Abstract
Precise and modular reconstruction of 3D tooth structures is crucial for creating interpretable, adaptable models for digital dental applications. To address the limitations of conventional segmentation approaches under conditions such as missing teeth, misalignment, or incomplete anatomical structures, we propose a process-oriented reconstruction pipeline composed of discrete yet integrated modules. The pipeline begins by decomposing 3D dental meshes into a series of 2D projections, allowing multi-view capture of morphological features. A fine-tuned Segment Anything Model (SAM), enhanced with task-specific bounding box prompts, performs segmentation on each view. T-Rex2, a general object detection module, enables automated prompt generation for high-throughput processing. Segmented 2D components are subsequently reassembled and mapped back onto the original 3D mesh to produce complete and anatomically faithful tooth models. This modular approach enables clear separation of tasks—view projection, segmentation, and reconstruction—enhancing flexibility and robustness. Evaluations on the MICCAI 3DTeethSeg’22 dataset show comparable or superior performance to existing methods, particularly in challenging clinical scenarios. Our method establishes a scalable, interpretable framework for 3D dental modeling, supporting downstream applications in simulation, treatment planning, and morphological analysis.
1. Introduction
In recent years, digital orthodontic technology has become a crucial assistance in dental treatments, offering advantages that surpass traditional methods. By leveraging advanced AI and 3D graphics technologies, digital orthodontics significantly enhances the precision of treatments and the ability to predict outcomes. Using digital orthodontic technology, dentists can provide patients with personalized, accurate, and efficient orthodontic solutions. In orthodontic procedures, a 3D oral scanner is typically used to reconstruct a comprehensive model of the patient’s teeth, providing a basis for subsequent treatments. However, accurately segmenting each tooth from a reconstructed tooth model still remains a challenging task. The accuracy of tooth segmentation directly impacts the effectiveness of subsequent treatment stages. Efficient and precise tooth segmentation can significantly reduce the time required for dentists to analyze and diagnose the patient’s dental condition.
Human teeth are classified into incisors, canines, and molars, each with different shapes and individual variations. Some patients may have cavities, incomplete crowns, or malformed teeth, which can make the boundaries between teeth and gums blurry and unpredictable. These special conditions increase the difficulty of tooth segmentation. Currently, many state-of-the-art three-dimensional tooth segmentation methods cannot effectively address automatic segmentation under non-ideal conditions [1], showing unstable segmentation results and lacking robustness. In cases of missing teeth, many algorithms confuse adjacent teeth, leading to incorrect tooth labels; in cases of malformed teeth, instance segmentation errors occur, resulting in misplaced tooth labels, which interfere with subsequent dental treatment stages.
To address these issues, this paper proposes a prompt-based three-dimensional tooth segmentation method using the SAM model [2]. By applying 3D-to-2D projection, the problem of 3D tooth segmentation is transformed into a 2D image segmentation problem. Then, the fine-tuned pre-trained image segmentation model SAM is used to segment two-dimensional tooth images. By using weakly supervised prompts through manual input, precise and efficient segmentation of these projected two-dimensional tooth images is achieved. Finally, these segmented two-dimensional images are mapped back to the three-dimensional tooth model, completing the three-dimensional tooth segmentation. This pipeline can be considered a structure-aware segmentation framework, as it incorporates not only the geometric features of individual teeth but also their spatial arrangement and contextual relationships within the jaw. This enables the model to better handle challenging scenarios such as tooth crowding, missing teeth, or asymmetries. Considering that in a single view, teeth in the two-dimensional images projected from three-dimensional models may occlude each other, it becomes impossible to completely segment all surfaces of the teeth. Therefore, we chose to project from multiple views, thereby obtaining multiple two-dimensional images for segmentation. For triangular patches appearing in multiple views, majority voting is used to determine their segmentation labels. To improve the efficiency of prompt-based segmentation, we introduce the latest generic object detection technology, “T-Rex2”. This allows for the segmentation of multiple similarly shaped teeth by drawing a single bounding box around one tooth, significantly accelerating the interactive segmentation process. Experiments show that our method maintains high accuracy in cases of missing and misaligned teeth and incomplete jaws, whereas other existing supervised methods exhibit various degrees of mis-segmentation or under-segmentation, demonstrating better zero-sample generalization and the flexibility of prompt-based segmentation.
The main contributions are as follows:
- We propose a prompt-based 3D tooth segmentation method that robustly adapts to both normal and malformed teeth, achieving accurate segmentation with minimal fine-tuning of the pre-trained SAM. We fully leverage the zero-shot capabilities of visual-prompted object detection to optimize the efficiency of prompt interactions.
- We construct a tooth segmentation images dataset (2D-TeethSeg) for SAM fine-tuning. Through a virtual camera, 131,400 images and ground truth masks with a resolution of are generated from the MICCAI 2022 Challenge publicly available dataset 3DTeethSeg22. Moreover, we will release it as a publicly available dataset.
- We present the results of multiple state-of-the-art tooth segmentation methods on the proposed dataset, providing a comprehensive comparison and analysis, particularly focusing on cases with missing teeth, misaligned teeth, and incomplete upper and lower jaws.
2. Related Work
2.1. Traditional and Deep Learning-Based Tooth Segmentation Methods
Traditional tooth segmentation methods are mostly based on geometry and topology, segmenting using two-dimensional images. Kondo et al. [3] utilize an algorithm that produces depth maps from three-dimensional data to enhance tooth segmentation. First, they obtain the dental arch from planar view range images as a reference and calculate panoramic range images. They then detect the gaps between teeth in both the planar and panoramic views and merge the results. Shah et al. [4] aim to identify individuals based on dental features. They propose extracting tooth contours using an active contour without edges based on the overall region intensity of tooth images. Nomir et al. [5] separate teeth from the background in dental X-ray images, then separate the upper and lower jaws, and finally use projection to isolate individual teeth. After isolating individual teeth, they propose a shape description and matching algorithm based on surface matching, extracting vector features from the tooth contours to match teeth with their contours. Similarly, based on processing dental X-rays, Said et al. [6] segment the ROIs in dental images as target objects, with each ROI representing a tooth, and employ mathematical morphology methods such as internal noise filtering and connected component labeling to improve segmentation results. And using grayscale contrast stretching transformation enhances the clarity between teeth and the background.
With the development of point clouds, the focus in the field of tooth segmentation has also shifted from 2D-based image segmentation methods to directly using 3D data for segmentation. The advent of deep learning has introduced many new approaches to the field of tooth segmentation. Xu et al. [7] use convolutional neural networks for the segmentation and labeling tasks of 3D tooth models and propose a boundary-aware tooth simplification method. In addition to segmenting teeth on two-dimensional images, directly segmenting on three-dimensional tooth models is also a more direct approach. Tian et al. [8] use a new method based on sparse voxel octrees and 3D CNNs for classification through two-stage hierarchical feature extraction and further optimize the results with a conditional random field model. Similarly, Cui et al. [9] construct a two-stage network using deep convolutional neural networks. The first stage extracts edge maps from input CBCT images using an edge extraction network, enhancing the contrast of image shape boundaries and serving as input for the second stage. The second stage constructs a 3D region proposal network (RPN), combining CBCT images and edge maps via using a similarity matrix for segmentation tasks, and adds spatial relation components to encode positional features, improving segmentation accuracy. Zhang et al. [10] propose a dual-stream graph convolutional network, namely TSCGNet, learning multi-view geometric information. This network independently learns multi-scale features from coordinates and normal vectors at the same time, using graph attention convolution and graph max-pooling to extract rough structures and fine details. Wu et al. [11] propose an end-to-end neural network directly based on meshes, namely MeshSeg, using the coordinates, normal vectors, and relative positional vectors of three vertices as inputs to reflect the positional relationships between points. This network selects multiple different scales of perception areas to expand the receptive field for learning multi-scale features, finally merging different scales to improve accuracy. Xiong et al. [12] introduce a 3D Transformer-based approach that leverages self-attention mechanisms to capture dependencies among different teeth, addressing the inherent complexity and irregular arrangement of dental structures. Their method incorporates a multi-task learning framework, which includes an auxiliary segmentation head to improve boundary delineation and employs a geometry-guided loss based on point curvature to enhance segmentation accuracy.
Deep learning methods often achieve excellent results, primarily relying on high-quality tooth datasets for network training. However, datasets related to tooth segmentation are currently scarce, and most tooth data are private, limiting the application of deep learning. Liu et al. [13] propose a self-supervised learning framework (STSNet) for this issue by combining unsupervised pre-training and supervised fine-tuning stages. The unsupervised pre-training stage adopts hierarchical contrastive learning, including point-level and region-level contrastive learning modules, which allow the use of unlabeled data for network training and enhance segmentation performance.
In addition to the issues mentioned above, deep learning methods also face the following challenges. Jana et al. [1] indicate that in some orthodontic, medical, and diagnostic cases, it is not necessary to obtain complete 3D information of the jaw. However, most existing deep learning-based 3D tooth segmentation methods rely on complete 3D information of the jaw and require consistency in shape and dental conditions between the test subjects and the training set. Additionally, most deep learning-based 3D tooth mesh segmentation methods are trained on tooth models with a relatively fixed number of mesh units. When the input mesh contains only partial tooth information, the inconsistency in numbering labels may hinder training and inference, leading to poor results.
2.2. Large Model-Based Medical Image Segmentation Methods
Recently, large segmentation models like SAM [2] have been proposed and widely applied in the field of medical imaging. SAM is an image segmentation model based on Transformer, utilizing a ViT (Vision Transformer) [14] pre-trained with MAEs (masked autoencoders) [15]. This model is trained on a massive dataset comprising 11 million images and over 1 billion mask images. It can generate high-quality object masks given any segmentation prompts (such as points or boxes) and can also directly generate masks for all objects in an image. SAM demonstrates strong generalization capabilities and can be used for new segmentation tasks through interactive prompts. Prompts are categorized into sparse (points, boxes, text) and dense (masks) prompts. Points and boxes use position encoding [16] to represent the corresponding extracted embeddings, while text uses the text encoder from CLIP [17].
SAM, with its powerful zero-shot segmentation capabilities, provides a pre-trained segmentation tool for numerous applications. Consequently, research on fine-tuning and lightweighting SAM has recently become a hot topic. Zhang et al. [18] propose a one-shot variant called PerSAM, building on SAM’s zero-shot capabilities. PerSAM extracts features from a specified input image and mask, then uses these features in subsequent image segmentation to generate foreground and background prompts, guiding the segmentation of specific targets. Zhao et al. [19] improve the segmentation efficiency of the SAM model from the perspective of reducing the computational overhead. In order to improve the efficiency of segmentation, the segmentation process is decoupled into the following two tasks: detecting object bounding boxes for all objects in the image and performing object segmentation according to these bounding boxes. The original ViT encoder structure of SAM is replaced by using the YOLOv8-seg backbone. Their framework reduces the amount of data required for SAM training and inference time. To create a lightweight deep learning network, Zhang et al. [20] apply knowledge distillation to substitute SAM’s image encoder with Tiny-ViT, significantly reducing the parameter count and enhancing inference speed. Ma et al. [21] adapt SAM specifically for medical image segmentation, fine-tuning SAM’s mask decoder and reducing input prompts to only bounding boxes. This method gives high accuracy and reliability in various medical imaging tests. Similarly, Wu et al. [22] adapt SAM for medical applications without training, instead of using lightweight adapters to fine-tune pre-trained SAM for the medical image segmentation task. Zhang et al. [23] focus on fine-tuning SAM by freezing the image encoder and using LoRA to adapt encoders for medical images. The same strategy is adopted for the decoder, with LoRA being utilized for fine-tuning. Inspired by the aforementioned work, we fine-tune the pre-trained model (SAM) on our own dataset to adapt to our tooth segmentation task. We only use bounding boxes as the input prompt. Ref. [24] shares a similar idea to ours; however, our method solely utilizes SAM for 2D segmentation, without integrating features from both 2D images and 3D point clouds as in their approach.
3. Method
To address the practical challenges of missing teeth, misaligned teeth, and incomplete upper and lower jaws in tooth segmentation, this paper proposes a prompt-based segmentation method that exhibits robustness for highly complex tooth shapes and conditions. Segmenting the two-dimensional projection images of the three-dimensional tooth model with the fine-tuned SAM model can provide various degrees of freedom for task segmentation of the teeth. With the aid of weak supervision through manually provided bounding boxes, accurate and efficient segmentation of the projected two-dimensional tooth images is achieved. Finally, the segmented two-dimensional images are mapped back to the three-dimensional tooth model to complete the 3D tooth annotation. Given that teeth in two-dimensional images projected from a single perspective may occlude one another, rendering it infeasible to fully segment all the surfaces of teeth. Hence, we opt to project from multiple perspectives, thereby generating several two-dimensional images for segmentation. The labeling of triangular facets appearing in more than one view is determined by voting, which is determined by the majority of pixels’ labels. Figure 1 illustrates the overall process of our prompt-based tooth segmentation method.
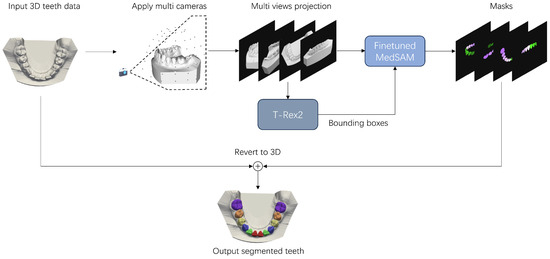
Figure 1.
The pipeline of prompt-based three-dimensional tooth segmentation. By using a virtual camera, the 3D tooth mesh is projected into a set of tooth images from multiple viewpoints. T-Rex2 is then used to optimize the selection of bounding box prompts. Finally, the masks generated by the fine-tuned MedSAM are mapped back to the 3D mesh to complete the 3D tooth segmentation.
3.1. 3D-to-2D Prompt-Based Tooth Segmentation
We propose a prompt-based 3D tooth segmentation based on converting three-dimensional dental annotations into two-dimensional images for processing. The specific operations are as follows: First, a virtual camera is used to project the three-dimensional tooth model from multiple angles, and then the generated images are annotated with bounding boxes and corresponding tooth labels (using the FDI tooth numbering system). The fine-tuned SAM model segments the teeth based on the bounding boxes, and the segmented tooth regions’ pixels are mapped back to the corresponding triangular patches through reverse 2D-to-3D mapping, achieving precise 3D tooth segmentation. By fine-tuning only the mask decoder, we can leverage the zero-shot inference capability of pre-trained large models to effectively segment unknown tooth samples. This method not only exhibits strong generalization ability but also offers rapid segmentation speed.
Since we use bounding boxes as input prompts for the SAM model, the accuracy of the bounding boxes affects the subsequent segmentation precision. Completely manual input of bounding boxes would be time-consuming and labor-intensive. Additionally, manually drawn bounding boxes are prone to errors and instability. To address these issues, we employ the T-Rex2 object detection model to obtain bounding boxes for the teeth. By fully utilizing the characteristics of teeth, such as type, size, and shape, which are quite similar, the visual similarity ensures that visual prompts work effectively. Compared to text prompts, providing visual examples as visual prompts can more intuitively and directly represent individual teeth, making them more suitable for tooth detection. Specifically, we use visual prompts as input for T-Rex2, which means using given tooth images to provide T-Rex2 with customized visual embeddings. These embeddings are then used for object detection in the two-dimensional tooth images obtained through virtual camera projection, resulting in the bounding boxes for the teeth.
In contrast, numerous tooth segmentation networks that directly operate on 3D data, such as point cloud-based segmentation networks, typically require resampling of the point cloud, using techniques like farthest point sampling (FPS). The quality of the resampled point cloud may affect segmentation accuracy. Additionally, these networks are sensitive to the coordinates of the 3D data, necessitating coordinate alignment of the dataset. Point clouds are also susceptible to noise and the nature of the 3D scanner, which limits their generalization capability.
Figure 2 illustrates the 3D-to-2D tooth segmentation process based on bounding box prompts. We segment multiple images projected from different viewpoints to ensure that all surfaces of each tooth can be segmented without being affected by occlusions between teeth. Given a teeth image projected from a viewpoint, manually drawing a bounding box around one tooth will automatically result in bounding boxes around other similar teeth. After inputting the label for each annotated tooth following FDI tooth notation, SAM performs segmentation for all the teeth using bounding box prompts. We then map each pixel in the segmented 2D image back to its corresponding triangular facet on the 3D mesh using an identifier map. This identifier map is generated during the rendering process using PyTorch3D version 0.7.4 (Facebook AI Research, Menlo Park, CA, USA), which records the association between each rendered pixel and a specific face on the original 3D mesh. Through this mechanism, we are able to accurately transfer 2D segmentation results back onto the 3D surface without altering the geometry. The specific details of camera viewpoint settings are provided in Section 4.1 and Figure 1. Since a given mesh face may be observed and labeled differently across multiple viewpoints, a majority voting strategy is adopted to aggregate all label candidates and assign the most frequent label to each mesh face. The existing method, FiboSeg, also performs automatic tooth segmentation using a 3D-to-2D approach. The main difference between FiboSeg and the proposed method is that FiboSeg employs a U-Net network for tooth segmentation, which lacks generalization capability for unseen samples. This is particularly problematic in cases of missing teeth, misaligned teeth, and incomplete upper and lower jaws, resulting in a high error rate. Thanks to the generalization capability of the pre-trained large model and the guidance provided by the bounding boxes, our method requires fewer projection viewpoints (73 viewpoints compared to FiboSeg’s 162 viewpoints) to generate the training set and still achieves accurate segmentation results. In terms of training approach, we fine-tune a pre-trained large model, whereas FiboSeg is trained from scratch. Therefore, our method has stronger capability and flexibility in handling complex and unseen new tasks.
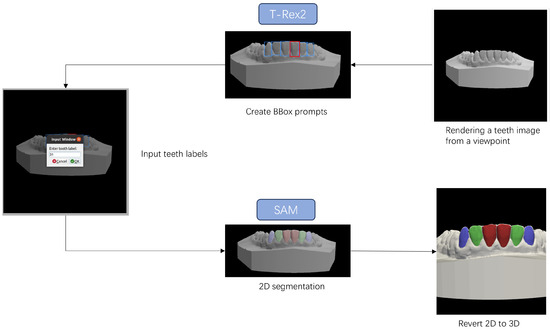
Figure 2.
Tooth segmentation process based on bounding box prompts. By performing a few selections on the 2D tooth images using T-Rex2, bounding boxes for most teeth can be obtained. Subsequently, the segmented pixels from MedSAM can be mapped back to the 3D mesh to complete the tooth segmentation.
3.2. Fine-Tuning Strategy for Pre-Trained Models
Using the SAM model directly for tooth segmentation usually does not achieve satisfactory results, it is necessitating fine-tuning of SAM. In this study, we fine-tuned the mask decoder of both SAM and its medical image-enhanced version, MedSAM, on our own teeth image dataset, while keeping other parameters unchanged. The dataset details used in this study are shown in Table 1 and will be further described in the Section 4. MedSAM is optimized for various medical images, including CT, MRI, and OCT. This model offers two fine-tuning approaches, as follows: one involves fine-tuning both the image encoder and the mask decoder, and the other involves fine-tuning only the mask decoder. Experimental results show that MedSAM’s fine-tuned effect outperforms SAM’s fine-tuned effect on our dataset. For a detailed comparison of the loss functions of SAM and MedSAM, refer to Table 2 in the Section 4. As the image encoder is based on the ViT structure, it typically requires a large amount of data for fine-tuning to achieve good results, and the training time is extensive. Therefore, we freeze the image encoder and fine-tune only the mask decoder, which has fewer parameters. A detailed analysis of MedSAM’s parameters is provided in Table 3. Experimental results confirm the effectiveness of our fine-tuning method.

Table 1.
Training and test sets of 2D-TeethSeg.

Table 2.
Results of fine-tuning mask decoders for SAM and MedSAM on teeth Images.

Table 3.
Parameter analysis of MedSAM.
In our research, we also tried fine-tuning the prompt encoder and used the LoRA technique to fine-tune the image encoder, as described in [23]. However, these methods did not yield satisfactory segmentation results. There is a degradation in performance compared to the results of the SAM model before fine-tuning. This degradation may be due to the numerous annotation categories in the images (with each tooth considered an individual category), which is not suitable for the SAM model’s original binary classification scenario that distinguishes between the target and the background. Therefore, further research is needed to optimize SAM’s fine-tuning strategy for multi-category target segmentation.
To evaluate the effect of fine-tuning the mask decoder, we used two metrics: Dice loss and BCE loss. Dice loss is a metric function for evaluating the similarity of two samples, and its formula is as follows:
where X represents the ground truth tooth segmentation image, and Y represents the predicted tooth segmentation image. The numerator represents the intersection of X and Y, which can be approximated by the dot product of the pixels in the predicted image and the ground truth image, summed over all pixels. The denominator approximates the sum of the pixels in each image.
The binary cross-entropy (BCE) loss function transforms the multiclass tooth segmentation into a binary classification for each tooth. The formula for BCE loss is:
where represents the predicted probability for class n, and represents the true value for class n.
Figure 3 illustrates the fine-tuning process and the network structure used in this work. The network structure of MedSAM is consistent with that of SAM, with the input prompts reduced to only bounding box prompts. In our segmentation framework, we adopt purely visual prompts in the form of bounding boxes, which are used as input to the prompt encoder of the MedSAM model. These bounding boxes are automatically generated using the T-Rex2 object detection model from 2D projections of the 3D tooth mesh, with manual adjustments allowed when necessary. This ensures high-quality prompt input for the segmentation task. A detailed explanation of the bounding box generation process can be found in Section 3.1. Moreover, the tooth bounding box prompts can be found in Figure 2 (“Create BBox prompts”), which visually illustrates how the prompts are constructed and applied.
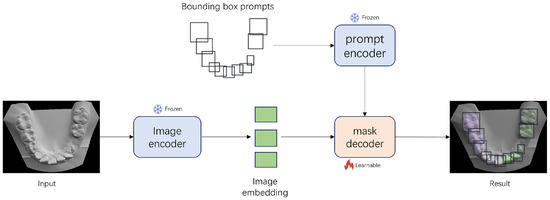
Figure 3.
The network structure of MedSAM and fine-tuning process on teeth images. Only the mask decoder is fine-tuned, while the other parameters are frozen. The prompt encoder uses only bounding box prompts.
4. Experiment
4.1. Dataset
We use the dataset from the 2022 MICCAI 3D Teeth Segmentation Challenge [25] as the training set for fine-tuning MedSAM and as the test set for tooth segmentation in the case of ideal teeth. A total of 900 upper and lower teeth were used for 3D viewpoint projection, and a 2D tooth image dataset was generated and split into training and test sets for the fine-tuning in this paper. In order to avoid the occlusion between the teeth, the viewpoint selection method with uniform sampling of elevation and azimuth angles is adopted, as shown in Figure 1, where the azimuth angles are selected at 30-degree intervals and the elevation angles at 15-degree intervals, resulting in 72 viewpoints. By adding the 90-degree elevation angle, the top view of the projected tooth directly above the tooth, we have 73 viewpoints for virtual camera viewpoint sampling. The ground truth label of the tooth corresponding to each viewpoint image was obtained by projection mapping from the real 3D tooth labels in the MICCAI teeth dataset, as shown in Figure 4.

Figure 4.
Some instances of the training set of 2D-TeethSeg. The top row shows the projected 2D tooth images, while the bottom row displays the corresponding ground truth masks. Each tooth is represented by a different color, and the tooth numbering follows the FDI notation.
Specifically, 3D tooth data from the MICCAI2022 Challenge public dataset 3DTeethSeg22 was used as the fine-tuning dataset (600 pairs of upper and lower teeth, totaling 16,004 teeth) and the test data (300 pairs of upper and lower teeth, totaling 7995 teeth). The MedSAM mask decoder was trained using projected 2D images, all 73 views, with an image resolution of , resulting in a total of 131,400 teeth images, namely 2D-TeethSeg, of which 87,600 were used as training images and 43,800 as test images, as shown in Table 1.
Similarly, we also collected data on teeth in non-ideal conditions as the test set, totaling 103 samples, including 34 cases of misaligned teeth, 21 cases of missing teeth, and 48 cases of incomplete jaw conditions.
4.2. Experimental Settings and Evaluation Metrics
The GPUs used for training were three RTX 3090 (24 GB, NVIDIA Corporation, Santa Clara, CA, USA), and the training time was 4 days, 3 h, 19 min, and 38 s. The test environment for the segmentation algorithm in this work includes an 11th Gen Intel® Core™ i7-11700 @ 2.50GHz processor (Intel Corporation, Santa Clara, CA, USA) with 32 GB of memory and a GeForce RTX 3060 graphics card (10 GB, NVIDIA Corporation, Santa Clara, CA, USA). The operating system is Ubuntu 20.04 (Canonical Ltd., London, UK), and the programming environment consists of Python 3.10 (Python Software Foundation, Wilmington, DE, USA) and PyTorch 2.0 (Meta Platforms, Inc., Menlo Park, CA, USA).
The experimental measures used in this study are consistent with those of the MICCAI 3DTeethSeg22 Challenge, specifically, tooth segmentation accuracy (TSA). TSA is calculated based on the average F1 score of all instances of the tooth point cloud. The F1 score for each tooth instance is calculated as follows:
4.3. Results and Analysis
In the test set containing 43,800 dental images, the fine-tuning results of MedSAM and SAM are compared as shown in Table 2. Although the loss after SAM fine-tuning is already very low, MedSAM performs better in both Dice loss and BCE loss. This indicates that fine-tuning based on MedSAM is more suitable for tooth image segmentation. Moreover, the number of parameters in MedSAM does not increase compared to SAM, and the network structure of both remains the same. The number of parameters in MedSAM is shown in Table 3.
We first test the performance of the proposed method under ideal conditions (no missing teeth, misaligned teeth, and incomplete jaws) by using the test set provided by the MICCAI 3DTeethSeg22 and comparing it with the top six methods in the MICCAI competition (CGIP, FiboSeg, IGIP, TeethSeg, OS, Chompers), and the experimental results are shown in Table 4. As shown in Table 4, the method proposed in this paper achieves the best segmentation results, comparable to other methods. In Table 4, CGIP, IGIP, TeethSeg, and Chompers are all 3D segmentation methods, FiboSeg and this paper’s method are based on 3D-to-2D projection segmentation, and the OS method is a 3D segmentation method that uses 2D dental images to locate the center of the teeth. The 3D tooth segmentation methods are often affected by point sampling. Due to the design of the 3D scanner, the dental mesh exhibits a high sampling density near the boundary, resulting in points that may correspond to multiple labels. This overlap complicates the extraction of precise tooth instance labels. To resolve this, CGIP applies a boundary-aware point sampling method to increase the density of sampled points at the boundary regions. By combining results from both farthest point sampling and boundary-aware point sampling, CGIP effectively achieves accurate tooth instance segmentation. The 2D-based FiboSeg method sometimes results in small holes within the segmented regions, which may be related to the prediction accuracy of U-Net, as the training dataset images have a relatively low resolution of . FiboSeg performs some preprocessing to achieve better results. FiboSeg employs an ‘island removal’ approach at the end of the prediction and a morphological closing operation to smooth the boundary of the segmented teeth.

Table 4.
Metrics of 3D tooth segmentation results.
Regarding the overall segmentation time of the proposed method, considering the human–computer interaction part, we invited five untrained users to participate in the testing, and each of them was asked to segment 5 dental models, and the average time of all users was taken as the segmentation time of this paper. According to the segmentation results of all users, the total time required for tooth segmentation varies significantly from person to person (4∼5 min). As shown in Figure 5, the main time consumption of the segmentation method in our method is in the human–computer interaction (Each upper/lower jaw set needs to select 6∼8 viewpoints, and there are 10∼16 teeth in each viewpoint. In addition, bounding box selection and teeth labels need to be input using the FDI tooth notation), and the inference of the MedSAM model and the T-Rex2 model occupies a small portion of the time. The human–computer interaction component can be further optimized in future work.
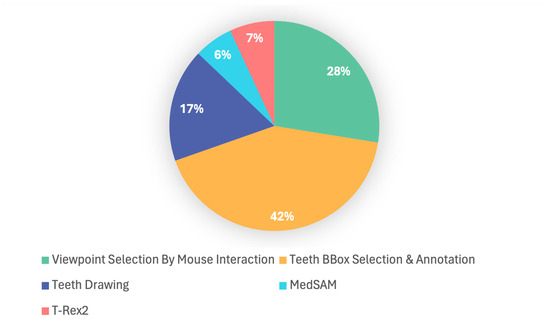
Figure 5.
Breakdown of our tooth segmentation time. Teeth BBox selection and annotation accounts for the largest share (42%), followed by the viewpoint selection by mouse interactions (28%) and teeth drawing (17%). T-Rex2 (7%) and MedSAM (6%) represent smaller shares; their inclusion demonstrates the integration of automated tools, contributing to improved annotation efficiency and accuracy.
Compared with traditional manual tooth segmentation, which typically takes between 10 and 30 min per model, depending on case complexity and operator experience, our method significantly reduces the time required by leveraging automation. The automatic generation of bounding boxes through the T-Rex2 module and the fast inference of the MedSAM model together minimize manual effort, making the entire segmentation process substantially more efficient.
In addition, to verify the stability and accuracy of our method under non-ideal conditions, we test it on patients with missing teeth, misaligned teeth, and diagnostic cases of incomplete jaws, comparing the results with those of the method proposed by the CGIP team [25], the FiboSeg team [25], and MeshSeg [11]. As shown in Figure 6, for the case of tooth misalignment, all the methods except ours have different degrees of under-segmentation and mis-segmentation, which leads to the confusion of the number of the final segmented teeth. However, the present method is almost unaffected. As shown in Figure 7, in cases of missing teeth, most methods have over-segmentation, which is caused by interference from the gingiva at the location of the missing tooth. This leads to errors such as recognizing the same tooth as two teeth, or misidentifying the gingiva in the missing tooth area as an actual tooth. Through the above tests, the following conclusion can be drawn: fully automated methods are highly dependent on the training dataset, resulting in limited generalization ability. Our method effectively avoids these errors in cases of missing and misaligned teeth, achieving more accurate and stable tooth segmentation.

Figure 6.
Comparison of segmentation results for misaligned teeth. Each row shows a misaligned case with outputs from CGIP, FiboSeg, MeshSeg, and our method. Existing methods exhibit label confusion or under-segmentation due to irregular spacing. Our method yields clearer boundaries and more accurate tooth separation, demonstrating better performance in complex alignments.

Figure 7.
Comparison of segmentation results for missing teeth. Each row shows a case with one or more missing teeth, with results from CGIP, FiboSeg, MeshSeg, and our method. Other methods often misidentify the gingiva as teeth or over-segment adjacent structures. Our method avoids such errors, producing cleaner and more accurate segmentation near missing regions.
In order to further verify the segmentation results in the case of incomplete jaws, some teeth were cropped and tested in this study, and the results are shown in Figure 8. In the case of incomplete jaws, the segmentation accuracy of most of the methods is greatly reduced, and in some cases, segmentation cannot be performed at all. However, our method can effectively overcome the challenges posed by jaw incompleteness, providing more accurate segmentation. It demonstrates strong generalization and offers greater flexibility for practical use by dentists. More segmentation comparison results are provided, including missing teeth, misaligned teeth, and incomplete jaws, as shown in Figure 9, Figure 10 and Figure 11.
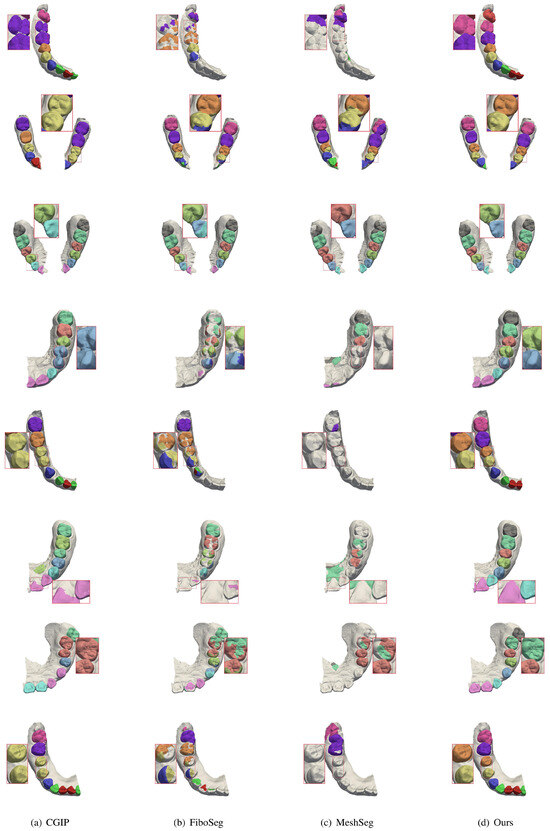
Figure 8.
Comparison of segmentation results for incomplete upper and lower jaws. Each row presents a case with partial jaw data. Competing methods struggle with missing context, leading to incomplete or fragmented segmentation. Our method maintains tooth integrity and delivers accurate results despite limited anatomical information.
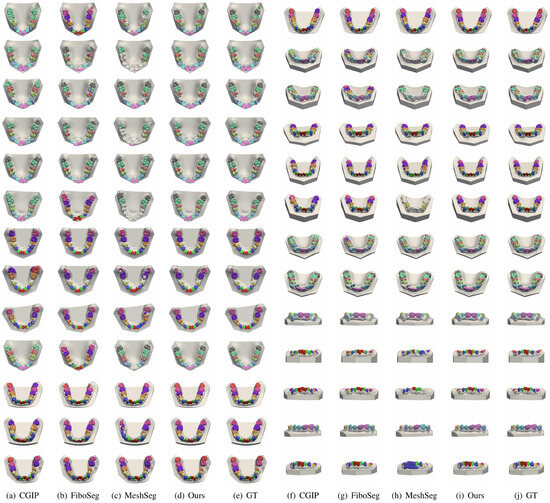
Figure 9.
More segmentation results for misaligned teeth. This figure shows more examples of severe misalignment. Our method consistently produces accurate and well-separated tooth boundaries, while other methods show errors such as tooth merging or boundary loss.

Figure 10.
More segmentation results for missing teeth. This figure shows more cases with one or more missing teeth. Other methods often over-segment or misclassify soft tissue. Our method produces cleaner outputs and accurately ignores missing regions.

Figure 11.
More segmentation results for incomplete upper and lower jaws. More examples with partial jaw scans are shown. Competing methods often fail to segment teeth fully or introduce artifacts. Our method remains robust, preserving tooth shape and completeness, despite missing structures.
5. Conclusions
In this paper, we propose a prompt-guided 3D tooth segmentation method that leverages multi-view 2D image projections and visual prompts to achieve accurate and efficient dental modeling. By using a fine-tuned SAM model in combination with automatically generated bounding boxes from the T-Rex2 module, we significantly reduce annotation time and improve segmentation robustness under challenging clinical conditions, including missing teeth, misalignment, and incomplete jaws. Extensive experiments on the MICCAI 3DTeethSeg’22 dataset validate the effectiveness of our approach. Our method demonstrates excellent generalization capabilities and outperforms existing techniques, particularly in cases that deviate from standard anatomical structures. Furthermore, the proposed framework can be potentially integrated into existing dental CAD/CAM workflows, as it supports fast and accurate segmentation from 3D scans. With appropriate adaptation, the method could also be used in clinical environments, such as pairing with intraoral scanners for real-time analysis or assisting in pathology detection and treatment planning in complex dental cases. Although the current state of human–computer interaction in this work is still at a preliminary stage, in future work, we could introduce additional contextual cues, such as center point prompts based on the dental arch, to accelerate the labeling process for bounding boxes in 2D images.
Author Contributions
Conceptualization, F.L. and R.F.; methodology, F.L. and C.W.; software, Y.C.; validation, C.W.; formal analysis, Y.C.; investigation, Y.C.; resources, R.F.; data curation, F.L. and C.W.; writing—Y.C. and C.W.; writing—review and editing, F.L.; visualization, C.W.; supervision, F.L.; project administration, F.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by the Zhejiang Provincial Natural Science Foundation of China under Grant No. LZ23F020002 and the National Natural Science Foundation of China under Grant No. 61972458.
Data Availability Statement
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
References
- Jana, A.; Maiti, A.; Metaxas, D.N. A Critical Analysis of the Limitation of Deep Learning based 3D Dental Mesh Segmentation Methods in Segmenting Partial Scans. In Proceedings of the 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Sydney, Australia, 24–27 July 2023; pp. 1–7. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; et al. Segment Anything. In Proceedings of the 2023 IEEE/CVF International Conference on Computer Vision (ICCV), Paris, France, 2–3 October 2023; pp. 3992–4003. [Google Scholar]
- Kondo, T.; Ong, S.H.; Foong, K.W.C. Tooth segmentation of dental study models using range images. IEEE Trans. Med. Imaging 2004, 23, 350–362. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Abaza, A.A.; Ross, A.; West, H.A. Automatic Tooth Segmentation Using Active Contour without Edges. In Proceedings of the 2006 Biometrics Symposium: Special Session on Research at the Biometric Consortium Conference, Baltimore, MD, USA, 19–21 September 2006; pp. 1–6. [Google Scholar]
- Nomir, O.; Abdel-Mottaleb, M. A system for human identification from X-ray dental radiographs. Pattern Recognit. 2005, 38, 1295–1305. [Google Scholar] [CrossRef]
- Said, E.; Nassar, D.; Fahmy, G.; Ammar, H. Teeth segmentation in digitized dental X-ray films using mathematical morphology. IEEE Trans. Inf. Forensics Secur. 2006, 1, 178–189. [Google Scholar] [CrossRef]
- Xu, X.; Liu, C.; Zheng, Y. 3D Tooth Segmentation and Labeling Using Deep Convolutional Neural Networks. IEEE Trans. Vis. Comput. Graph. 2019, 25, 2336–2348. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Dai, N.; Zhang, B.; Yuan, F.; Yu, Q.; Cheng, X. Automatic Classification and Segmentation of Teeth on 3D Dental Model Using Hierarchical Deep Learning Networks. IEEE Access 2019, 7, 84817–84828. [Google Scholar] [CrossRef]
- Cui, Z.; Li, C.; Wang, W. ToothNet: Automatic Tooth Instance Segmentation and Identification From Cone Beam CT Images. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 6361–6370. [Google Scholar]
- Zhang, L.; Zhao, Y.; Meng, D.; Cui, Z.; Gao, C.; Gao, X.; Lian, C.; Shen, D. TSGCNet: Discriminative Geometric Feature Learning with Two-Stream Graph Convolutional Network for 3D Dental Model Segmentation. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 19–25 June 2021; pp. 6695–6704. [Google Scholar]
- Wu, T.H.; Lian, C.; Lee, S.; Pastewait, M.F.; Piers, C.; Liu, J.; Wang, F.; Wang, L.; Chiu, C.Y.; Wang, W.; et al. Two-Stage Mesh Deep Learning for Automated Tooth Segmentation and Landmark Localization on 3D Intraoral Scans. IEEE Trans. Med Imaging 2021, 41, 3158–3166. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Li, K.; Tan, K.; Feng, Y.; Zhou, J.T.; Hao, J.; Liu, Z. TFormer: 3D tooth segmentation in mesh scans with geometry guided transformer. arXiv 2022, arXiv:2210.16627. [Google Scholar]
- Liu, Z.; He, X.; Wang, H.; Xiong, H.; Zhang, Y.; Wang, G.; Hao, J.; Feng, Y.; Zhu, F.; Hu, H. Hierarchical Self-Supervised Learning for 3D Tooth Segmentation in Intra-Oral Mesh Scans. IEEE Trans. Med. Imaging 2022, 42, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16×16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- He, K.; Chen, X.; Xie, S.; Li, Y.; Doll’ar, P.; Girshick, R.B. Masked Autoencoders Are Scalable Vision Learners. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognitionm, Nashville, TN, USA, 20–25 June 2021; pp. 15979–15988. [Google Scholar]
- Tancik, M.; Srinivasan, P.; Mildenhall, B.; Fridovich-Keil, S.; Raghavan, N.; Singhal, U.; Ramamoorthi, R.; Barron, J.; Ng, R. Fourier features let networks learn high frequency functions in low dimensional domains. Adv. Neural Inf. Process. Syst. 2020, 33, 7537–7547. [Google Scholar]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J.; et al. Learning transferable visual models from natural language supervision. In Proceedings of the International Conference on Machine Learning, Vienna, Austria, 18–24 July 2021; pp. 8748–8763. [Google Scholar]
- Zhang, R.; Jiang, Z.; Guo, Z.; Yan, S.; Pan, J.; Ma, X.; Dong, H.; Gao, P.; Li, H. Personalize segment anything model with one shot. arXiv 2023, arXiv:2305.03048. [Google Scholar]
- Zhao, X.; Ding, W.; An, Y.; Du, Y.; Yu, T.; Li, M.; Tang, M.; Wang, J. Fast segment anything. arXiv 2023, arXiv:2306.12156. [Google Scholar]
- Zhang, C.; Han, D.; Qiao, Y.; Kim, J.U.; Bae, S.H.; Lee, S.; Hong, C.S. Faster segment anything: Towards lightweight sam for mobile applications. arXiv 2023, arXiv:2306.14289. [Google Scholar]
- Ma, J.; He, Y.; Li, F.; Han, L.J.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Z.; Hong, M.; Ji, W.; Fu, H.; Xu, Y.; Xu, M.; Jin, Y. Medical sam adapter: Adapting segment anything model for medical image segmentation. Med. Image Anal. 2025, 102, 103547. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Liu, D. Customized segment anything model for medical image segmentation. arXiv 2023, arXiv:2304.13785. [Google Scholar]
- Liu, Y.; Li, W.; Wang, C.; Chen, H.; Yuan, Y. When 3D partial points meets sam: Tooth point cloud segmentation with sparse labels. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; pp. 778–788. [Google Scholar]
- Ben-Hamadou, A.; Smaoui, O.; Rekik, A.; Pujades, S.; Boyer, E.; Lim, H.; Kim, M.; Lee, M.; Chung, M.; Shin, Y.G.; et al. 3DTeethSeg’22: 3D teeth scan segmentation and labeling challenge. arXiv 2023, arXiv:2305.18277. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).