrial succinate production. BMC Genom. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Park, D.H.; Zeikus, J.G. Utilization of electrically reduced neutral red by Actinobacillus succinogenes: Physiological function of neutral red in membrane-driven fumarate reduction and energy conservation. J. Bacteriol. 1999, 181, 2403–2410. [Google Scholar] [PubMed]
- Bradfield, M.F.A.; Mohagheghi, A.; Salvachúa, D.; Smith, H.; Black, B.A.; Dowe, N.; Beckham, G.T.; Nicol, W. Continuous succinic acid production by Actinobacillus succinogenes on xylose-enriched hydrolysate. Biotechnol. Biofuels 2015, 8, 181. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Song, H.; Lee, S.Y. Genome-based metabolic engineering of Mannheimia succiniciproducens for succinic acid production. Appl. Environ. Microbiol. 2006, 72, 1939–1948. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Yi, J.; Kim, T.Y.; Choi, S.; Ahn, J.H.; Song, H.; Lee, M.-H.; Lee, S.Y. Homo-succinic acid production by metabolically engineered Mannheimia succiniciproducens. Metab. Eng. 2016, 38, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Johns, M.R.; Greenfield, P.F.; Doelle, H.W. Byproducts from Zymomonas mobilis. In Bioreactor Systems and Effects; Springer: Berlin/Heidelberg, Germany, 1991; pp. 97–121. ISBN 978-3-540-47400-5. [Google Scholar]
- Swings, J.; De Ley, J. The biology of Zymomonas. Bacteriol. Rev. 1977, 41, 1–46. [Google Scholar] [PubMed]
- Beer, L.L.; Boyd, E.S.; Peters, J.W.; Posewitz, M.C. Engineering algae for biohydrogen and biofuel production. Curr. Opin. Biotechnol. 2009, 20, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Martín, H.G.; Ivanova, N.; Kunin, V.; Warnecke, F.; Barry, K.W.; McHardy, A.C.; Yeates, C.; He, S.; Salamov, A.A.; Szeto, E.; et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 2006, 24, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Lee, S.Y. Towards systems metabolic engineering of microorganisms for amino acid production. Curr. Opin. Biotechnol. 2008, 19, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Rossouw, D.; Næs, T.; Bauer, F.F. Linking gene regulation and the exo-metabolome: A comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast. BMC Genom. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.Z.; Wu, C.Y.; Chen, T.; Chen, X.; Zhao, X.M. Integrating metabolomics into systems biology framework to exploit metabolic complexity: Strategies and applications in microorganisms. Appl. Microbiol. Biotechnol. 2006, 70, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Mienda, B.S.; Shamsir, M.S.; Illias, M.R. Model-assisted formate dehydrogenase-O (fdoH) gene knockout for enhanced succinate production in Escherichia coli from glucose and glycerol carbon sources. J. Biomol. Struct. Dyn. 2016, 34, 2305–2316. [Google Scholar] [CrossRef] [PubMed]
- Khodayari, A.; Chowdhury, A.; Maranas, C.D. Succinate Overproduction: A Case Study of Computational Strain Design Using a Comprehensive Escherichia coli Kinetic Model. Front. Bioeng. Biotechnol. 2015, 2. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Zeng, B.; Qian, X. Adaptive bi-level programming for optimal gene knockouts for targeted overproduction under phenotypic constraints. BMC Bioinform. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, K.; Yang, L.; Cluett, W.R.; Mahadevan, R. Dynamic strain scanning optimization: An efficient strain design strategy for balanced yield, titer, and productivity. DySScO strategy for strain design. BMC Biotechnol. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Patil, K.R.; Åkesson, M.; Nielsen, J. Use of genome-scale microbial models for metabolic engineering. Curr. Opin. Biotechnol. 2004, 15, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Alper, H. Systems metabolic engineering: Genome-scale models and beyond. Biotechnol. J. 2010, 5, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Teusink, B.; Van Enckevort, F.H.J.; Francke, C.; Wiersma, A.; Wegkamp, A.; Smid, E.J.; Siezen, R.J. In silico reconstruction of the metabolic pathways of Lactobacillus plantarum: Comparing predictions of nutrient requirements with those from growth experiments. Appl. Environ. Microbiol. 2005, 71, 7253–7262. [Google Scholar] [CrossRef] [PubMed]
- Burgard, A.P.; Pharkya, P.; Maranas, C.D. OptKnock: A Bilevel Programming Framework for Identifying Gene Knockout Strategies for Microbial Strain Optimization. Biotechnol. Bioeng. 2003, 84, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Pharkya, P.; Burgard, A.P.; Maranas, C.D. OptStrain: A computational framework for redesign of microbial production systems. Genome Res. 2004, 14, 2367–2376. [Google Scholar] [CrossRef] [PubMed]
- Patil, K.R.; Rocha, I.; Förster, J.; Nielsen, J. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinform. 2005, 6. [Google Scholar] [CrossRef]
- Ranganathan, S.; Suthers, P.F.; Maranas, C.D. OptForce: An optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLoS Comput. Biol. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.K.S.; Lee, D.Y. Flux-sum analysis: A metabolite-centric approach for understanding the metabolic network. BMC Syst. Biol. 2009, 3, 117. [Google Scholar] [CrossRef] [PubMed]
- King, Z.A.; Feist, A.M. Optimizing Cofactor Specificity of Oxidoreductase Enzymes for the Generation of Microbial Production Strains—OptSwap. Ind. Biotechnol. 2013, 9, 236–246. [Google Scholar] [CrossRef]
- Lakshmanan, M.; Chung, B.K.-S.; Liu, C.; Kim, S.-W.; Lee, D.-Y. Cofactor modification analysis: A computational framework to identify cofactor specificity engineering targets for strain improvement. J. Bioinform. Comput. Biol. 2013, 11, 1343006. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, M.; Lee, N.-R.; Lee, D.-Y. Genome-Scale Metabolic Modeling and In silico Strain Design of Escherichia coli. In Systems Biology; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2017; pp. 109–137. ISBN 9783527696130. [Google Scholar]
- Widiastuti, H.; Kim, J.Y.; Selvarasu, S.; Karimi, I.A.; Kim, H.; Seo, J.S.; Lee, D.Y. Genome-scale modeling and in silico analysis of ethanologenic bacteria Zymomonas mobilis. Biotechnol. Bioeng. 2011, 108, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Yun, H.; Park, S.; Lee, S.Y. MetaFluxNet: The management of metabolic reaction information and quantitative metabolic flux analysis. Bioinformatics 2003, 19, 2144–2146. [Google Scholar] [CrossRef] [PubMed]
- McCarl, B.A.; Meeraus, A.; van der Eijk, P.; Bussieck, M.; Dirkse, S.; Steacy, P. McCarl GAMS User Guide (2008). Available online: http//gams.com/docs/document.htm (accessed on 14 February 2004).
- Sang, Y.L.; Lee, D.Y.; Tae, Y.K. Systems biotechnology for strain improvement. Trends Biotechnol. 2005, 23, 349–358. [Google Scholar]
- Seo, J.S.; Chong, H.; Park, H.S.; Yoon, K.O.; Jung, C.; Kim, J.J.; Hong, J.H.; Kim, H.; Kim, J.H.; Kil, J.; et al. The genome sequence of the ethanologenic bacterium Zymomonas mobil is ZM4. Nat. Biotechnol. 2005, 23, 63–68. [Google Scholar] [CrossRef] [PubMed]
- DiMarco, A.A.; Romano, A.H. D-Glucose transport system of Zymomonas mobilis. Appl. Environ. Microbiol. 1985, 49, 151–157. [Google Scholar] [PubMed]
- Lee, K.Y.; Park, J.M.; Kim, T.Y.; Yun, H.; Lee, S.Y. The genome-scale metabolic network analysis of Zymomonas mobilis ZM4 explains physiological features and suggests ethanol and succinic acid production strategies. Microb. Cell Fact. 2010, 9. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-S.; Chong, H.-Y.; Kim, J.H.; Kim, J.-Y. Method for Mass Production of Primary Metabolites, Strain for Mass Production of Primary Metabolites, and Method for Preparation Thereof. U.S. Patent Application 20090162910 A1, 25 June 2009. [Google Scholar]
- Hong, S.H.; Lee, S.Y. Importance of redox balance on the production of succinic acid by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2002, 58, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Meijer, S.; Nielsen, M.L.; Olsson, L.; Nielsen, J. Gene deletion of cytosolic ATP: Citrate lyase leads to altered organic acid production in aspergillus niger. J. Ind. Microbiol. Biotechnol. 2009, 36, 1275–1280. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).