1. Introduction
In recent decades, conventional agricultural practices using agrochemical inputs have significantly increased global per capita agricultural production. At the same time, per capita income is estimated to multiply by 2050 compared with its current level [
1,
2]. However, this effort not only increases production costs but also causes environmental damage, such as soil pollution and degradation, fertilizer and pesticide emissions, biodiversity loss, and several negative impacts on human health [
2]. In this sense, alternative methods are needed to improve agricultural production or reduce dependence on agrochemicals and balance productivity with sustainability [
3]. The proliferation of environmental concerns and the concomitant desire for chemical-free foods have precipitated an increase in demand for sustainable agricultural methods, specifically organic agriculture. [
4].
Wittwer et al. in [
5] state that organic agriculture is a production system that avoids or excludes the use of fertilizers, pesticides, growth regulators, and animal feed additives. To the extent possible, organic farming systems rely on crop rotation, the use of animal manure, legumes, green manures, and organic waste from off-farm sources, natural minerals, and biological pest control to maintain soil structure and productivity, provide nutrients to plants, and control insects, weeds, and other pests. Organic farming systems are predicated, to the greatest extent feasible, on the principles of crop rotation, the utilization of animal manure, legumes, green manures, organic waste originating from external sources, natural minerals, and biological pest control. These systems are designed to maintain soil structure and productivity, provide plant nutrients, and control insects, weeds, and other pests [
6].
The dynamic and attractive market for organic food has stimulated the conversion of conventional agriculture to organic farming. It is estimated that more than 80% of the current organic acreage was incorporated into this system during the final decade of the previous century [
7]. This gradual rise can be attributed to several factors, including the robust political and economic support for conventional agriculture, the underestimation of the adverse effects of chemical-intensive agriculture, and the pervasive denial of conventional production alternatives [
8]. The global organic beverage market is segmented by product type: alcoholic (wine, beer, spirits) and non-alcoholic (fruit and vegetable juice, dairy, coffee, tea). The global beverage market is expected to expand at a compound annual growth rate (CAGR) of 12.3% during the forecast period, which extends through 2025. The North American region is currently the most substantial market for organic beverages, while the Asia-Pacific region is projected to exhibit a more rapid rate of growth. The growth of the organic beverage market has been attributed to two primary factors. Firstly, there has been a marked increase in consumer awareness regarding organic products. Secondly, the market has seen a continuous introduction of new product variants [
7]. The market size for organic beverages was estimated at 50.19 billion U.S. dollars in 2024. Projections indicate that this figure will reach 63.79 billion U.S. dollars by 2029, indicating a compound annual growth rate (CAGR) of 4.91% during the forecast period (2024–2029) [
9,
10].
The continued increase in global energy demand requires the development of sustainable alternatives to non-renewable energy sources. This positions bioethanol as a vital part of future energy portfolios [
11]. A rigorous analysis of the fermentation stage is necessary to determine the overall economic viability and environmental impact of bioethanol synthesis [
12]. While traditional fermentation methods are effective, they often present challenges related to process efficiency, yield optimization, and energy consumption, which limit their potential for large-scale industrial applications [
13]. Consequently, advanced statistical optimization techniques coupled with metabolic modeling are being used more frequently to improve productivity and reduce operating costs at the fermentation stage [
14].
The production of organic ethanol presents a unique set of challenges, including limited nutrient availability, a deficiency of inorganic nitrogen, and constraints in the utilization of fermentation enhancers. The aforementioned factors frequently culminate in diminished fermentation efficiency and cell viability. The production of organic alcohol entails the utilization of certified organic ingredients, which are cultivated without the employment of synthetic pesticides, herbicides, or other deleterious chemicals. Moreover, the production process does not involve the use of artificial additives. Instead, natural fermentation methods are employed. [
15].
Two categories of ethanol are produced on a global scale: fermented and synthetic. The production of bioethanol is possible through the utilization of biomass that is readily available and contains fermentable free sugars [
16]. From a technical perspective, the production of organic alcohol is characterized by its sustainability and social responsibility, thereby satisfying multiple criteria. For instance, the process under discussion eliminates the addition of synthetic chemicals, including but not limited to sulfuric acid, urea, ammonium phosphate, and antifoam agents [
17].
A notable illustration of this commitment is evidenced by CADO (Consorcio Agroartesanal Dulce Orgánico), a company established by small-scale sugarcane farmers. This enterprise engages in the production of granulated panela and organic alcohol in Ecuador, exemplifying a commitment to sustainable and community-driven agricultural practices. The company’s product line includes panela, ethanol, and lacto-alcoholic beverages (cocoa cream and coffee) under the Wilkay brand. Additionally, it produces potable alcohol for individuals who engage in the production of liqueurs. The company holds the internationally recognized organic certification from BCS Öko-Garantie of Germany, which is currently under renewal. Fermented ethanol, also known as bioethanol, is produced through the fermentation of corn or other biomass materials. This fuel is primarily utilized in the production of energy, with a smaller percentage being employed in the beverage industry [
18]. Synthetic ethanol is produced from ethylene, a petroleum byproduct, and is primarily used in industrial applications [
19].
Among biofuels, the most developed and widely implemented process at the industrial level is the production of bioethanol through anaerobic fermentation of carbohydrates by yeast microorganisms [
18,
20,
21]. The most conventional method for producing ethanol involves the fermentation of sugar sources, which are subsequently degraded by the action of microorganisms. The process is comprised of three fundamental stages: propagation, fermentation, and distillation. Among these, fermentation is the primary stage [
22]. In the fermentation stage, ethanol is produced, along with other byproducts such as higher alcohols, organic acids, esters, and aldehydes, which give the characteristic aroma and flavor of spirits, rums, and spirits [
21]. In the field of biofuel production, the fourth stage of the process is the preparation of the raw material, depending on the source. For example, starches from corn are used for first-generation alcohol, and lignocellulosic materials are used for second-generation ethanol. In both cases, the action of catalysts such as exogenous enzymes is required for hydrolysis. In the process of producing ethanol as a fuel, a fifth stage of dehydration is incorporated.
Other authors, such as Asmamaw in 2021 [
23], include a fourth stage, that of raw material preparation, depending on the source. This phenomenon is exemplified by corn starches utilized in the production of first-generation alcohol and lignocellulosic materials employed in the second-generation ethanol process. Both of these processes necessitate the utilization of catalysts, such as exogenous enzymes, to facilitate the hydrolysis reaction. In the process of producing ethanol as a fuel, a fifth stage of dehydration is incorporated.
Research such as Ojeda (2005) [
24] and Rodríguez (2016) [
25] has addressed fermentation studies for the development of organic ethanol in Cuba. However, the results obtained are not conclusive enough to introduce this product as a new offering for the alcohol industry. Taking all this into account, the present research aims to study the production of organic ethanol using yeast culture. This study is novel in its exclusive use of certified organic final molasses with statistical optimization to enhance additive-free ethanol production. The organic ethanol under consideration is derived from molasses produced by the sole company in Cuba that is certified as a producer of organic sugar. The production of this alcohol does not involve the addition of chemical nutrients or acids during the process. The yeast utilized in Cuban distilleries, designated as Ethanol Red, is employed exclusively.
Although numerous studies have examined ethanol production from molasses and other substrates, most of these studies have employed inorganic nutrients, chemical additives, or optimized fermentation conditions using conventional inputs [
16,
17,
20,
26]. Few studies have addressed ethanol production under strict organic criteria by completely ignoring inorganic sources of nitrogen and phosphorus and using only certified raw materials.
Furthermore, the scientific literature reports only a few examples of combining advanced statistical designs, such as Plackett–Bürman and Box–Behnken, with organic fermentation processes, particularly in Latin American and Caribbean contexts. Most previous studies have focused on optimizing one variable at a time or comparing strains rather than on systematically integrating statistical optimization into real, pilot-scale organic processes [
20,
26,
27].
The main novelty of this work is its sequential application of statistical optimization methodologies to maximize yeast proliferation and fermentation efficiency in a completely organic system without chemical additives, using certified molasses and realistic industrial conditions. This approach contributes to the literature on sustainable bioprocesses and offers a replicable methodological foundation for the organic ethanol industry in regions with limited resources. Given these constraints, the scope of this contribution is intentionally limited to demonstrating feasibility and providing a replicable methodology for practitioners pursuing organic certification, not to delivering generalizable predictive tools.
Accordingly, this work is scoped as a feasibility-oriented investigation under organic-only constraints, primarily targeting researchers and practitioners focused on organic bioethanol and statistical optimization under additive-free conditions.
5. Statistical Analysis of the Results Obtained
The mean cell count was calculated for each factor level to identify those with the greatest impact on final cell count and determine the optimal configuration that maximizes their value.
Table 4 presents the experimental matrix, which contains the final results for the response variable under consideration.
The primary effects of the independent variables were estimated as the difference between the mean cell count response when the factor is at its high level
and its low level
, divided by half of the runs, as illustrated in
Table 5.
As illustrated in
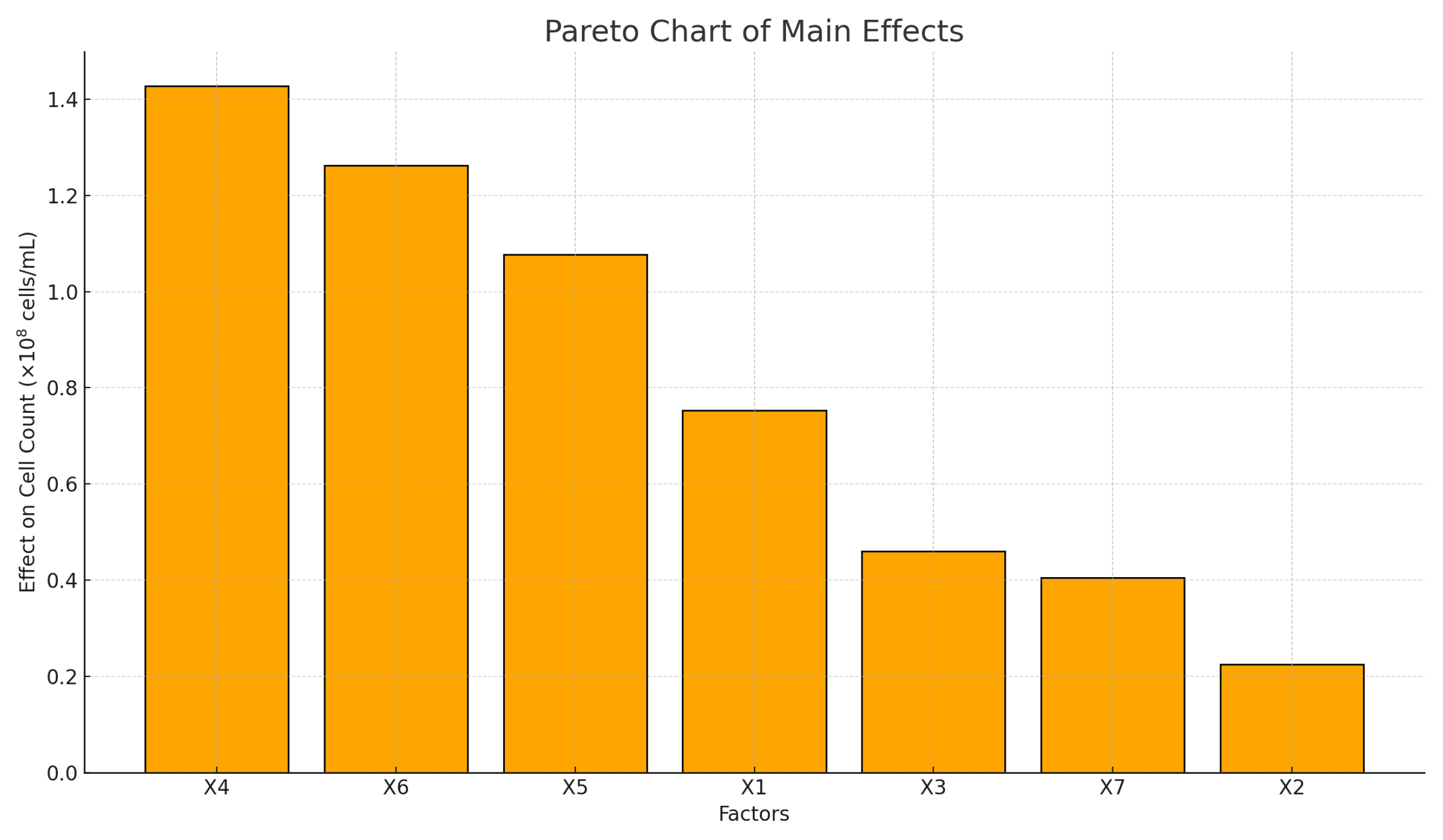
Figure 3, the Pareto chart provides a graphical representation of the estimated main effects on cell count for each factor considered in the Plackett–Bürman design. In this chart, the factors are arranged in descending order according to their absolute impact, thereby facilitating a clear and hierarchical interpretation of the relative importance of each factor.
The Pareto chart reveals the presence of four significant variables, of which the first three are deemed false variables. The sequence begins with X4, X6, and X5, followed by X1, which is a physical variable. Factor X4 is particularly noteworthy, as it exerts the most significant influence, with an effect exceeding 70 million cells. This finding suggests that the variation in Factor X4 has the greatest impact on cell count. Subsequent to these, the effects of X6 and X5 also exceed 50 million cells/mL, thereby consolidating them as critical variables for system optimization. In the experimental design, X4, X5, and X6 were designated as false variables, suggesting that other factors, not previously considered, influence cell count. As mentioned in [
41,
42], factors such as substrate concentration, microbial cell concentration, pH, and nitrogen source influence ethanol production. These authors add that the impact of these factors on ethanol production yield depends, in turn, on the microbial species, fermentation conditions, and types of raw materials used.
In this case, the false variables may correspond to the interaction of X1 with the other two variables (sugar concentration over time and yeast concentration as well as the interaction of these three variables in combination). As noted by Nuanpeng et al. [
31], factors such as substrate concentration, microbial cell concentration, pH, and the nitrogen source have been identified as influential in the context of ethanol production. The authors further posit that the impact of these factors on ethanol production yield is contingent upon the microbial species, the fermentation conditions, and the types of raw materials utilized. Alternatively, these false variables may be other variables not considered in the experiment that affect the process, such as pH, aeration, or agitation. Nutrition is another critical factor that must be taken into consideration. To ensure an optimal fermentation process, yeast requires specific nutrients in precise proportions. To ensure an optimal fermentation process, yeast requires the presence of macro- and micronutrients in precise proportions, including minerals or vitamins [
23]. These elements were not directly incorporated into the fermentation process due to the necessity of producing organic ethanol. In the present study, the pseudofactors (X4, X5, X6) were incorporated exclusively to complete the Plackett–Bürman statistical design, since the number of real factors was lower than required by the design. These pseudofactors do not correspond to physical or experimental variables but rather allow estimating experimental error and detecting the possible influence of uncontrolled factors. Therefore, any interpretation of their physical significance should be considered speculative, and it is not recommended to draw practical conclusions from their statistical significance.
At a secondary level of relevance, the concentration of sugars in solution (X1) is associated with the reducing sugars that constitute the substrate source for fermentation, with an effect close to 40 million, suggesting a moderate influence. Finally, the yeast concentration, time, and variable X7 exhibited minor effects, below 25 million, indicating that their contribution to the model is limited or even dispensable for further analysis.
This graphical representation is two-fold: it complements the numerical analysis and facilitates decision-making by visually highlighting which factors should be prioritized in fine-tuning or expanding the experimental design. The necessity to regulate and enhance the levels of X4, X5, and X6 is further accentuated, as these factors significantly influence the variability observed in cell count.
Given the absence of dummy columns—that is, additional columns in the experimental design that do not represent any real factors in the system—the standard error (SE) was estimated as the standard deviation of the estimated effects. This calculation yielded a value of .
The significance of the main effects on cell count enables the identification of factors that exert a statistically relevant influence, which extends beyond their absolute magnitude. To accomplish this objective, the standard error (SE) was estimated from the three minor effects. The Student’s t-values and p-values were calculated for each factor with three degrees of freedom, under the assumption of a Student’s t-value distribution. The t-values (estimate/standard error) are displayed in
Table 6, and the effect is employed to ascertain significance.
Factors X4, X5, X6, and X1 exhibit p-values below the standard significance threshold of 5% (p < 0.05), signifying that their effects on the cell count variable are statistically significant. Factor X4 is the most significant, with a negative effect of −142.75, a t-value of −6.81, and a p-value of 0.0065, establishing it as the most potent and pertinent factor in the model. X5 and X6 exhibit extremely significant beneficial effects (t = 5.14 and 6.02, respectively), affirming their importance as critical variables for optimizing cell count. X1 exhibits a positive effect of 75.25 and a t-value of 3.59 (p = 0.037), indicating significance, albeit with a diminished relative influence.
Conversely, factors X2, X3, and X7 lack adequate statistical evidence to be deemed relevant in the model, as their p-values (0.363, 0.116, and 0.149, respectively), beyond the necessary threshold. X3 exhibits a moderate negative effect; nevertheless, its insignificance indicates that its impact may be attributable to random variation or unexplained sources of variability within the model. The impacts of X2 and X7 are minimal and statistically insignificant, justifying their removal from streamlined iterations of the predictive model.
The statistical analysis endorses the prioritization of components X4, X5, and X6 in the optimization of cell culture processes. This also rationalizes the exclusion of components X2, X3, and X7, as their contributions are statistically insignificant within the assessed model.
5.1. Linear Regression Model
The full linear regression model is
where the estimated coefficients are
Therefore, the final equation is
5.2. Full Model Metrics
Total Sum of Squares (SST):
Regression Sum of Squares (SSR):
Error Sum of Squares (SSE):
Coefficient of Determination (
):
Adjusted
(
):
The adjusted for the full model is not reported here, given a clear case of overfitting (p = 7 predictors, n = 8 observations). Due to the limited size of the dataset and the lack of replications, the statistical models presented may be subject to overfitting. Therefore, the results should be interpreted as a preliminary approximation and require further validation with larger, replicated datasets. Caution is advised when interpreting the robustness and predictive capacity of the models until further experimental evidence is available. These are conditions that would not provide much confidence in model predictions. Therefore, further analysis was done by reducing the model to include only the significant variables (X4, X5, X6), with an adjusted of 0.699. The high and adjusted values observed describe a good in-sample fit; however, given the small sample size and lack of external validation, there is a risk of overfitting. Cross-validation was not applied due to sample size limitations. The predictive capacity of the models should be confirmed with new replicated data before proposing prediction or process control tools.
The Plackett–Bürman design was employed to construct a linear regression model, which was then utilized to eliminate non-significant variables and predict the cell count response as a function of the primary factors. The model is depicted in Equation (
4):
As shown in
Table 7, X4 exerts a substantial negative influence, with a magnitude of −71.375 million cells/mL. This observation suggests that its presence should be minimized to optimize cell count. This would correspond to working with a lower Brix and less time. Conversely, X5 and X6 exhibited substantial positive effects (53,875 and 63,125 million cells/mL, respectively), suggesting that their levels should be maintained at a high (+1) setting, which would entail the amalgamation of two negative variables and one positive variable.
Total Sum of Squares (SST):
Regression Sum of Squares (SSR):
Error Sum of Squares (SSE):
Coefficient of Determination (
):
Adjusted
(
):
where
(number of observations) and
(number of predictors).
The linear regression analysis identified that the factors , , and are the primary determinants of final cell count (FCC) during the yeast cultivation stage for organic ethanol production. The fitted model explains approximately 81.4% of the observed variability in FCC, with an adjusted of 0.699, indicating a good fit given the small sample size and number of predictors.
The resulting equation enables FCC prediction under various combinations of the significant factors, supporting process optimization. Results show that minimizing and maximizing and leads to a substantial increase in cell count, improving fermentation outcomes.
Excluding non-significant factors simplifies the model and focuses experimental efforts on truly influential variables. Furthermore, the use of organic molasses as the sole nutrient source and the absence of chemical additives reinforce the feasibility of sustainably producing organic ethanol.
This work lays the groundwork for future research aimed at validating and expanding the model, exploring potential factor interactions, and incorporating new relevant variables such as pH, aeration, and nutrient content that could further optimize the process. The methodology and findings presented here represent a significant contribution toward more efficient, sustainable, and competitive bioprocesses in the organic ethanol industry.
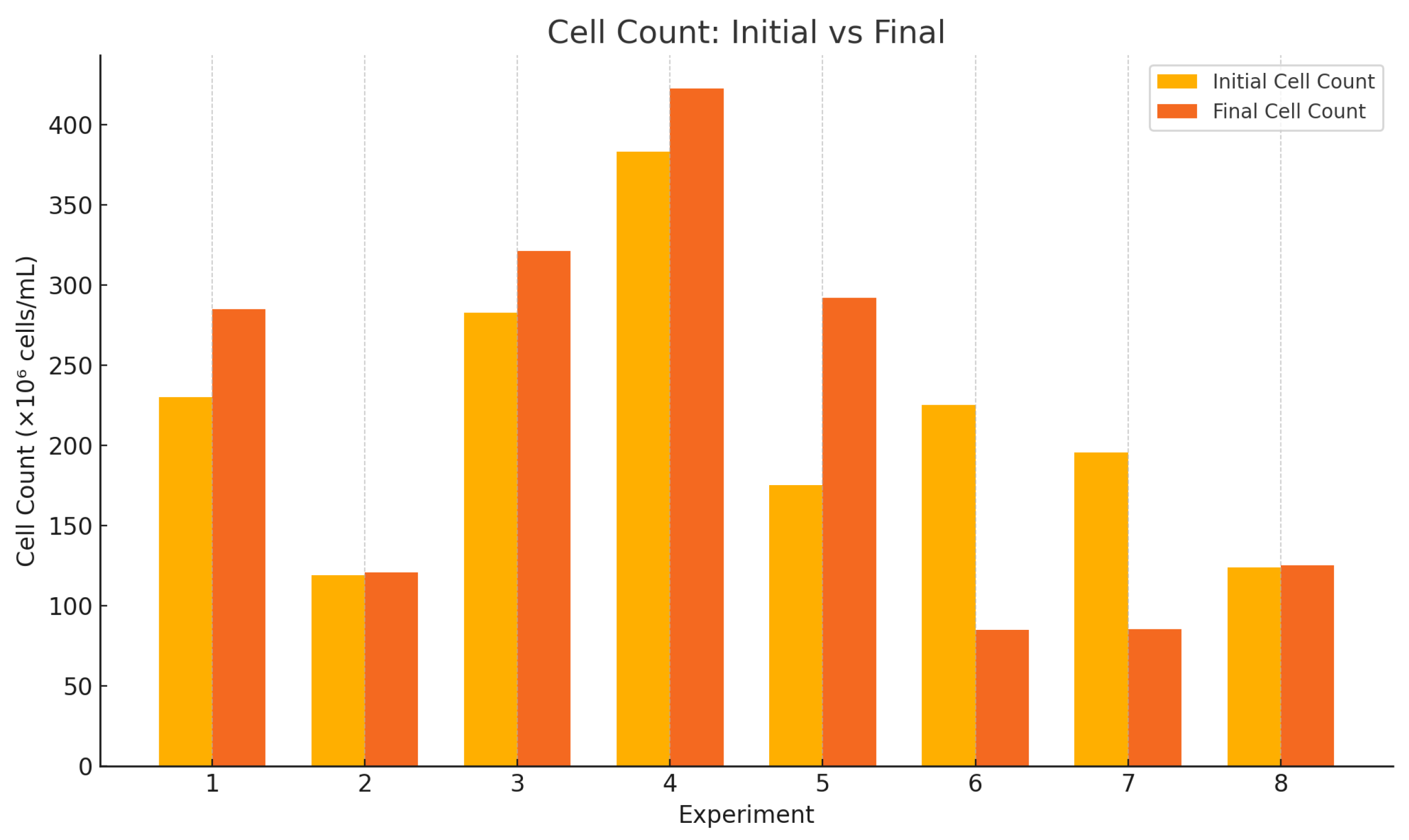
Subsequent to the determination of the coefficients, the cell count is maximized through the implementation of a combination of coded levels of X4 = −1, X5 = +1, and X6 = +1. The predicted cell number under these conditions is approximately 221,027,074.31 cells/mL in the yeast culture for pre-fermentation.
5.3. Interaction Analysis in a Box–Behnken Design
Since the original Plackett–Bürman design identified three pseudo-factors (X4, X5, X6) as significant, this indicates that there are important factors that were not directly measured.
The report presents the fit of a quadratic model using a Box–Behnken design with three factors: concentration of sugars in solution (X1), yeast concentration (X2), and time (X3). A total of 13 experimental runs were utilized, employing authentic data from the study on fermentation for the purpose of producing organic ethanol.
5.3.1. Experimental Design
As summarized in
Table 8, three factors with two coded levels were considered for the Box–Behnken design.
5.3.2. Adjusted Quadratic Model
The estimated regression model was
where
Y = Cell concentration ( cells/mL);
= Coded variables of experimental factors;
= Model coefficients estimated by multiple regression.
5.3.3. Estimated Coefficients
The model coefficients, estimated using the least squares method, are
As summarized in
Table 9, the RSM model includes linear, quadratic, and interaction effects with their estimated coefficients and interpretation.
5.3.4. Statistical Validation Through ANOVA
Statistical validation of the model was performed through Analysis of Variance (ANOVA), with results presented in
Table 10.
Statistical interpretation:
;
;
Conclusion: The model is highly significant.
However, this apparent accuracy is constrained by the small sample size and the absence of biological/technical replicates; thus, the model should be considered provisional until confirmed with replicated experiments and out-of-sample validation.
5.3.5. Model Statistics
As summarized in
Table 11, the goodness-of-fit metrics indicate an adequate model fit and high explained variance.
Table 11.
Goodness-of-fit statistics for the RSM model.
Table 11.
Goodness-of-fit statistics for the RSM model.
| Statistic | Value | Interpretation |
|---|
| Coefficient of determination () | 0.9774 | 97.7% of variability explained |
| Adjusted | 0.9504 | 95.0% predictive capacity |
| Residual standard error | 15.5 | High model precision |
| Coefficient of variation (CV) | 8.7% | Low experimental variability |
| Error degrees of freedom | 5 | Sufficient for validation |
5.3.6. Statistical Assumptions Validation
Validation Tests
The model was subjected to rigorous tests to verify compliance with statistical assumptions:
As summarized in
Table 12, the statistical assumptions for the validated RSM model are satisfied.
Residual Analysis
The model residuals present the following characteristics:
Residual mean: (centered);
Standard deviation: ;
Range: ;
Distribution: Normal ().
5.3.7. Model Comparison
Optimal Model Selection
As summarized in
Table 13, we compared alternative RSM models by parsimony, adjusted
, and overall ANOVA significance.
Selection criteria:
- 1.
Statistical significance ();
- 2.
Maximum adjusted ;
- 3.
Sufficient degrees of freedom ();
- 4.
Model parsimony;
- 5.
Statistical assumptions compliance.
5.3.8. Model Interpretation
Main Effects
Factor : Very strong positive effect ();
Factor : Strong positive effect ();
Factor : Moderate negative effect ().
5.3.9. Curvature Effects
Negative quadratic terms indicate the presence of optimal maxima:
Interactions
The
interaction presents the strongest effect in the model:
This interaction indicates that the effect of significantly depends on the level of , and vice versa.
Biochemical Interpretation
The results show that:
The validated response surface model () reveals fundamental aspects of microbial physiology during ethanolic fermentation. The constant term ( cells/mL) represents the basal cell growth capacity under standard conditions, reflecting the intrinsic metabolic potential of the microorganism. The positive coefficient of () indicates that this factor acts as a growth promoter, possibly related to the availability of essential nutrients, dissolved oxygen, or pH conditions that favor enzymatic activity and biomass synthesis. Conversely, the negative coefficient of () suggests an inhibitory effect on growth, which could be associated with the accumulation of toxic metabolites, osmotic stress, or environmental conditions that compromise cellular viability. The interaction term () is particularly significant from a biochemical perspective, as it indicates a synergistic effect where the combination of both factors produces a result superior to that expected from their individual effects, suggesting complex regulatory mechanisms such as activation of complementary metabolic pathways, modulation of gene expression, or compensatory effects that optimize fermentative efficiency. This positive interaction implies that, although is individually inhibitory, its presence in combination with can activate stress response systems or alternative metabolic routes that result in a net increase in biomass, which is characteristic of the metabolic plasticity of fermentative microorganisms and their capacity for adaptation to variable culture medium conditions. The synergistic interaction between sugar concentration (X1) and incubation time (X3) indicates that high substrate levels offset the decrease in fermentative activity caused by prolonged incubation, thereby maintaining cell viability and productivity.
The subsequent section will describe the experimental application of the model that was obtained. The results of this study are presented in
Table 14.
According to [
16], the following parameters are utilized to assess the dynamics of fermentations: cell density, ethanol yield, final molasses concentration, pH, fermentation efficiency, and residual sugar concentration. The fermented must exhibits an alcohol percentage of 6.4%, which exceeds the values obtained by [
27,
34,
43] in fermentation processes with inorganic molasses. These researchers achieved concentrations of 5.15%, 6.38%, and 5.48%, respectively, through the fermentation of molasses using traditional sugarcane molasses. The final reducing sugar concentration was measured at 66.117 g/L, enabling the calculation of fermentation yield and efficiency. As illustrated in
Table 15, the results obtained demonstrate the efficacy with which the fermentation stage is executed.
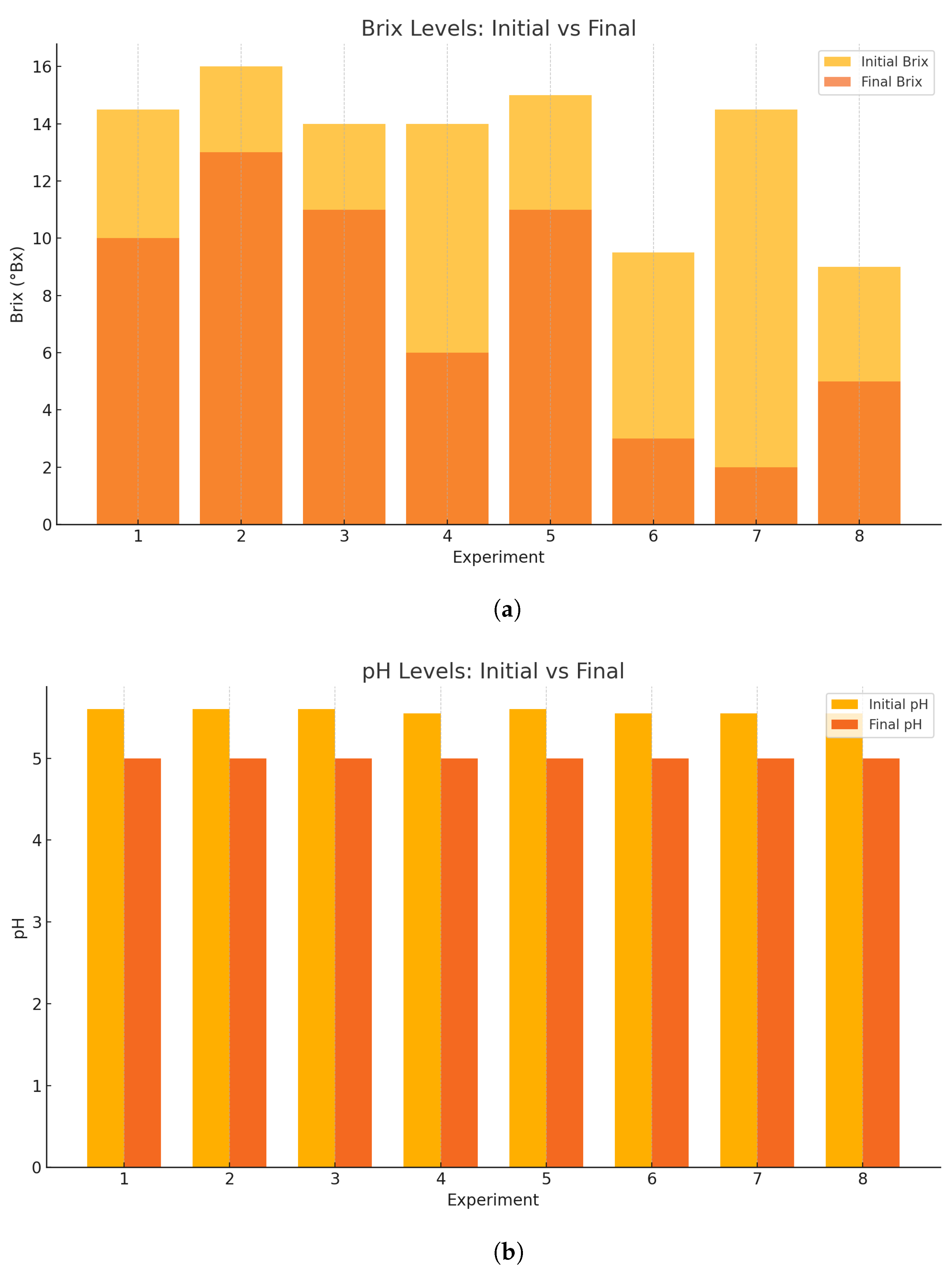
The implementation of the RSM-optimized fermentation conditions resulted in substantial improvements across all critical performance parameters, demonstrating the effectiveness of statistical optimization in bioprocess enhancement. The fermentation process was initiated with an optimized sugar concentration of 13°Brix (125.40 g/L ART), which was strategically adjusted to provide adequate substrate availability while avoiding inhibitory effects associated with excessive sugar concentrations. The enhanced microbial activity under optimized conditions achieved superior sugar conversion efficiency, reducing the final Brix to 6.5 (62.70 g/L ART), representing a 50% sugar utilization rate compared with the original 35.7% conversion. The fermentation time was successfully reduced from 48 to 42 h, achieving a 12.5% time reduction while simultaneously increasing alcohol production from 6.4% to 7.8%, demonstrating the synergistic effects identified through the RSM model. The theoretical fermentation efficiency experienced a remarkable improvement from 63.04% to 78.52%, indicating enhanced metabolic performance and better utilization of available substrates under the optimized factor combinations. Similarly, the alcohol-to-substrate yield increased significantly from 47.43% to 62.15%, reflecting improved conversion stoichiometry and reduced metabolic losses. The overall productivity enhanced from 1.69 to 1.86 g/L·h, representing a 10.1% improvement that combines the benefits of higher alcohol yields with reduced processing time. These comprehensive improvements validate the RSM model’s predictive capability and confirm that the identified optimal factor interactions successfully maximized both fermentation efficiency and economic viability, establishing a robust foundation for scaled-up organic ethanol production processes.
As a literature benchmark (not a within-study control), conventional molasses fermentations typically report final alcohol contents in the 5–6% v/v range under comparable scales and substrates. Our optimized run achieved 7.8% v/v; however, because no inorganic-nutrient control was run side-by-side, these figures should be read only as contextual reference rather than evidence of superiority. A direct benchmark with a conventional nutrient-supplemented process under identical operating conditions will be included in follow-up work.
Among the key challenges for developing this process on a large scale are high investment and capital costs, the technological maturity of biofuels, the large-scale supply of feedstock, and political and regulatory issues, according to [
44]. Organic ethanol production can be carried out using the same technological framework as traditional processes for this product. In this case, the main difference lies in the raw materials used and their characteristics. The average difference between organic and conventional products applied to the final consumer ranges between 45 and 55% according to Molina et al. [
45]. The use of organic molasses as the main substrate increases the total production cost of this process, in turn obtaining an organic product that will increase the value of production.
6. Conclusions
This work presents a methodological proof-of-concept showing that sequential PB→BBD can improve performance metrics in a fully organic system. The resulting models reflect in-sample behavior only and require replicated validation and side-by-side conventional benchmarking before any general claims of predictive utility or industrial competitiveness can be made. The identification of significant pseudo-factors (X4, X5, X6) in the screening phase revealed the presence of important uncontrolled variables, highlighting the complexity of organic fermentation processes and the need for comprehensive factor consideration. The validated quadratic response surface model explained 97.7% of the experimental variability (, adjusted ), indicating good in-sample fit for the explored design space; however, the absence of replication limits generalizability and calls for external validation. The high values obtained for the models, such as 0.977 for the Box–Behnken model, should be interpreted with caution. Given the absence of experimental replicates and the limited number of degrees of freedom in the dataset, these values may be inflated and may not fully reflect the predictive capacity or generalizable robustness of the models. Despite the acknowledged risk of overfitting, it is imperative to emphasize the necessity of cross-validation or the acquisition of new experimental data with replicates. These methodologies are indispensable in confirming the generalizability and predictive capabilities of the proposed models. The synergistic interaction between sugar concentration and incubation time () proved to be the most significant factor, indicating that optimal fermentation performance requires a careful balance of substrate availability and processing time rather than simple maximization of individual parameters.
Experimental validation under optimized conditions achieved remarkable improvements across all performance metrics: ethanol content increased from 6.4% to 7.8% v/v (21.9% improvement), theoretical fermentation efficiency rose from 63.04% to 78.52% (24.6% enhancement), alcohol-to-substrate yield improved from 47.43% to 62.15% (31.0% increase), and productivity enhanced from 1.69 to 1.86 g/L·h (10.1% improvement). Simultaneously, the fermentation time was reduced from 48 to 42 h (12.5% reduction), demonstrating that optimization can achieve both improved quality and enhanced efficiency. The maximum cell density of cells/mL was consistently achieved and maintained throughout the fermentation process, confirming the model’s predictive capability and the sustainability of the optimized conditions.
The demonstration that organic ethanol can be effectively produced from sugarcane molasses alone without chemical additives shows that the technical and economic viability of sustainable bioethanol production is possible. The statistical method offers a useful and systematic tool to optimize the processes and is suitable for a variety of organic substrates and fermentation parameters. Further work is still required to scale up the optimized process to industrial scale, to evaluate the long-term stability of the optimized parameters, and to implement additional organic substrates in the process to increase flexibility and economic competitiveness. The developed methodology and results form a strong basis for further development of competitive organic ethanol production processes fulfilling the performance requirements as well as the environmental sustainability. Accordingly, this work is primarily intended for producers operating under organic certification or in resource-constrained settings who require additive-free fermentation. Broad industrial claims require replication and direct, side-by-side benchmarking for validation. Overall, this study provides proof of concept under organic-only constraints. The generalizability and predictive utility of the results depend on independent replication, external validation, and direct comparison with conventional processes.
7. Limitations and Future Prospects
One of the main limitations of this study is the lack of biological or technical replicates, which prevents an accurate estimation of experimental variability and limits the statistical robustness of the generated models. Similarly, the absence of a control group based on conventional fermentation with inorganic nutrients hinders direct comparisons and evaluations of the competitiveness of the proposed organic process. The absence of a side-by-side conventional control remains a key limitation to assessing practical competitiveness and will be addressed in the next experimental iteration. Another relevant limitation is the absence of quantified critical byproducts, such as acetic acid, glycerol, and other organic acids. These byproducts are essential for evaluating the purity and safety of the produced ethanol. Furthermore, the interpretation of the pseudofactors used in the experimental design is purely statistical and does not correspond to directly measured or controlled physical variables.
Moving forward, it is recommended that experiments be conducted with adequate replicates to strengthen statistical validity and allow for the inclusion of error bars in the results. Priority should also be given to incorporating a control group with conventional fermentation and performing quantitative byproduct analyses to ensure ethanol quality. Finally, the experimental design should be expanded to include and control for variables such as pH, aeration, and agitation. The proposed models should also be validated on larger scales and under different industrial conditions. These actions will consolidate the technical and commercial viability of the organic ethanol production process and contribute to the development of more sustainable and competitive bioprocesses.
In the next phase, we will re-run the Plackett–Burman screening with duplicate runs and include triplicated center points in the Box–Behnken design to obtain independent estimates of pure error and improve statistical robustness. Future work will include a side-by-side benchmark against conventional fermentation with inorganic nutrients under matched substrate, inoculum, pH/temperature, and time profiles to quantify industrial competitiveness (yield, productivity, efficiency, and cost).