Abstract
This study aims to elucidate the genomic characteristics of Bacillus subtilis MGE 2012, a strain isolated from Korean traditional fermented food, meju, which contributes to its high enzyme activity and potential applications. The whole genome sequence of B. subtilis MGE 2012 was assembled using MEGAHIT, annotated using RAST and BLASTKOALA v3.1. Phylogenetic analysis placed MGE 2012 within the Bacillus clade, showing high similarity to B. subtilis NCIB 3610 and B. subtilis ATCC 6051. AntiSMASH analysis identified 14 biosynthetic gene clusters (BGCs) capable of producing various secondary metabolites, including subtilosin, bacillibactin, fengycin, bacilysin, plipastatin, and surfactin. This study provides an overview of the whole genome and secondary metabolite profile of B. subtilis MGE 2012, emphasizing its potential applications in biotechnology. While the primary focus of this study was to explore the genomic characteristics and secondary metabolite profile, future research could delve deeper into genome mining for enzyme activities and their applications.
1. Introduction
The Bacillus genus is a highly versatile group that includes both pathogenic and plant growth-promoting bacteria [1]. Strains classified as B. subtilis and Bacillus amyloliquefaciens exhibit characteristics beneficial to the biotechnology industry, such as the production of extracellular enzymes like cellulase, protease, lipase, xylanase, and phytase, as well as bioactive compounds like antibiotics [2]. Bacillus species effectively control pathogens through various mechanisms, including antibiotic production, the induction of host–plant resistance, and the promotion of plant growth [3]. Antibiotic production is the primary mechanism behind their biological control effects [4].
Formerly known as Vibrio subtilis, its current nomenclature was devised by Ferdinand Cohn in 1872. Upon sequencing its genome, the first Gram-positive bacterium was discovered to have a single 4.2 Mbp chromosome with a 43% G+C content. Only 253 of the roughly 4200 genes in the genome are necessary for laboratory cultivation [5]. Surfactin, plipastatin, and bacillaene are examples of bioactive secondary metabolites that provide B. subtilis its antibacterial and antifungal characteristics [6]. During World War II, it was discovered that B. subtilis could treat dysentery. It secretes exopolysaccharides with probiotic properties that help reduce inflammation and illnesses brought on by enteric pathogens [7]. B. subtilis demonstrates a variety of spreading behaviors in the laboratory, such as multicellular swarming, sliding enabled by exopolysaccharides, surfactin, and hydrophobin, and single-cell motility via peritrichous flagella [8]. B. subtilis produces up to 20–25 g of proteins per liter of media and is a great platform for manufacturing a variety of enzymes due to its simple cultivation and effective secretion mechanism. These enzymes are used in dairy, baking, animal feed, textile, washing, and pharmaceutical industries [9]. The matrix components of the B. subtilis biofilm, which comprise amyloid fibers, exopolysaccharides, and the tiny hydrophobin protein BslA, make it more repellant than Teflon [10].
Due to its innate ability to absorb extracellular DNA, which enables relatively straightforward genetic alteration and the occurrence of sporulation—one of the earliest bacterial cell differentiation processes to be studied—B. subtilis rose to prominence as the most researched species in the genus Bacillus. The latent spores can withstand extreme weather conditions (high temperatures, desiccation, UV, and γ-radiation), microbial and macroorganism predation, and even alien environments. From soil to marine ecosystems, B. subtilis can be isolated and used for a variety of purposes, including food fermentation, enzyme synthesis, antimicrobial biosynthetic gene clusters production, and plant biocontrol. In addition, cell division, protein secretion, surface motility (swimming, swarming, and sliding), biofilm formation, attachment to fungal hyphae or plant roots, secondary metabolite production, cytoplasmic exchange via intercellular nanotubes, extracellular vesicle release, and kin-discrimination can all be studied using B. subtilis as a model microbe [11].
In our previous study, we identified a highly active strain of B. subtilis from the Korean traditional fermented food, meju, which exhibited significant alpha-amylase and protease enzyme activities [12]. The existence of superior alleles—genetic variations that increase the expression or activity of important biosynthetic genes—is frequently the cause of variations in the biosynthetic efficiency of different B. subtilis populations. The strain’s ability to produce enzymes and secondary metabolites may be impacted by these allelic differences. While this study’s main goal is to use whole genome sequence analysis to investigate the secondary metabolite profile of the MGE 2012 strain, it also provides a foundation for future allele-level comparisons that could help explain the strain’s enhanced functional potential.
2. Materials and Methods
2.1. Genome Assembly and Annotation
B. subtilis MGE 2012 was isolated from meju in previous research [12]. Genomic DNA extraction kit (G-spinTM) was used for bacterial DNA isolation. The isolated genomic DNA was then subjected to whole genome sequencing using iSeq100 (Illumina, Inc., San Diego, CA, USA) using standard protocols. The sequencing reads were then processed for the quality by trimming adapter sequences using Trimmomatic v0.39 [13]. The good quality reads were then assembled using MEGAHIT v1.2.9 [14] assembler. RAST (Rapid Annotation using Subsytem technology) [15], Prokka v1.14.5 [16], and BLASTKOALA v3.1 [17] were used to perform the functional annotation of the MGE 2012 sequence because of their ability to provide accurate and comprehensive annotation for the prokaryotes. EggNOG-mapper v2 [18] was used for orthology assignment and predicted COGs were then categorized into functional classes to provide insights into the metabolic potential of B. subtilis MGE 2012.
2.2. Phylogenetic Analysis
For a whole genome sequence-based taxonomic analysis, the genome sequence of B. subtilis MGE 2012 was uploaded on Type (Strain) Genome Server (TYGS) [19]. The List of Prokaryotic names with Standing in Nomenclature (LPSN, https://lpsn.dsmz.de, accessed on 7 January 2025) [20] supplied information on nomenclature, synonymy, and related taxonomic literature. The GGDC server (https://ggdc.dsmz.de/, accessed on 7 January 2025) was used for performing in silico DNA–DNA hybridization [21]. Two complementary methods were used to determine the closest type of strain genomes: First, the MASH [22] algorithm was used to compare the MGE 2012 genome to all type strain genomes in the TYGS database, and the 10 type strains with the shortest MASH distances were selected. Second, using the 16S rRNA gene sequences, ten more closely related type strains were identified. This was accomplished by using RNAmmer [23] to extract the 16S rRNA gene sequence from the MGE 2012 genome, followed by BLASTed [24] against the 16S rRNA gene sequences found in the TYGS database. Using this as a proxy, the top 50 matching type strains (based on bitscore) for the MGE 2012 genome were identified. Then, using the Genome BLAST Distance Phylogeny method (GBDP), using the distance formula d4 [21] and algorithm “coverage”, exact distances were determined. Ultimately, the ten closest type strain genomes for the MGE 2012 genome were identified using these distances. Confidence intervals and digital DDH values were computed using the GGDC 3.0’s [20] recommended settings. Average Nucleotide Identity was calculated using OAT (Orthologous Average Nucleotide Identity Tool) from EzBioCloud (https://www.ezbiocloud.net/, accessed on 8 January 2025) [25]. OAT calculates the total similarity between two genomic sequences using OrthoANI. Large comparative studies have shown that the algorithms ANI and OrthoANI create nearly identical reciprocal similarities, and they both have the same species delineation cut-off of 95–96%.
2.3. Prediction of Biosynthetic Gene Cluster
The presence of biosynthetic gene clusters in the B. subtilis MGE 2012 genome was examined using AntiSMASH v7.1.0 [26], a tool widely utilized to identify and characterize secondary metabolite gene clusters. For analysis, the entire genome sequence was submitted to the AntiSMASH web server (https://antismash.secondarymetabolites.org, accessed on 20 January 2025), and the prediction was made using the default settings. BGCs were found and annotated by AntiSMASH using integrated methods for domain discovery, cluster type prediction, and comparison analysis. These algorithms were based on sequence homology to known clusters in its database. Results were grouped according to their potential for the biosynthesis of secondary metabolites.
3. Results
3.1. Genome Annotation
MGE 2012’s genome is made up of a single circular chromosome that is 3,759,743 bp long and contains 43.5% G+C. There were 4468 predicted coding sequences (CDS), among which 2632 were unique functional genes identified in the MGE 2012 genome, while 39 predicted RNAs were found (Table S1). The whole genome of MGE 2012 was sequenced, assembled, and processed for the deposit in the GenBank database (accession number JBNAYZ000000000).
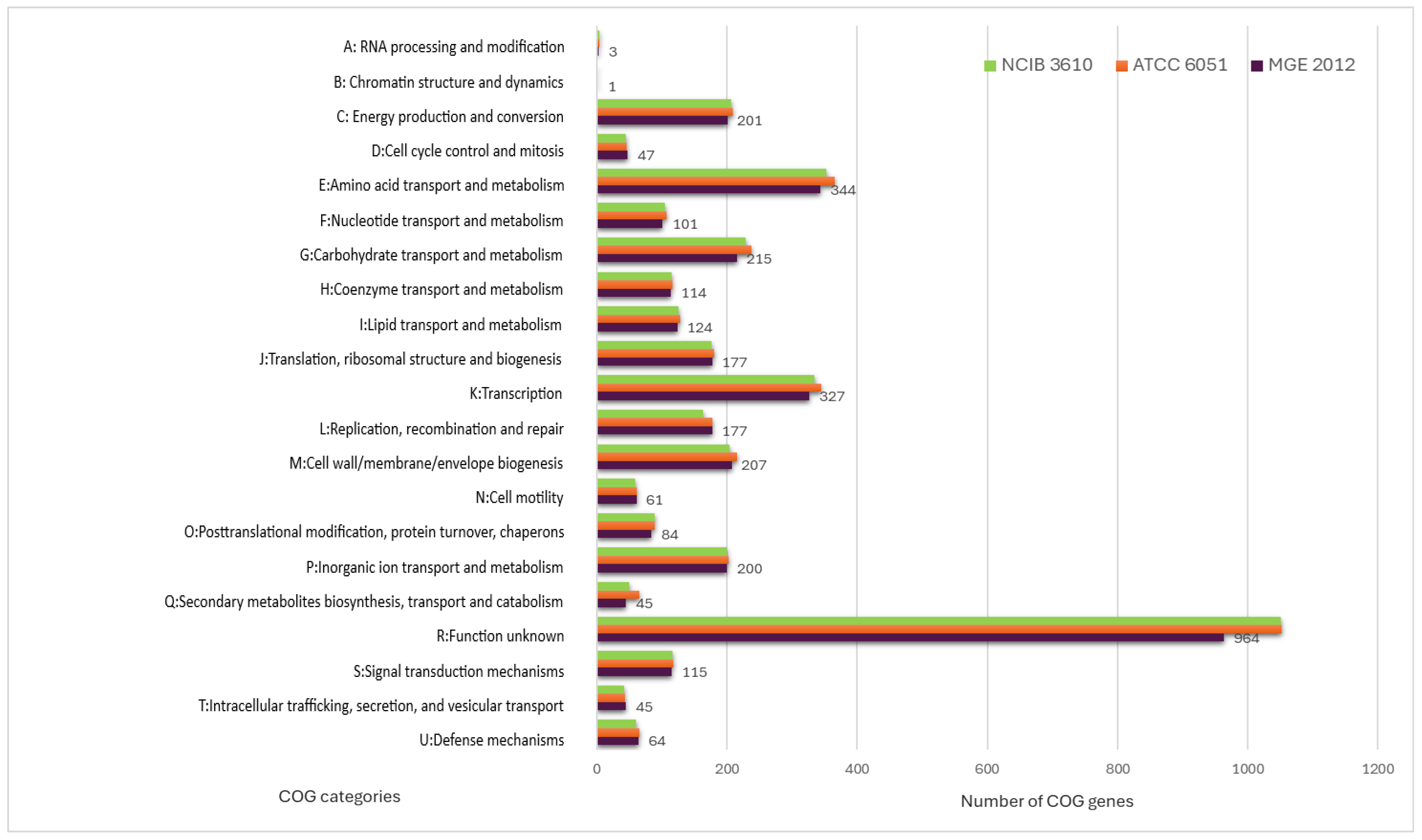
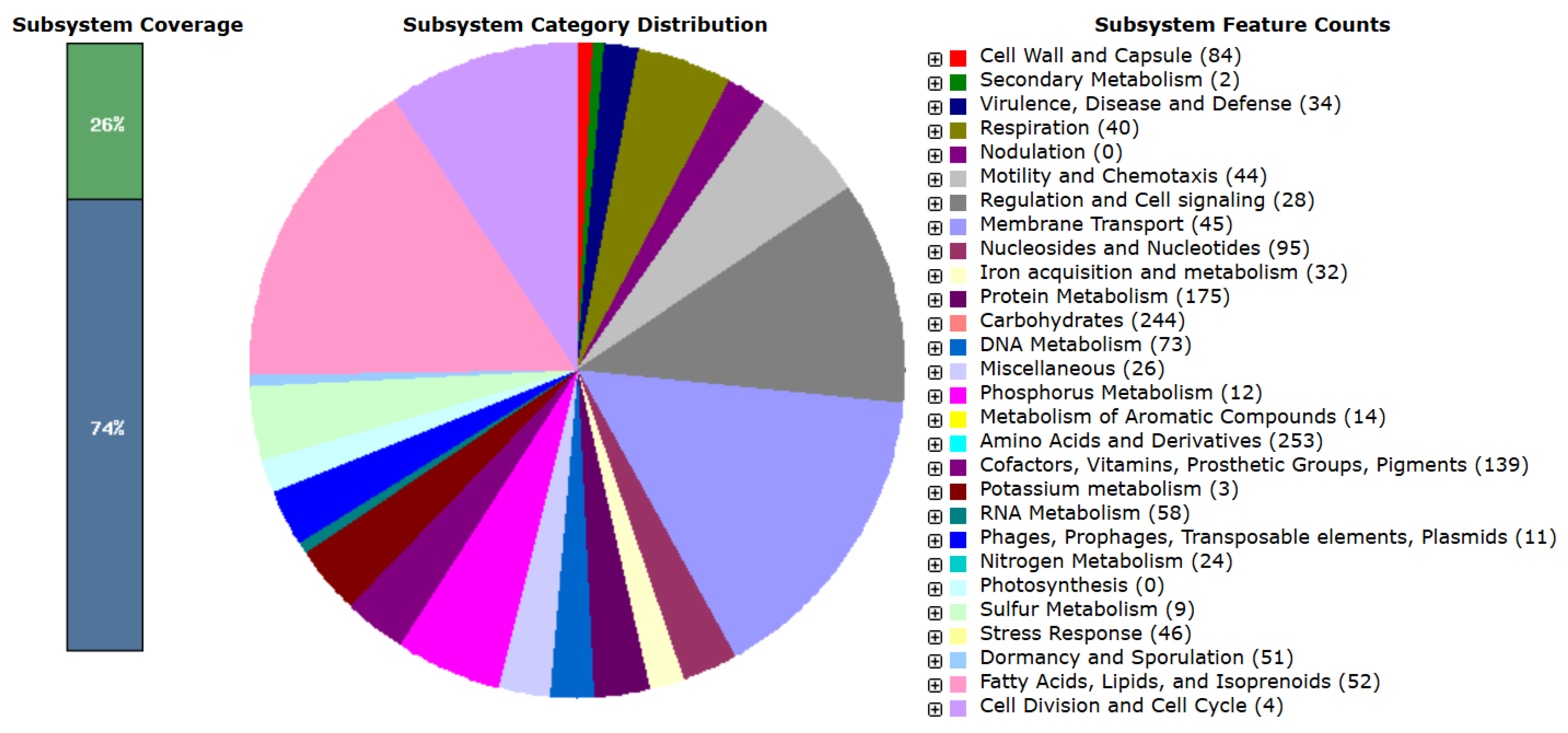
The eggNOG-mapper algorithm was used to functionally classify the predicted genes in the MGE 2012 genome (Table S2) [18]. The distribution within the COG categories is shown in Figure 1. When analyzing B. subtilis ATCC 6051 and B. subtilis NCIB 3610 together, a slight difference in the number of genes was observed, even though they are the same species. A total of 21 functional annotations was recorded, with the highest number of genes (964) associated with an unknown function, followed by amino acid transport and metabolism (344), transcription (327), carbohydrate transport and metabolism (215), cell wall/membrane/envelope biogenesis (207), energy production and conversion (201), and inorganic ion transport and metabolism (200). The 45 genes associated with secondary metabolite biosynthesis are highlighted, while the rest of the genes remain to be annotated (Figure 1). Specifically, 253 coding sequences in strain MGE 2012 were annotated as “amino acids and derivatives” based on the RAST subsystem statistics (Table S3 and Figure 2). This detailed RAST sub-categorization suggests that the analyzed strains assimilate glutamine, aspartate, asparagine, ammonia, and so on, which are essential for histidine metabolism and amino acid metabolisms. The subsystem feature counts from the RAST system showed 34 features associated with virulence, disease, and defense with genes related to invasion and intracellular resistance (12 genes), antibiotic resistance (15 genes), and bacteriocin production (7 genes). No genes were found in important virulence categories like adhesion, detection, toxins and superantigens, and general virulence factors without any subcategory. The strain does not possess the genetic characteristics often linked to active pathogenesis, as seen by the lack of recognized adhesins and toxins. The idea that this strain is unlikely to induce infection under typical circumstances is further supported by the non-specific virulence markers. These results suggest that the strain may have low virulence, making it suitable for safe use.
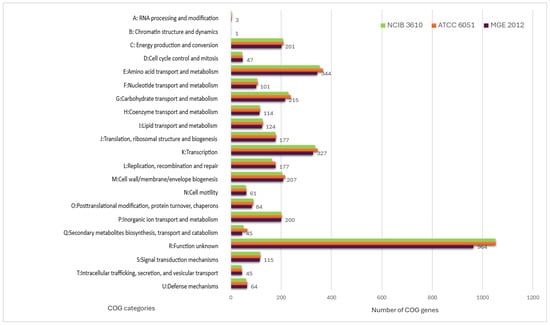
Figure 1.
Categorization of the MGE 2012, B. subtilis ATCC 6051 and B. subtilis NCIB 3610 genome’s orthologous functional gene cluster. The number of genes allocated to each COG functional category is shown by different bars.
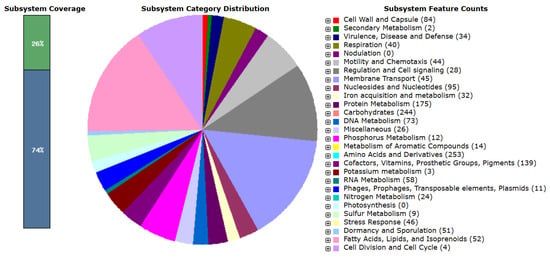
Figure 2.
Feature counts associated with different cellular subsystems based on RAST annotation.
Additionally, multiple antibiotic and cobalt–zinc–cadmium resistance genes, which presumably enable these bacteria to live in toxic environments, were detected.
3.2. Phylogenetic Relationship of B. subtilis MGE2012
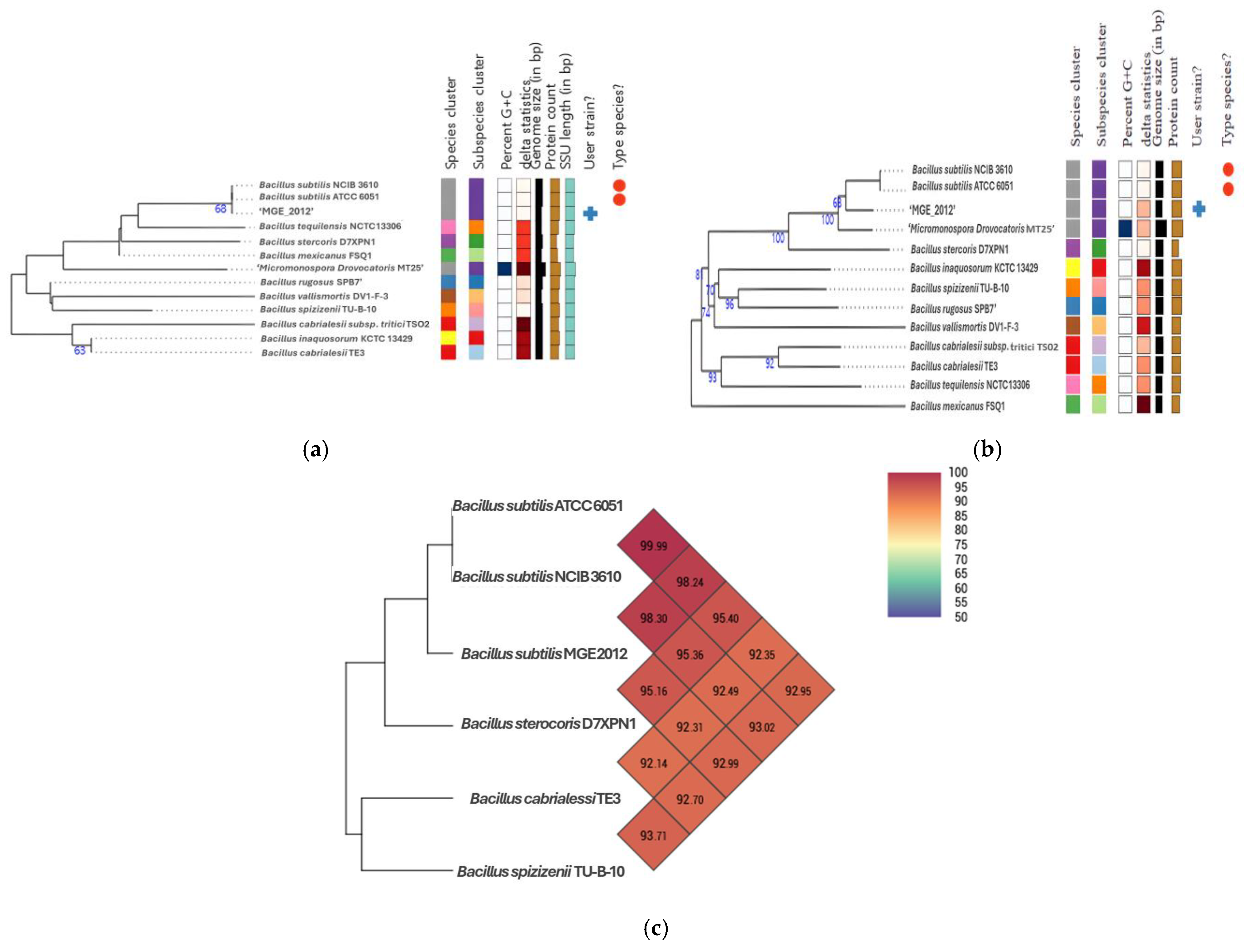
After the whole genome sequencing, average nucleotide identity (ANI) calculations and digital DNA-DNA hybridization (dDDH) were performed to further establish its taxonomic place. Phylogenetic trees generated by the type genome server (TYGS) classified MGE 2012 within the Bacillus clade, showing high similarity to B. subtilis NCIB 3610 and B. subtilis ATCC 6051 (Figure 3a,b). ANI and dDDH analyses revealed that MGE 2012 belongs to the same species as B. subtilis ATCC 6051 (ANI: 98.24%, dDDH: 86.6%) and B. subtilis NCIB 3610 (ANI: 98.30%, dDDH: 86.4%) (Figure 3c, Tables S4 and S5). These values exceed the typical species thresholds of ANI 95–96% and dDDH 70%.
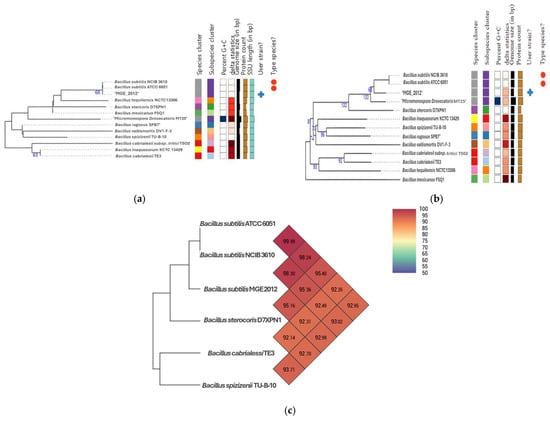
Figure 3.
GBDP distances computed from (a) 16S rDNA gene sequences, (b) genome sequences were used to estimate a tree using FastME 2.1.6.1 [27]. The GBDP distance formula d5 is used to scale the branch lengths. With an average branch support of 38.2/82.5% apiece, the statistics above represent GBDP pseudo-bootstrap support values > 60% from 100 replications. The midpoint was where the tree was rooted [28]. (c) Heatmap of MGE 2012 and other relatives generated with OrthoANI values calculated from the OAT software v0.93.1 [29].
3.3. Genomic Identification and Functional Annotation of Amylases and Proteases
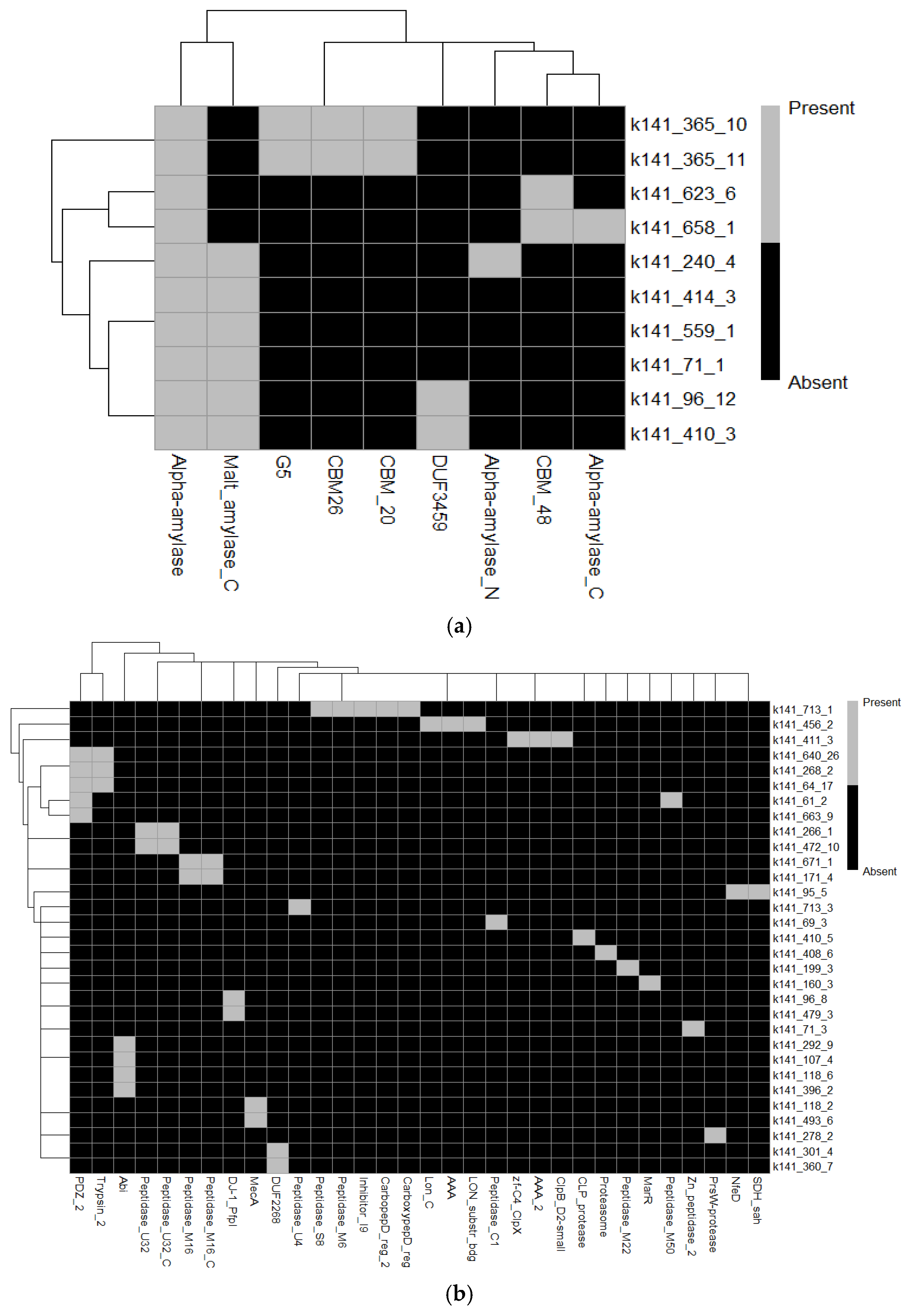
Using eggNOG to annotate the B. subtilis MGE 2012 genome, 10 amylase-related sequences were found (Table S6). These were mainly linked to the glycoside hydrolase (GH13) family, which is well-known for its alpha-amylase activity. These sequences were found to possess a variety of enzymatic activities, such as trehalose-6-phosphate hydrolase activity (EC 3.2.1.93), malt-amylase activity (EC 3.2.1.133, 3.2.1.135), and alpha-amylase activity (EC 3.2.1.1, 3.2.1.10, 3.2.1.20). According to a functional analysis, their association with KEGG pathways like map00500, map01100, and map01110 demonstrated their involvement in important metabolic pathways like the metabolism of starch and sucrose, glycolysis/gluconeogenesis, and other metabolic activities. Furthermore, their inclusion in CAZy families like CBM48, GH13, and GH31 emphasizes their important functions in the metabolism and hydrolysis of carbohydrates. As the biggest glycoside hydrolase family, the GH13 family contains enzymes that aid in the hydrolysis of starch and related polysaccharides, including pullulanases, α-amylases, and maltogenic amylases. Enzymes having α-glucosidase and α-xylosidase activity, which are part of the GH31 family, help break down oligosaccharides into fermentable sugars. Furthermore, starch-binding domains are frequently associated with CBM48 (Carbohydrate-Binding Module 48), which improves the substrate specificity of glycoside hydrolases. The existence of several CAZymes indicates that B. subtilis MGE 2012 has a robust enzyme system for energy metabolism, starch breakdown, and possible industrial uses in food processing and bioconversion. Their functional relevance was further supported by the identification of specific reactions, such as R02108, R02112, and R11262. Strong sequence alignment scores and low e-values indicated great confidence in the annotation and functional relevance of notable amylase-related genes, including amyE, amyX, treC, and malL. These results highlight the existence of several glycoside hydrolase enzymes in B. subtilis and highlight their possible functions in the hydrolysis of starch, the metabolism of carbohydrates, and industrial activities including food processing and bioconversion (Table S6 and Figure 4a).
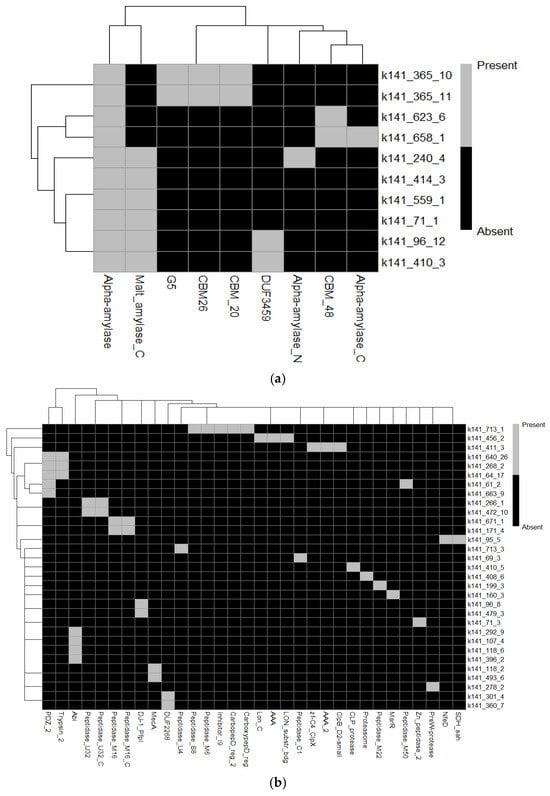
Figure 4.
(a): Heatmap showing the distribution of protein functional domains (x-axis) across identified amylase genes (y-axis) in Bacillus subtilis MGE2012. (b): Heatmap showing the distribution of protein functional domains (x-axis) across identified protease genes (y-axis) in Bacillus subtilis MGE2012.
Numerous proteases and related proteins with different e-values and functional annotations were found in the dataset, mostly from Bacillus species. PrsW (k141_278_2), a member of the COG2339 family, is implicated in the breakdown of anti-sigma factors. Strong matches in the COG5504 family were found for YjfC (k141_301_4) and YjaZ (k141_360_7), which were predicted to be Zn-dependent proteases (DUF2268). A membrane-bound serine protease (ClpP class) with a KEGG orthology (K07403) was identified as YqeZ (k141_95_5). Furthermore, MecA (k141_118_2 and k141_493_6), a protein that targets proteolysis and is a negative regulator of competence development, is linked to KEGG K16511. According to KEGG K07052, YpbD (k141_118_6) and YdiL (k141_396_2) were found to be metal-dependent membrane proteases and CAAX protease self-immunity proteins, respectively.
Additionally, YfkM (k141_96_8) and YugP (k141_71_3) were designated as general proteases (DJ-1/PfpI class) and Zn-dependent proteases with KEGG orthology (K06973 and K05520). While YrrO (k141_266_1 and k141_472_10) was discovered to be a collagenase-like protease (COG0826) with KEGG K08303, YmfH (k141_671_1 and k141_171_4) also matched zinc proteases. Notably, the zinc metalloprotease RasP (k141_61_2) mapped to K11749 and was linked to pathways like ko02024 and ko0411. HtrA (k141_268_2 and k141_64_17), another trypsin-like serine protease, matched KEGG K04771 and took part in proteolytic pathways such as ko01503 and ko02020. Hpr (k141_160_3), a negative regulator of sporulation and protease synthesis, was finally linked to KEGG K09682 (Table S7 and Figure 4b).
Collectively, these regulatory proteins and proteases are essential for Bacillus species’ cellular control, stress response, and protein breakdown. They also reveal possible targets for biotechnological applications and provide further understanding of the adaptability and survival mechanisms of bacteria.
3.4. Biosynthetic Gene Clusters
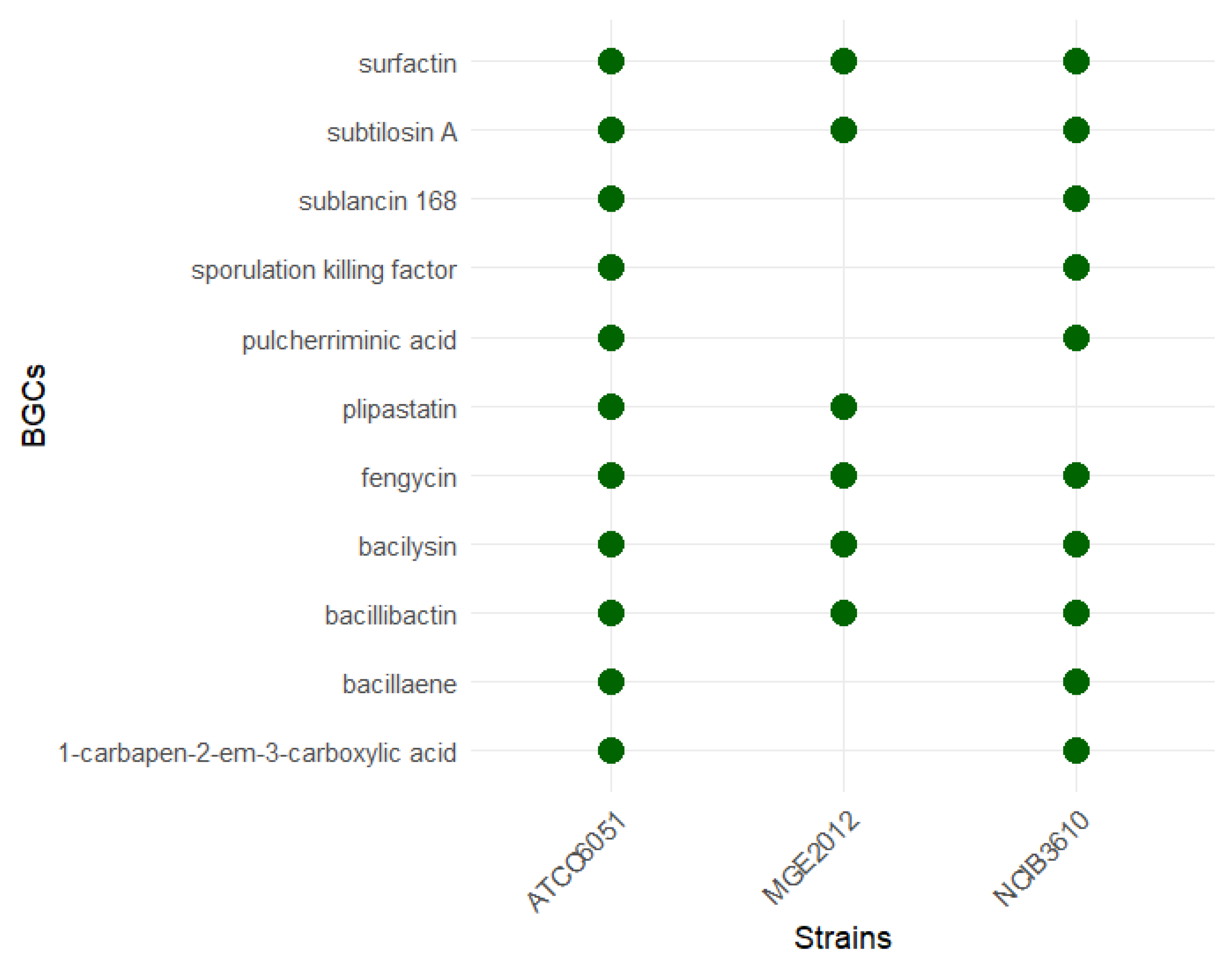
Out of a total of 494 contigs of the bacterial genome, antiSMASH 7.1 predicted a total of 14 BGCs on 14 contigs (Table 1 and Figure 5). Among the 14 BGCs, 7 were annotated as NRPS or NRP-like genes, while the remaining 7 were annotated as RiPP-like, T3PKS, sactipeptide, betalactone, terpene, and other genes. Six BGCs identified in the MGE 2012 genome shared homology with known BGCs, including subtilosin A, bacillibactin, fengycin, bacilysin, plipastatin, and surfactin.

Table 1.
Using antiSMASH 7.1, putative gene clusters of B. subtilis MGE 2012 secondary metabolites were identified.

Figure 5.
Organization of putative novel BGCs coding for (A) bacillibactin (NRPS); (B) bacilysin (other); (C) fengycin (betalactone); (D) plipastatin (NRPS-like); (E) surfactin (NRPS); (F) plipastatin (NRPS); (G) surfactin (NRPS); (H) subtilosin (sactipeptide); and (I) plipastatin (NRPS), in B. subtilis MGE 2012 genome.
Comparative analysis of MGE2012 with the genomes of other prevalent B. subtilis strains; e.g., NCIB3610 and ATCC6051 using antiSMASH revealed that MGE2012 has fewer BGCs and no unique BGCs were identified in MGE2012 compared to the reference genomes (Figure 6).
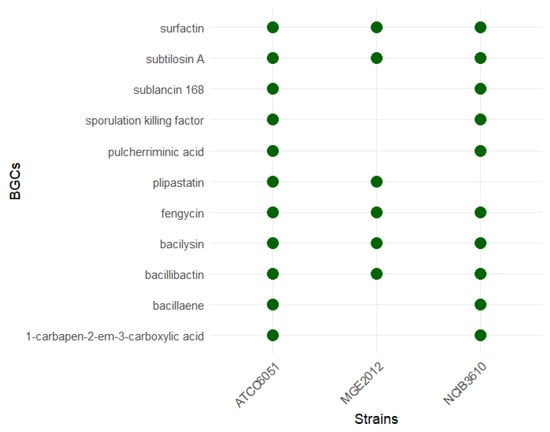
Figure 6.
Presence of biosynthetic gene clusters (BGCs) per strain.
4. Discussion
The actual ecological function of specialized metabolites is only now becoming clear to us. The generating strains have a competitive advantage in establishing themselves inside an ecological niche thanks to secondary metabolites, which are mainly regarded as biological weapons [30]. However, at subinhibitory concentrations, secondary metabolites are also identified as agents that influence cellular differentiation [31], signaling molecules within microbial communities [32,33], or substances that change the nutrient uptake, thereby reducing the niche overlap among competing organisms [34].
A significant number of genes (about 964) were classified as having “function unknown” in the COG annotation results. Bacterial genomes frequently exhibit this, particularly in strains that are less well characterized. A comparative study with closely related strains of B. subtilis would be a more robust method, even though additional annotations against curated databases might enhance some functional predictions. To assign probable activities to these uncharacterized genes, future research will need deeper homology searches using programs like BLASTp (https://blast.ncbi.nlm.nih.gov/, accessed on 20 January 2025) and OrthoFinder v2.5.5.
The B. subtilis species complex has been found to produce a wide variety of natural products, but the ecological roles of these products have not been fully investigated due to the variability in secondary metabolite production among isolates from the same niche. Bacillus species have also shown remarkable biocontrol skills by promoting plant development and reducing illnesses brought on by bacteria and fungi that cause plant pathogenicity [35]. Their secondary metabolite patterns are mostly responsible for these abilities. Non-ribosomally produced lipopeptides are particularly a strong class of secondary metabolites with a number of antibacterial characteristics. Several lipopeptide isoforms from the surfactin, fengycin, and iturin families are produced by Bacillus species [36]. Eleven putative biosynthetic gene clusters in B. subtilis strains were found by a comparative examination of different Bacillus genomes [37]. Nevertheless, the synthesis of the natural product is not verified by the existence of a predicted BGC. A lack of BGC expression may be caused by gene silence or the absence of particular environmental stimuli [38].
Polyketide synthases (PKS), modular non-ribosomal peptide synthetases (NRPS), and particularly multi-modular enzyme complexes of PKS-NRPS hybrid systems generate a variety of intriguing secondary metabolites. Conversely, it has been less common to isolate natural compounds with different structural frameworks, like alkaloids, terpenoids, and phenylpropanoids [39]. Surfactin, plipastatin, and bacillibactin are the three NRPS gene clusters found in B. subtilis, together with bacillaene, a hybrid nonribosomal peptide synthetase–polyketide synthetase (NRPS-PKS) gene cluster [40].
The srfAA-srfAD gene cluster produces the well-known and adaptable secondary metabolite surfactin. By reducing the surface tension, this biosurfactant promotes sliding and swarming motility [8,41]. Its surfactant qualities are the main source of its cytolytic action; it penetrates bacterial lipid bilayer membranes and creates ion-conducting channels, which causes cell lysis [42,43]. According to studies, surfactin disrupts the Staphylococcus aureus membrane and exhibits bioactivity against Listeria monocytogenes and several Legionella species in vitro and at low concentrations [44,45,46]. Surfactin was recently found to facilitate the exploitation of nonpreferred carbon sources in B. amyloliquefaciens and improve the availability of oxygen for B. subtilis in liquid cultures [47,48].
The ppsA-ppsE gene cluster synthesizes the powerful antifungal lipopeptide plipastatin, which is chemically similar to fengycin, but differs in where d-tyrosine is located within the peptide backbone. Recent research has demonstrated that the fengycin BGC is found in the B. amyloliquefaciens and B. velezensis clades, whereas the plipastatin BGC is located in the B. subtilis clade [49]. Nevertheless, we found both plipastatin and fengycin in the B. subtilis MGE 2012 BGC profile. This suggests that fengycin or other secondary metabolites with comparable roles can be produced by MGE 2012. It is thought that plipastatin inhibits phospho-lipase A2, creating pores and altering the morphology of fungal membranes and cell walls; however, the precise mechanism of action is still unclear [38,50,51]. Numerous investigations have demonstrated the bioactivity of plipastatin and fengycin against a variety of filamentous fungi [46,52,53,54,55,56,57].
A broad-spectrum antibiotic that primarily works by preventing bacterial protein synthesis, bacillaene, is expressed from the pksB-pksS gene cluster. It has also been demonstrated to shield cells and spores from bacterial predators [58,59]. The dhbACEBF gene cluster synthesizes bacillibactin, a siderophore that carries iron from the environment into the cell [60]. Nevertheless, no research on its direct antimicrobial qualities has been reported.
AntiSMASH analysis of the type III PKS genes in MGE 2012 reveals low sequence similarity with known clusters, but a comparison of the core biosynthetic genes using NCBI BlastP (GenBank: ASV00624.1; 100% identity, 97% coverage) showed high sequence similarity to the sequence produced by most B. subtilis T3PKSs. Therefore, it is worthwhile to further investigate the potential of MGE 2012 to produce useful alkylresorcinols, alkylresorcinos, alkylpyrones, or other substances.
Only a small number of antibiotics are produced by domesticated B. subtilis because certain biosynthetic pathways are malfunctioning. Other wild-type strains, on the other hand, generate a distinctive mixture of several peptide antibiotics [61]. Three intramolecular bridges make up the macrocyclic bacteriocin subtilosin [62]. Under anaerobic and oxygen-limited conditions, subtilosin transcription rises [63]. Thus, it is anticipated that the facultative anaerobe MGE 2012 (87% similarity) will produce subtilosin A at a high level. Additionally, B. subtilis-based solutions are already being utilized in Europe to fight fire blight for biological control objectives. The dipeptide bacilysin and the polyketide difficidin have been found to be the two substances causing this potent antagonistic effect. Plant-associated strains of B. amyloliquefaciens produce both chemicals [64]. Given that the MGE 2012 BGC profile shows 100% similarity to bacilysin, it could potentially be used for biological control purposes.
In MGE 2012, the BGC count was reduced, which could indicate genetic refinement for particular environmental adaptation, potentially prioritizing important and conserved secondary metabolite pathways over novel ones. Regulation, expression dynamics, or metabolic interactions may, nevertheless, reveal functional potential, even when this restricts novelty at the genomic level.
5. Conclusions
B. subtilis isolates that have not been domesticated produce a range of secondary metabolites that characterize their biocontrol capabilities. The growth and differentiation of bacteria and fungi are influenced by these secondary metabolites, which may also have an impact on other microorganisms and macroorganisms. An overview of B. subtilis MGE 2012’s entire genome and secondary metabolite profile is given by our investigation.
In the process of finding a strain with excellent alpha amylase and protease activity to support the growth of probiotics, we discovered B. subtilis MGE 2012. This strain not only exhibits outstanding enzyme activities, but also has the potential to produce powerful antimicrobial substances. It would be very interesting to confirm by biochemical assays and gene expression profiling whether the strain produces the predicted secondary metabolites and characterize the expression and functional activity of its conserved BGCs under different environmental conditions to evaluate their biotechnological potential. Additionally, B. subtilis is already found as a normal symbiotic bacterium in the human gut, and its potential as a probiotic is being researched. Thus, testing the potential use of MGE 2012 as a probiotic in the future would be a worthwhile challenge.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pr13051503/s1, Table S1_Annotation; Table_S2_eggNOG annotation; Table S3_RAST annotation; Table S4_dDDH; Table S5_ANI; Table S6_amylase genes; Table S7_protease genes.
Author Contributions
J.K. and H.H.B. contributed equally to this work. Conceptualization, J.K. and G.-S.M.; methodology, J.K. and H.H.B.; software, H.H.B.; validation, J.K., H.H.B., J.H. and G.-S.M.; data curation, J.K. and H.H.B.; writing—original draft preparation, J.K. and H.H.B.; funding acquisition, G.-S.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2021R1A6A1A03046418).
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Iqbal, S.; Begum, F.; Rabaan, A.A.; Aljeldah, M.; Al Shammari, B.R.; Alawfi, A.; Alshengeti, A.; Sulaiman, T.; Khan, A. Classification and Multifaceted Potential of Secondary Metabolites Produced by Bacillus subtilis Group: A Comprehensive Review. Molecules 2023, 28, 927. [Google Scholar] [CrossRef] [PubMed]
- Medeot, D.; Sannazzaro, A.; Estrella, M.J.; Torres Tejerizo, G.; Contreras-Moreira, B.; Pistorio, M.; Jofré, E. Unraveling the Genome of Bacillus Velezensis MEP218, a Strain Producing Fengycin Homologs with Broad Antibacterial Activity: Comprehensive Comparative Genome Analysis. Sci. Rep. 2023, 13, 22168. [Google Scholar] [CrossRef]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological Control of Plant Pathogens by Bacillus Species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Penha, R.O.; Vandenberghe, L.P.S.; Faulds, C.; Soccol, V.T.; Soccol, C.R. Bacillus Lipopeptides as Powerful Pest Control Agents for a More Sustainable and Healthy Agriculture: Recent Studies and Innovations. Planta 2020, 251, 70. [Google Scholar] [CrossRef]
- Chen, I.; Christie, P.J.; Dubnau, D. The Ins and Outs of DNA Transfer in Bacteria. Science 2005, 310, 1456–1460. [Google Scholar] [CrossRef] [PubMed]
- Benoit, I.; van den Esker, M.H.; Patyshakuliyeva, A.; Mattern, D.J.; Blei, F.; Zhou, M.; Dijksterhuis, J.; Brakhage, A.A.; Kuipers, O.P.; de Vries, R.P.; et al. Bacillus subtilis Attachment to Aspergillus niger Hyphae Results in Mutually Altered Metabolism. Environ. Microbiol. 2015, 17, 2099–2113. [Google Scholar] [CrossRef]
- Jones, S.E.; Paynich, M.L.; Kearns, D.B.; Knight, K.L. Protection from Intestinal Inflammation by Bacterial Exopolysaccharides. J. Immunol. 2014, 192, 4813–4820. [Google Scholar] [CrossRef]
- Kearns, D.B.; Losick, R. Swarming Motility in Undomesticated Bacillus subtilis. Mol. Microbiol. 2003, 49, 581–590. [Google Scholar] [CrossRef]
- Tjalsma, H.; Antelmann, H.; Jongbloed, J.D.H.; Braun, P.G.; Darmon, E.; Dorenbos, R.; Dubois, J.-Y.F.; Westers, H.; Zanen, G.; Quax, W.J.; et al. Proteomics of Protein Secretion by Bacillus subtilis: Separating the “Secrets” of the Secretome. Microbiol. Mol. Biol. Rev. 2004, 68, 207–233. [Google Scholar] [CrossRef]
- Beauregard, P.B.; Chai, Y.; Vlamakis, H.; Losick, R.; Kolter, R. Bacillus subtilis Biofilm Induction by Plant Polysaccharides. Proc. Natl. Acad. Sci. USA 2013, 110, E1621–E1630. [Google Scholar] [CrossRef]
- Kovács, Á.T. Bacillus subtilis. Trends Microbiol. 2019, 27, 724–725. [Google Scholar] [CrossRef]
- Lee, J.-H.; Moon, G.-S. Development of Functional Material by Using Bacillus subtilis Harboring α-Amylase and Protease Enzyme Activity. Curr. Top. Lact. Acid Bact. Probiotics 2023, 9, 81–85. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS Is an Automated High-Throughput Platform for State-of-the-Art Genome-Based Taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-Based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Öker, M.G. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast Genome and Metagenome Distance Estimation Using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and Rapid Annotation of Ribosomal RNA Genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Chalita, M.; Kim, Y.O.; Park, S.; Oh, H.S.; Cho, J.H.; Moon, J.; Baek, N.; Moon, C.; Lee, K.; Yang, J.; et al. EzBioCloud: A Genome-Driven Database and Platform for Microbiome Identification and Discovery. Int. J. Syst. Evol. Microbiol. 2024, 74, 6421. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Farris, J.S. Estimating Phylogenetic Trees from Distance Matrices. Am. Nat. 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: An Improved Algorithm and Software for Calculating Average Nucleotide Identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Foster, K.R.; Bell, T. Competition, Not Cooperation, Dominates Interactions among Culturable Microbial Species. Curr. Biol. 2012, 22, 1845–1850. [Google Scholar] [CrossRef]
- Straight, P.D.; Willey, J.M.; Kolter, R. Interactions between Streptomyces coelicolor and Bacillus subtilis: Role of Surfactants in Raising Aerial Structures. J. Bacteriol. 2006, 188, 4918–4925. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; Traxler, M.F.; López, D.; Kolter, R. Antibiotics as Signal Molecules. Chem. Rev. 2011, 111, 5492–5505. [Google Scholar] [CrossRef]
- Linares, J.F.; Gustafsson, I.; Baquero, F.; Martinez, J.L. Antibiotics as Intermicrobiol Signaling Agents Instead of Weapons. Proc. Natl. Acad. Sci. USA 2006, 103, 19484–19489. [Google Scholar] [CrossRef]
- Vaz Jauri, P.; Bakker, M.G.; Salomon, C.E.; Kinkel, L.L. Subinhibitory Antibiotic Concentrations Mediate Nutrient Use and Competition among Soil Streptomyces. PLoS ONE 2013, 8, e81064. [Google Scholar] [CrossRef]
- Fan, B.; Blom, J.; Klenk, H.P.; Borriss, R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis Form an “Operational Group B. amyloliquefaciens” within the B. subtilis Species Complex. Front. Microbiol. 2017, 8, 22. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P. Bacillus Lipopeptides: Versatile Weapons for Plant Disease Biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Grubbs, K.J.; Bleich, R.M.; Santa Maria, K.C.; Allen, S.E.; Farag, S.; Shank, E.A.; Bowers, A.A. Large-Scale Bioinformatics Analysis of Bacillus Genomes Uncovers Conserved Roles of Natural Products in Bacterial Physiology. mSystems 2017, 2, e00040-17. [Google Scholar] [CrossRef]
- Harwood, C.R.; Mouillon, J.M.; Pohl, S.; Arnau, J. Secondary Metabolite Production and the Safety of Industrially Important Members of the Bacillus subtilis Group. FEMS Microbiol. Rev. 2018, 42, 721–738. [Google Scholar] [CrossRef] [PubMed]
- Zha, W.; Rubin-Pitel, S.B.; Zhao, H. Characterization of the Substrate Specificity of PhlD, a Type III Polyketide Synthase from Pseudomonas fluorescens. J. Biol. Chem. 2006, 281, 32036–32047. [Google Scholar] [CrossRef]
- Kiesewalter, H.T.; Lozano-Andrade, C.N.; Wibowo, M.; Strube, M.L.; Maróti, G.; Snyder, D.; Jørgensen, T.S.; Larsen, T.O.; Cooper, V.S.; Weber, T.; et al. Genomic and Chemical Diversity of Bacillus subtilis Secondary Metabolites against Plant Pathogenic Fungi. mSystems 2021, 6, e00770-20. [Google Scholar] [CrossRef]
- Grau, R.R.; De Oña, P.; Kunert, M.; Leñini, C.; Gallegos-Monterrosa, R.; Mhatre, E.; Vileta, D.; Donato, V.; Hölscher, T.; Boland, W.; et al. A Duo of Potassium-Responsive Histidine Kinases Govern the Multicellular Destiny of Bacillus subtilis. mBio 2015, 6, e00581. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, J.D.; Jumarie, C.; Cooper, D.G.; Laprade, R. Ionic Channels Induced by Surfactin in Planar Lipid Bilayer Membranes. BBA Biomembr. 1991, 1064, 13–23. [Google Scholar] [CrossRef]
- Heerklotz, H.; Seelig, J. Leakage and Lysis of Lipid Membranes Induced by the Lipopeptide Surfactin. Eur. Biophys. J. 2007, 36, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Sabaté, D.C.; Audisio, M.C. Inhibitory Activity of Surfactin, Produced by Different Bacillus subtilis Subsp. subtilis Strains, against Listeria monocytogenes Sensitive and Bacteriocin-Resistant Strains. Microbiol. Res. 2013, 168, 125–129. [Google Scholar] [CrossRef]
- Loiseau, C.; Schlusselhuber, M.; Bigot, R.; Bertaux, J.; Berjeaud, J.M.; Verdon, J. Surfactin from Bacillus subtilis Displays an Unexpected Anti-Legionella Activity. Appl. Microbiol. Biotechnol. 2015, 99, 5083–5093. [Google Scholar] [CrossRef]
- Gao, L.; Han, J.; Liu, H.; Qu, X.; Lu, Z.; Bie, X. Plipastatin and Surfactin Coproduction by Bacillus subtilis PB2-L and Their Effects on Microorganisms. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2017, 110, 1007–1018. [Google Scholar] [CrossRef]
- Arjes, H.A.; Vo, L.; Dunn, C.M.; Willis, L.; DeRosa, C.A.; Fraser, C.L.; Kearns, D.B.; Huang, K.C. Biosurfactant-Mediated Membrane Depolarization Maintains Viability during Oxygen Depletion in Bacillus subtilis. Curr. Biol. 2020, 30, 1011–1022.e6. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Wen, J.; Zhao, X.; Ding, J.; Qi, G. Surfactin: A Quorum-Sensing Signal Molecule to Relieve CCR in Bacillus amyloliquefaciens. Front. Microbiol. 2020, 11, 631. [Google Scholar] [CrossRef]
- Steinke, K.; Mohite, O.S.; Weber, T.; Kovács, Á.T. Phylogenetic Distribution of Secondary Metabolites in the Bacillus subtilis Species Complex. mSystems 2021, 6, e00057-21. [Google Scholar] [CrossRef]
- Umezawa, H.; Aoyagi, T.; Nishikiori, T.; Okuyama, A.; Yamagishi, Y.; Hamada, M.; Takeuchi, T. Plipastatins: New Inhibitors of Phospholipase A2, Produced by Bacillus cereus BMG302-FF67 I. Taxonomy, Production, Isolation and Preliminary Characterization. J. Antibiot. 1986, 39, 737–744. [Google Scholar] [CrossRef]
- Deleu, M.; Paquot, M.; Nylander, T. Fengycin Interaction with Lipid Monolayers at the Air-Aqueous Interface—Implications for the Effect of Fengycin on Biological Membranes. J. Colloid Interface Sci. 2005, 283, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; De Vicente, A.; Rakotoaly, R.H.; Dufour, S.E.; Veening, J.W.; Arrebola, E.; Cazorla, F.M.; Kuipers, O.P.; Paquot, M.; Pérez-García, A. The Iturin and Fengycin Families of Lipopeptides Are Key Factors in Antagonism of Bacillus subtilis toward Podosphaera fusca. Mol. Plant Microbe Interact. 2007, 20, 430–440. [Google Scholar] [CrossRef]
- Alvarez, F.; Castro, M.; Príncipe, A.; Borioli, G.; Fischer, S.; Mori, G.; Jofré, E. The Plant-Associated Bacillus amyloliquefaciens Strains MEP218 and ARP23 Capable of Producing the Cyclic Lipopeptides Iturin or Surfactin and Fengycin Are Effective in Biocontrol of Sclerotinia Stem Rot Disease. J. Appl. Microbiol. 2012, 112, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Falardeau, J.; Wise, C.; Novitsky, L.; Avis, T.J. Ecological and Mechanistic Insights Into the Direct and Indirect Antimicrobial Properties of Bacillus subtilis Lipopeptides on Plant Pathogens. J. Chem. Ecol. 2013, 39, 869–878. [Google Scholar] [CrossRef]
- Roy, A.; Mahata, D.; Paul, D.; Korpole, S.; Franco, O.L.; Mandal, S.M. Purification, Biochemical Characterization and Self-Assembled Structure of a Fengycin-like Antifungal Peptide from Bacillus thuringiensis Strain SM. Front. Microbiol. 2013, 4, 332. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Bie, X.; Lu, Z.; Lv, F.; Tao, Y.; Qu, X. Effects of Fengycin from Bacillus subtilis FmbJ on Apoptosis and Necrosis in Rhizopus Stolonifer. J. Microbiol. 2014, 52, 675–680. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, C. Fengycins, Cyclic Lipopeptides from Marine Bacillus subtilis Strains, Kill the Plant-Pathogenic Fungus Magnaporthe grisea by Inducing Reactive Oxygen Species Production and Chromatin Condensation. Appl. Environ. Microbiol. 2018, 84, e00445-18. [Google Scholar] [CrossRef]
- Mayerl, F.; Fisher, S.; Pirnik, D.; Aklonis, C.; Dean, L.; Meyers, E.; Fernandes, P. Bacillaene, a Novel Inhibitor of Procaryotic Protein Synthesis Produced by Bacillus subtilis: Production, Taxonomy, Isolation, Physico-Chemical Characterization and Biological Activity. J. Antibiot. 1995, 48, 997–1003. [Google Scholar] [CrossRef]
- Müller, S.; Strack, S.N.; Hoefler, B.C.; Straight, P.D.; Kearns, D.B.; Kirby, J.R. Bacillaene and Sporulation Protect Bacillus subtilis from Predation by Myxococcus Xanthus. Appl. Environ. Microbiol. 2014, 80, 5603–5610. [Google Scholar] [CrossRef]
- May, J.J.; Wendrich, T.M.; Marahiel, M.A. The Dhb Operon of Bacillus subtilis Encodes the Biosynthetic Template for the Catecholic Siderophore 2,3-Dihydroxybenzoate-Glycine-Threonine Trimeric Ester Bacillibactin. J. Biol. Chem. 2001, 276, 7209–7217. [Google Scholar] [CrossRef]
- Stein, T.; Düsterhus, S.; Stroh, A.; Entian, K.D. Subtilosin Production by Two Bacillus subtilis Subspecies and Variance of the Sbo-Alb Cluster. Appl. Environ. Microbiol. 2004, 70, 2349–2353. [Google Scholar] [CrossRef] [PubMed]
- Kawulka, K.; Sprules, T.; McKay, R.T.; Mercier, P.; Diaper, C.M.; Zuber, P.; Vederas, J.C. Structure of Subtilosin A, an Antimicrobial Peptide from Bacillus subtilis with Unusual Posttranslational Modifications Linking Cysteine Sulfurs to α-Carbons of Phenylalanine and Threonine. J. Am. Chem. Soc. 2003, 125, 4726–4727. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.M.; Zheng, G.; Zuber, P. Dual Control of Sbo-Alb Operon Expression by the Spo0 and ResDE Systems of Signal Transduction under Anaerobic Conditions in Bacillus subtilis. J. Bacteriol. 2000, 182, 3274–3277. [Google Scholar] [CrossRef]
- Chen, X.H.; Scholz, R.; Borriss, M.; Junge, H.; Mögel, G.; Kunz, S.; Borriss, R. Difficidin and Bacilysin Produced by Plant-Associated Bacillus amyloliquefaciens Are Efficient in Controlling Fire Blight Disease. J. Biotechnol. 2009, 140, 38–44. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).