Efficient Bioreduction of Cr(VI) by a Halotolerant Acinetobacter sp. ZQ-1 in High-Salt Environments: Performance and Metabolomic Mechanism
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Isolation and Identification of Strain
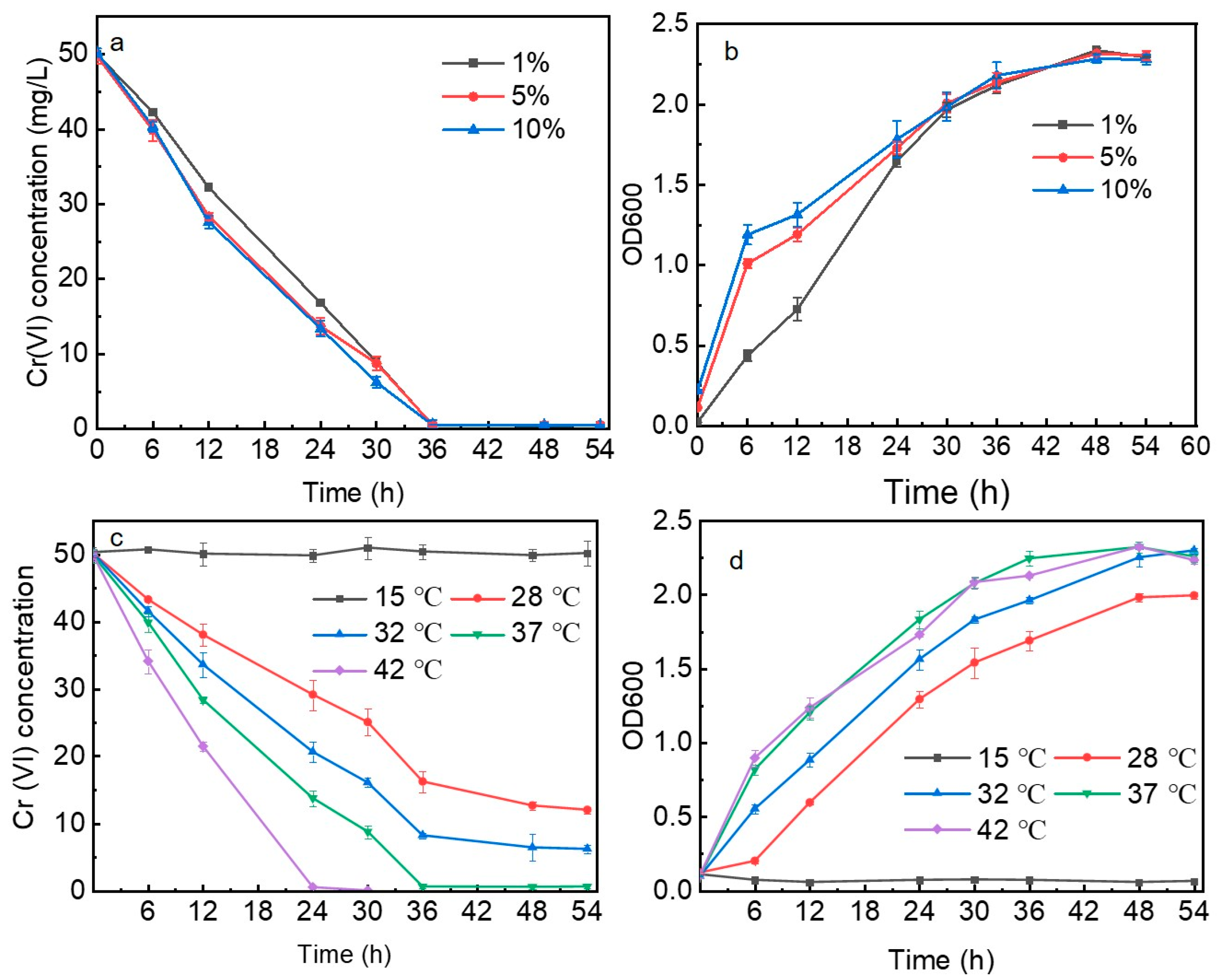
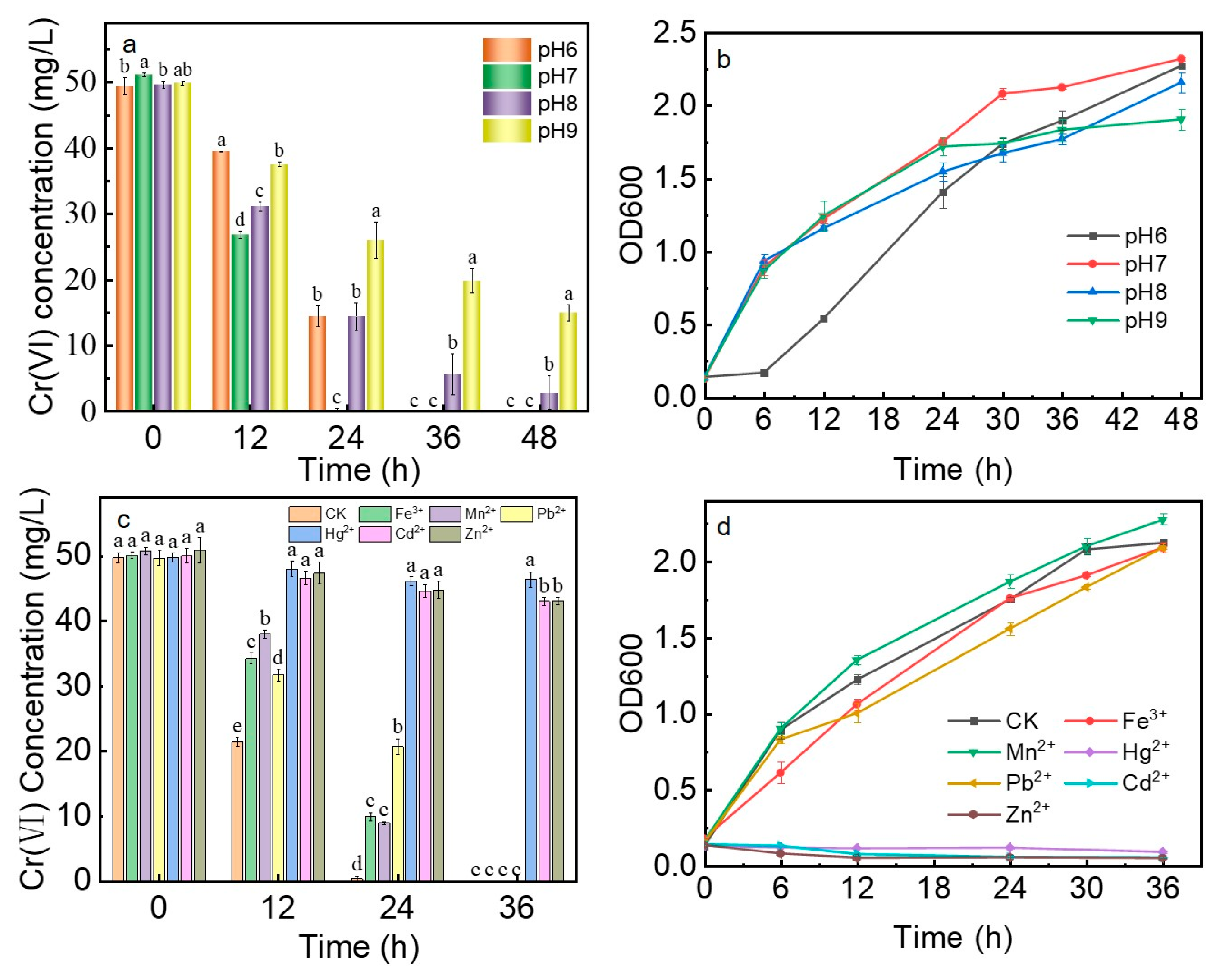
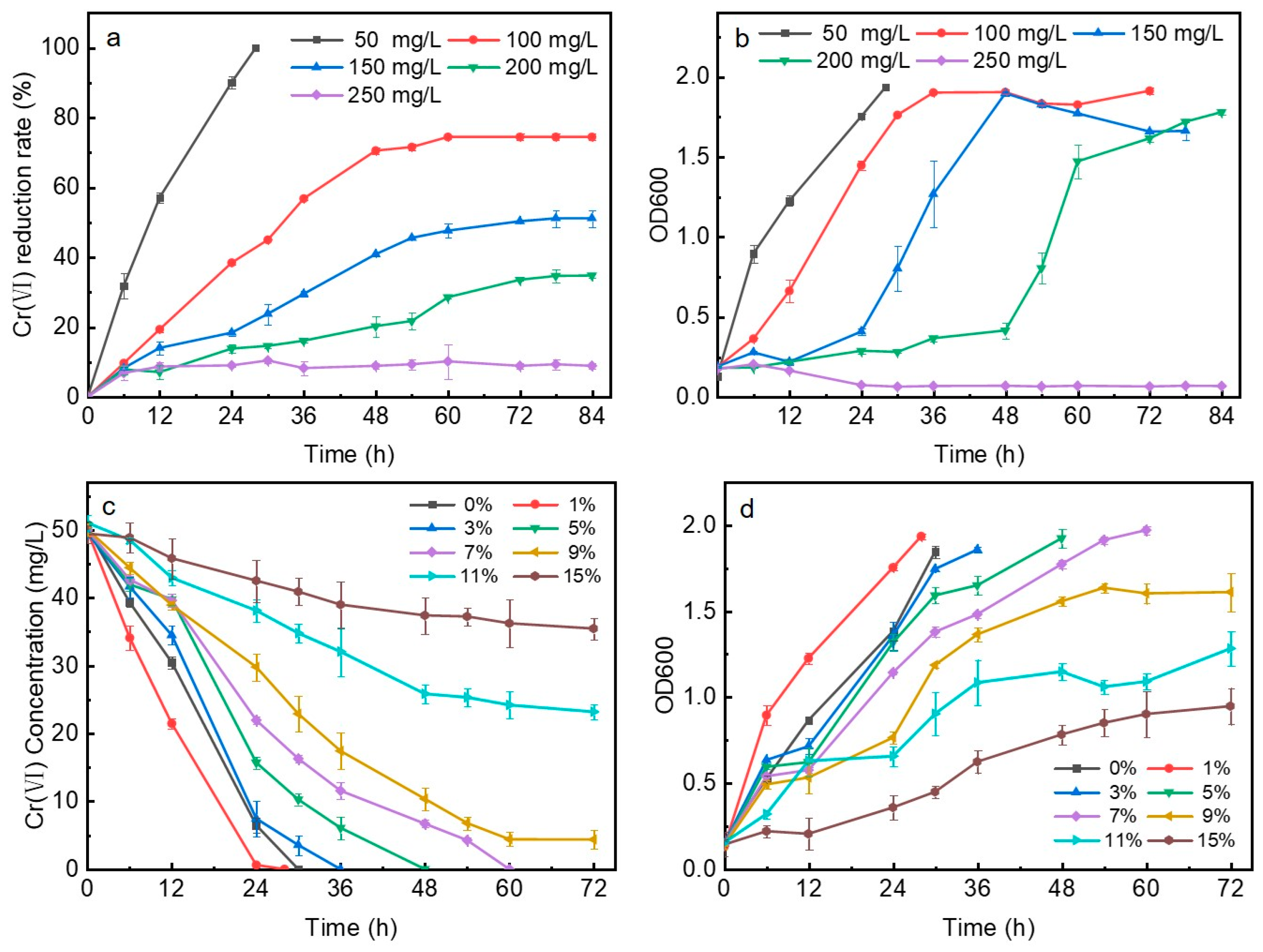
2.3. Influencing Factors of Bioreduction of Cr(VI)
2.4. Detection of Metabolites
2.5. Measurements
3. Results and Discussion
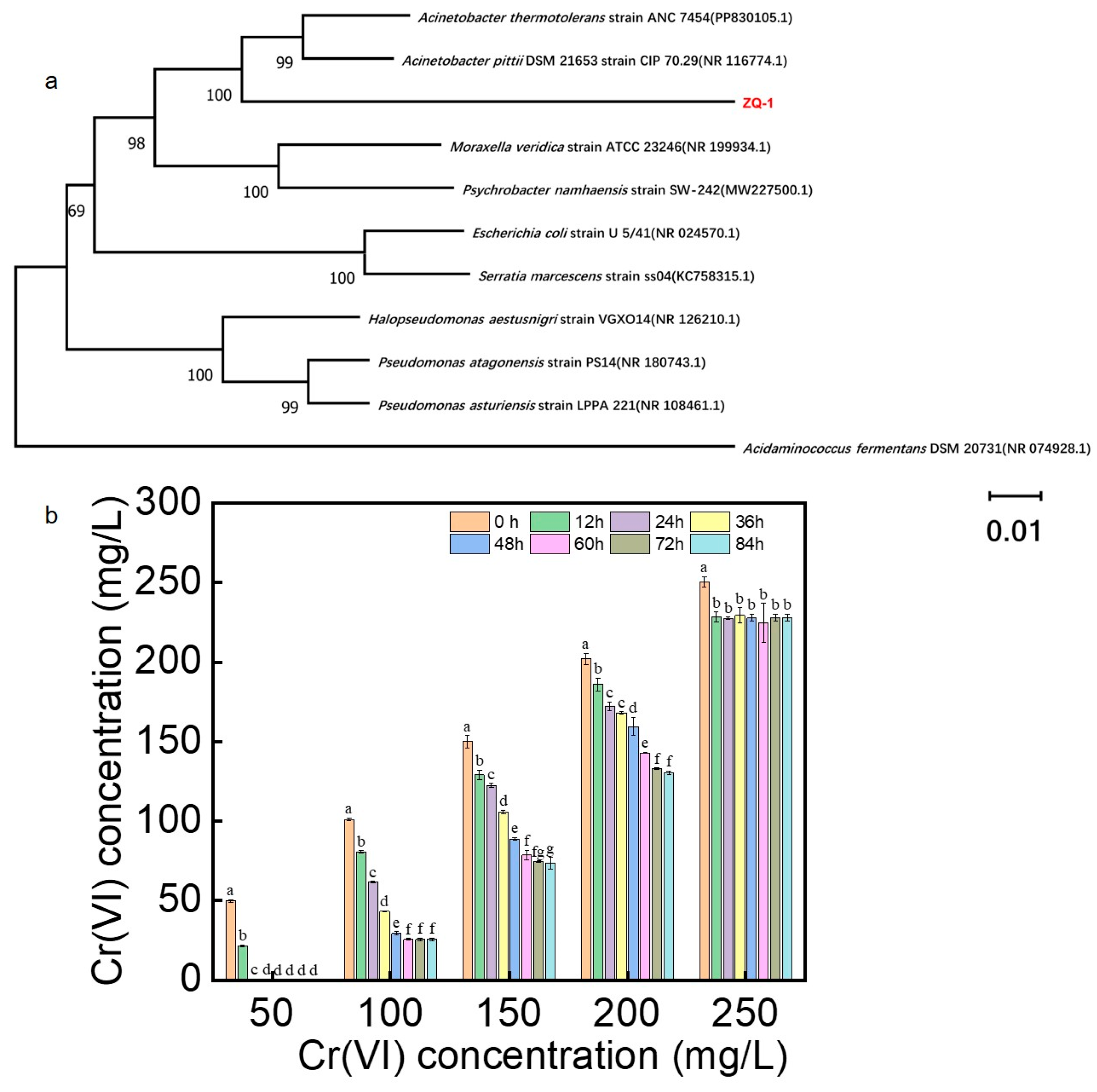
3.1. Isolation and Identification of Strain ZQ-1
3.2. Bioreduction of Cr(VI)
3.3. Deciphering of Strain ZQ-1 on Salt-Tolerance
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Elahi, A.; Rehman, A.; Zajif Hussain, S.; Zulfiqar, S.; Shakoori, A.R. Isolation and characterization of a highly effective bacterium Bacillus cereus b-525k for hexavalent chromium detoxification. Saudi J. Biol. Sci. 2022, 29, 2878–2885. [Google Scholar] [CrossRef]
- Das, S.; Mishra, J.; Das, S.K.; Pandey, S.; Rao, D.S.; Chakraborty, A.; Sudarshan, M.; Das, N.; Thatoi, H. Investigation on mechanism of Cr(VI) reduction and removal by Bacillus amyloliquefaciens, a novel chromate tolerant bacterium isolated from chromite mine soil. Chemosphere 2014, 96, 112–121. [Google Scholar] [CrossRef]
- Banerjee, S.; Joshi, S.R.; Mandal, T.; Halder, G. Insight into Cr6+ reduction efficiency of Rhodococcus erythropolis isolated from coalmine waste water. Chemosphere 2017, 167, 269–281. [Google Scholar] [CrossRef]
- Banerjee, S.; Misra, A.; Chaudhury, S.; Dam, B. A Bacillus strain TCL isolated from Jharia coalmine with remarkable stress responses, chromium reduction capability and bioremediation potential. J. Hazard. Mater. 2019, 367, 215–223. [Google Scholar] [CrossRef]
- Bharagava, R.N.; Mishra, S. Hexavalent chromium reduction potential of Cellulosimicrobium sp. isolated from common effluent treatment plant of tannery industries. Ecotoxicol. Environ. Saf. 2018, 147, 102–109. [Google Scholar] [CrossRef]
- Das, S.; Chandra Behera, B.; Mohapatra, R.K.; Pradhan, B.; Sudarshan, M.; Chakraborty, A.; Thatoi, H. Reduction of hexavalent chromium by Exiguobacterium mexicanum isolated from chromite mines soil. Chemosphere 2021, 282, 131135. [Google Scholar] [CrossRef]
- Ma, Y.; Zhong, H.; He, Z. Cr(VI) reductase activity locates in the cytoplasm of Aeribacillus pallidus BK1, a novel Cr(VI)-reducing thermophile isolated from Tengchong geothermal region, China. Chem. Eng. J. 2019, 371, 524–534. [Google Scholar] [CrossRef]
- Xiao, W.; Ye, X.; Yang, X.; Zhu, Z.; Sun, C.; Zhang, Q.; Xu, P. Isolation and characterization of chromium(VI)-reducing Bacillus sp. FY1 and Arthrobacter sp. WZ2 and their bioremediation potential. Bioremediation J. 2017, 21, 100–108. [Google Scholar] [CrossRef]
- Gu, Y.; Chen, X.; Liu, L.; Wang, S.; Yu, X.; Jia, Z.; Zhou, X. Cr(VI)-bioremediation mechanism of a novel strain Bacillus paramycoides Cr6 with the powerful ability to remove Cr(VI) from contaminated water. J. Hazard. Mater. 2023, 455, 131519. [Google Scholar] [CrossRef]
- Zhu, Y.; Yan, J.; Xia, L.; Zhang, X.; Luo, L. Mechanisms of Cr(VI) reduction by Bacillus sp. CRB-1, a novel Cr(VI)-reducing bacterium isolated from tannery activated sludge. Ecotoxicol Env. Saf 2019, 186, 109792. [Google Scholar] [CrossRef]
- Dhal, B.; Thatoi, H.N.; Das, N.N.; Pandey, B.D. Chemical and microbial remediation of hexavalent chromium from contaminated soil and mining/metallurgical solid waste: A review. J. Hazard. Mater. 2013, 250–251, 272–291. [Google Scholar] [CrossRef]
- Karthik, C.; Barathi, S.; Pugazhendhi, A.; Ramkumar, V.S.; Thi, N.B.D.; Arulselvi, P.I. Evaluation of Cr(VI) reduction mechanism and removal by Cellulosimicrobium funkei strain AR8, a novel haloalkaliphilic bacterium. J. Hazard. Mater. 2017, 333, 42–53. [Google Scholar] [CrossRef]
- Ashraf, S.; Naveed, M.; Afzal, M.; Ashraf, S.; Rehman, K.; Hussain, A.; Zahir, Z.A. Bioremediation of tannery effluent by Cr- and salt-tolerant bacterial strains. Environ. Monit. Assess. 2018, 190, 716. [Google Scholar] [CrossRef]
- Kumar, M.; Saini, H.S. Reduction of hexavalent chromium (VI) by indigenous alkaliphilic and halotolerant Microbacterium sp. M5: Comparative studies under growth and nongrowth conditions. J. Appl. Microbiol. 2019, 127, 1057–1068. [Google Scholar] [CrossRef]
- Amoozegar, M.; Ghasemi, A.; Razavi, M.; Naddaf, S.R. Evaluation of hexavalent chromium reduction by chromate-resistant moderately halophile, Nesterenkonia sp. strain MF2. Process Biochem. 2007, 42, 1475–1479. [Google Scholar] [CrossRef]
- He, G.; Wu, C.; Huang, J.; Zhou, R. Metabolic response of Tetragenococcus halophilus under salt stress. Biotechnol. Bioprocess Eng. 2017, 22, 366–375. [Google Scholar] [CrossRef]
- Lin, W.H.; Chien, C.C.; Ou, J.H.; Yu, Y.L.; Chen, S.C.; Kao, C.M. Cleanup of Cr(VI)-polluted groundwater using immobilized bacterial consortia via bioreduction mechanisms. J. Environ. Manag. 2023, 339, 117947. [Google Scholar] [CrossRef]
- Meng, Y.; Ma, X.X.; Luan, F.B.; Zhao, Z.W.; Li, Y.; Xiao, X.; Wang, Q.Q.; Zhang, J.D.; Thandar, S.M. Sustainable enhancement of Cr(VI) bioreduction by the isolated Cr(VI)-resistant bacteria. Sci. Total Environ. 2022, 812, 152433. [Google Scholar] [CrossRef]
- Zhang, F.; Xu, W.Q.; Zhang, L.W.; Xi, L.Y.; Du, Y.G.; Ma, L.Y.; Chen, S.H.; Du, D.Y. Riboflavin as a non-quinone redox mediator for enhanced Cr(VI) removal by Shewanella putrefaciens. J. Mol. Liq. 2022, 351, 118622. [Google Scholar] [CrossRef]
- Pan, X.; Liu, Z.; Chen, Z.; Cheng, Y.; Pan, D.; Shao, J.; Lin, Z.; Guan, X. Investigation of Cr(VI) reduction and Cr(III) immobilization mechanism by planktonic cells and biofilms of Bacillus subtilis ATCC-6633. Water Res. 2014, 55, 21–29. [Google Scholar] [CrossRef]
- Reddy, G.K.K.; Kavibharathi, K.; Singh, A.; Nancharaiah, Y.V. Growth-dependent Cr(VI) reduction by Alteromonas sp ORB2 under haloalkaline conditions: Toxicity removal mechanism effect of heavy metals. World J. Microbiol. Biotechnol. 2024, 40, 165. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, W.; Nie, T.; Wang, L.; Ni, Y. Extracellular Cr(VI) Reduction by the Salt-Tolerant Strain Bacillus safensis BSF-4. Microorganisms 2025, 13, 1961. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Z.; Li, H.; Yan, Z.; Meng, Z.; Liu, C.; Chen, J.; Duan, C. Resistance mechanisms and remediation potential of hexavalent chromium in Pseudomonas sp. strain AN-B15. Ecotoxicol. Environ. Saf. 2023, 250, 114498. [Google Scholar] [CrossRef]
- Sun, Y.; Jin, J.; Li, W.; Zhang, S.; Wang, F. Hexavalent chromium removal by a resistant strain Bacillus cereus ZY-2009. Environ. Technol. 2021, 44, 1926–1935. [Google Scholar] [CrossRef]
- Chen, W.; Li, W.; Wang, T.; Wen, Y.; Shi, W.; Zhang, W.; Guo, B.; Yang, Y. Isolation of functional bacterial strains from chromium-contaminated site and bioremediation potentials. J. Environ. Manag. 2022, 307, 114557. [Google Scholar] [CrossRef]
- Chen, J.; Li, X.; Gan, L.; Jiang, G.; Zhang, R.; Xu, Z.; Tian, Y. Mechanism of Cr(VI) reduction by Lysinibacillus sp. HST-98, a newly isolated Cr (VI)-reducing strain. Environ. Sci. Pollut. Res. 2021, 28, 66121–66132. [Google Scholar] [CrossRef]
- Zhao, R.; Wang, B.; Cai, Q.T.; Li, X.X.; Liu, M.; Hu, D.; Guo, D.B.; Wang, J.; Fan, C. Bioremediation of Hexavalent Chromium Pollution by Sporosarcina saromensis M52 Isolated from Offshore Sediments in Xiamen, China. Biomed. Environ. Sci. 2016, 29, 127–136. [Google Scholar]
- Li, Y.; Wang, H.; Wu, P.; Yu, L.; Rehman, S.; Wang, J.; Yang, S.; Zhu, N. Bioreduction of hexavalent chromium on goethite in the presence of Pseudomonas aeruginosa. Environ. Pollut. 2020, 265, 114765. [Google Scholar] [CrossRef]
- Knapp, B.D.; Huang, K.C. The Effects of Temperature on Cellular Physiology. Annu. Rev. Biophys. 2022, 51, 499–526. [Google Scholar] [CrossRef]
- Cheng, H.; Yuan, M.; Zeng, Q.; Zhou, H.; Zhan, W.; Chen, H.; Mao, Z.; Wang, Y. Efficient reduction of reactive black 5 and Cr(VI) by a newly isolated bacterium of Ochrobactrum anthropi. J. Hazard. Mater. 2021, 406, 124641. [Google Scholar] [CrossRef]
- Ge, S.; Dong, X.; Zhou, J.; Ge, S. Comparative evaluations on bio-treatment of hexavalent chromate by resting cells of Pseudochrobactrum sp. and Proteus sp. in wastewater. J. Environ. Manag. 2013, 126, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Ge, S.; Zhou, M.; Dong, X. Bioremediation of hexavalent chromate using permeabilized Brevibacterium sp. and Stenotrophomonas sp. cells. J. Environ. Manag. 2015, 157, 54–59. [Google Scholar] [CrossRef]
- He, Z.; Gao, F.; Sha, T.; Hu, Y.; He, C. Isolation and characterization of a Cr(VI)-reduction Ochrobactrum sp. strain CSCr-3 from chromium landfill. J. Hazard. Mater. 2009, 163, 869–873. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, N.; Liu, T.; Feng, C.; Ma, L.; Chen, S.; Li, M. The mechanism of nitrate-Cr(VI) reduction mediated by microbial under different initial pHs. J. Hazard. Mater. 2020, 393, 122434. [Google Scholar] [CrossRef]
- Cárdenas González, J.F.; Acosta Rodríguez, I.; Terán Figueroa, Y.; Lappe Oliveras, P.; Martínez Flores, R.; Rodríguez Pérez, A.S. Biotransformation of Chromium (VI) via a Reductant Activity from the Fungal Strain Purpureocillium lilacinum. J. Fungi 2021, 7, 1022. [Google Scholar] [CrossRef]
- Ma, L.; Xu, J.; Chen, N.; Li, M.; Feng, C. Microbial reduction fate of chromium (Cr) in aqueous solution by mixed bacterial consortium. Ecotoxicol. Environ. Saf. 2019, 170, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Murugavelh, S.; Mohanty, K. Bioreduction of hexavalent chromium by free cells and cell free extracts of Halomonas sp. Chem. Eng. J. 2012, 203, 415–422. [Google Scholar]
- Shi, Y.; Chai, L.; Yang, Z.; Jing, Q.; Chen, R.; Chen, Y. Identification and hexavalent chromium reduction characteristics of Pannonibacter phragmitetus. Bioprocess Biosyst. Eng. 2012, 35, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Wang, Q.; Lv, M.; Chen, L. Microorganism remediation strategies towards heavy metals. Chem. Eng. J. 2019, 360, 1553–1563. [Google Scholar] [CrossRef]
- Priya, A.K.; Gnanasekaran, L.; Dutta, K.; Rajendran, S.; Balakrishnan, D.; Soto-Moscoso, M. Biosorption of heavy metals by microorganisms: Evaluation of different underlying mechanisms. Chemosphere 2022, 307, 135957. [Google Scholar] [CrossRef]
- Liang, J.; Li, X.; Zheng, Y.; Pang, Y.; Zhao, Y.; Wang, Y.; Zhang, J.F. A novel advanced reduction process for the reduction of Cr(VI): Assistance of microbial metabolites. J. Hazard. Mater. 2024, 480, 136121. [Google Scholar] [CrossRef]
- Gong, Y.; Werth, C.J.; He, Y.; Su, Y.; Zhang, Y.; Zhou, X. Intracellular versus extracellular accumulation of Hexavalent chromium reduction products by Geobacter sulfurreducens PCA. Environ. Pollut. 2018, 240, 485–492. [Google Scholar] [CrossRef]
- Banerjee, S.; Kamila, B.; Barman, S.; Joshi, S.R.; Mandal, T.; Halder, G. Interlining Cr(VI) remediation mechanism by a novel bacterium Pseudomonas brenneri isolated from coalmine wastewater. J. Environ. Manag. 2019, 233, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Pan, Y.; Yu, H.; Zhang, X.; Shen, Y.; Jiao, S.; Wu, K.; La, G.; Yuan, Y.; et al. Mechanisms of Cd and Cr removal and tolerance by macrofungus Pleurotus ostreatus HAU-2. J. Hazard. Mater. 2017, 330, 1–8. [Google Scholar] [CrossRef]
- Okeke, B.C. Bioremoval of hexavalent chromium from water by a salt tolerant bacterium, Exiguobacterium sp. GS1. J. Ind. Microbiol. Biotechnol. 2008, 35, 1571–1579. [Google Scholar] [CrossRef]
- Liu, W.; Liu, C.; Liu, L.; You, Y.; Jiang, J.; Zhou, Z.; Dong, Z. Simultaneous decolorization of sulfonated azo dyes and reduction of hexavalent chromium under high salt condition by a newly isolated salt-tolerant strain Bacillus circulans BWL1061. Ecotoxicol. Environ. Saf. 2017, 141, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Y.; Ma, Y.; Wang, X.; Zhang, B.; Zhang, G.; Bahadur, A.; Chen, T.; Liu, G.; Zhang, W.; et al. Research progress regarding the role of halophilic and halotolerant microorganisms in the eco-environmental sustainability and conservation. J. Clean. Prod. 2023, 418, 138054. [Google Scholar] [CrossRef]
- Li, H.; Meng, F.; Duan, W.; Lin, Y.; Zheng, Y. Biodegradation of phenol in saline or hypersaline environments by bacteria: A review. Ecotoxicol. Environ. Saf. 2019, 184, 109658. [Google Scholar] [CrossRef]
- Vreeland, R.H. Mechanisms of halotolerance in microorganisms. Crit. Rev. Microbiol. 1987, 14, 311–356. [Google Scholar] [CrossRef]
- Peng, X.; Wang, Y.; Zhu, G.; Zhu, W. Fatty acid derivatives from the halotolerant fungus Cladosporium cladosporioides. Magn. Reson. Chem. MRC 2018, 56, 18–24. [Google Scholar] [CrossRef]
- Das, P.; Manna, I.; Sil, P.; Bandyopadhyay, M.; Biswas, A.K. Exogenous silicon alters organic acid production and enzymatic activity of TCA cycle in two NaCl stressed indica rice cultivars. Plant Physiol. Biochem. PPB 2019, 136, 76–91. [Google Scholar] [CrossRef]
- Li, C.; Liu, Q.; Wang, Y.; Yang, X.; Chen, S.; Zhao, Y.; Wu, Y.; Li, L. Salt stress improves thermotolerance and high-temperature bioethanol production of multi-stress-tolerant Pichia kudriavzevii by stimulating intracellular metabolism and inhibiting oxidative damage. Biotechnol. Biofuels 2021, 14, 222–238. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; He, Z.; Liu, M.; Jin, Y.; Zhao, J.; Zhou, R.; Wu, C.; Qin, J. Exogenous fatty acid renders the improved salt tolerance in Zygosaccharomyces rouxii by altering lipid metabolism. LWT 2023, 177, 114579. [Google Scholar] [CrossRef]
- Bertrand, A.; Bipfubusa, M.; Dhont, C.; Chalifour, F.P.; Drouin, P.; Beauchamp, C.J. Rhizobial strains exert a major effect on the amino acid composition of alfalfa nodules under NaCl stress. Plant Physiol. Biochem. PPB 2016, 108, 344–352. [Google Scholar] [CrossRef]
- Hefnawy, M.A.; Nasr, M.I.; El-Mongy, M. Growth and soluble amino acid composition in Penicillium corylophilum and Halobacterium halobium grown under salt stress. Folia Microbiol. 1999, 44, 25–31. [Google Scholar] [CrossRef]
- Chun, B.H.; Han, D.M.; Kim, K.H.; Jeong, S.E.; Park, D.; Jeon, C.O. Genomic and metabolic features of Tetragenococcus halophilus as revealed by pan-genome and transcriptome analyses. Food Microbiol. 2019, 83, 36–47. [Google Scholar] [CrossRef]
- Trevino, S.R.; Scholtz, J.M.; Pace, C.N. Amino acid contribution to protein solubility: Asp, Glu, and Ser contribute more favorably than the other hydrophilic amino acids in RNase Sa. J. Mol. Biol. 2007, 366, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, D.; Qiao, Y.; Song, Q.; Guan, Z.; Qiu, K.; Cao, J.; Huang, L. Salt tolerance mechanism of a hydrocarbon-degrading strain: Salt tolerance mediated by accumulated betaine in cells. J. Hazard. Mater. 2020, 392, 122326. [Google Scholar] [CrossRef]
- Roder, A.; Hoffmann, E.; Hagemann, M.; Berg, G. Synthesis of the compatible solutes glucosylglycerol and trehalose by salt-stressed cells of Stenotrophomonas strains. FEMS Microbiol. Lett. 2005, 243, 219–226. [Google Scholar] [CrossRef] [PubMed]
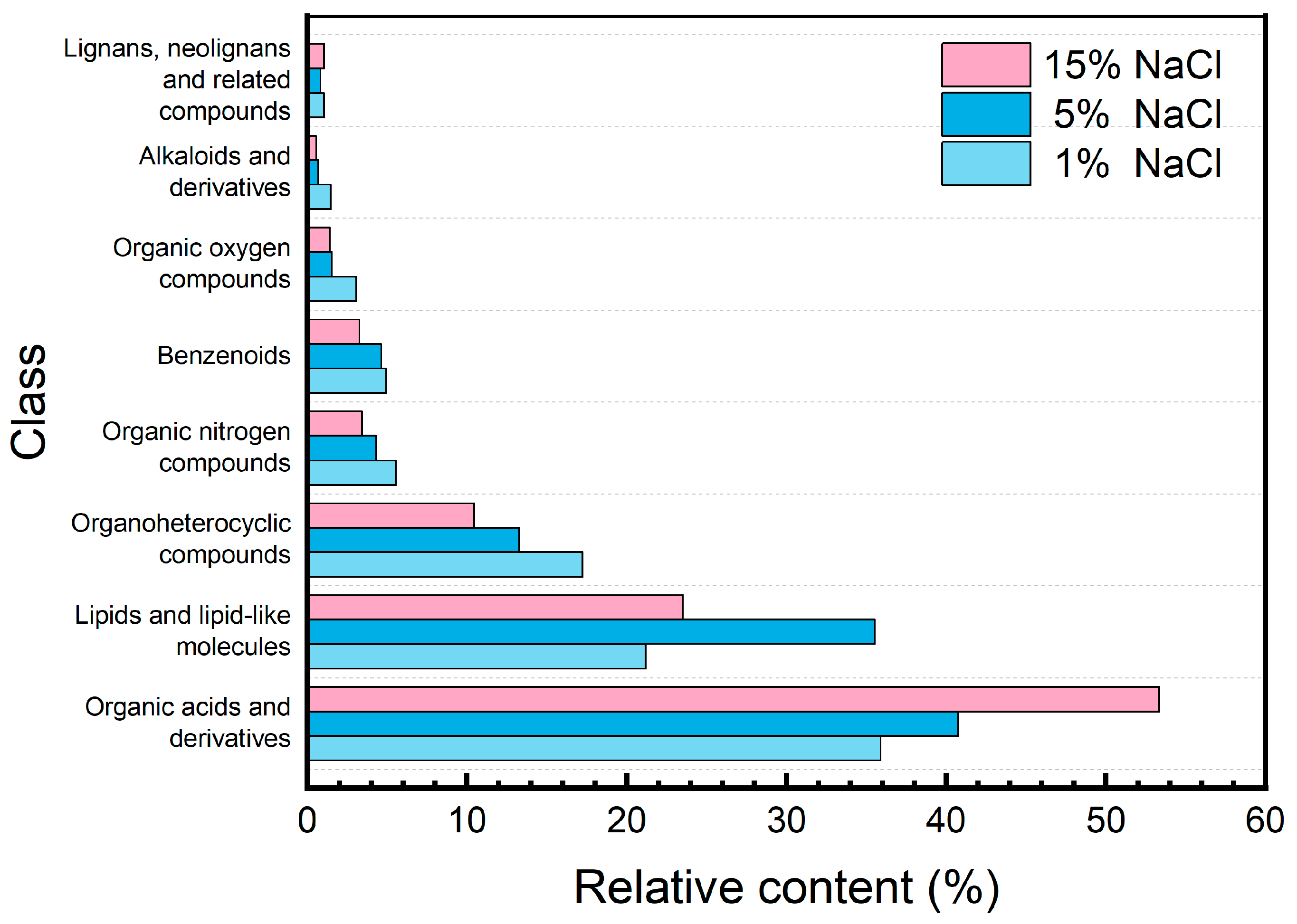
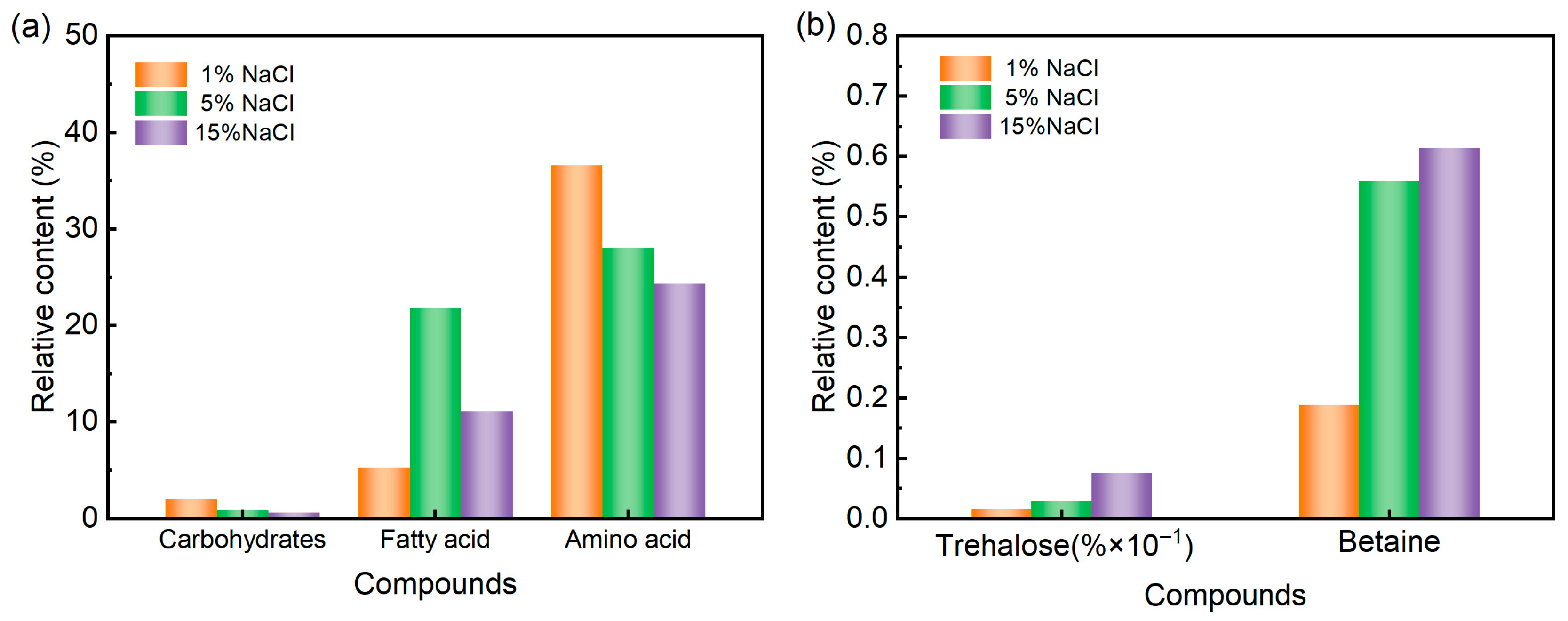
| m/z | rt (s) | Compounds | Relative Content (% × 10−2) | ||
|---|---|---|---|---|---|
| 1% NaCl | 5% NaCl | 15% NaCl | |||
| 529.08 | 175.12 | Arginine | 12.45 | 14.15 | 456.43 |
| 401.70 | 146.04 | Glutamic acid | 39.40 | 11.60 | 8.61 |
| 320.02 | 145.06 | Glutamine | 0.96 | 0.82 | 0.24 |
| 401.43 | 156.08 | Histidine | 50.17 | 41.96 | 20.69 |
| 247.27 | 173.13 | Isoleucine | 0.61 | 1.38 | 0.17 |
| 330.15 | 132.10 | Leucine | 4.78 | 38.32 | 3.30 |
| 529.08 | 147.11 | Lysine | 118.45 | 137.97 | 108.37 |
| 291.57 | 150.06 | Methionine | 33.79 | 26.03 | 15.80 |
| 261.62 | 166.09 | Phenylalanine | 0.59 | 0.48 | 0.26 |
| 318.31 | 116.07 | Proline | 29.57 | 164.52 | 10.62 |
| 94.39 | 102.05 | Threonine | 1.85 | 1.53 | 1.28 |
| 268.24 | 205.10 | Tryptophan | 6.34 | 20.77 | 35.88 |
| 311.66 | 182.08 | Tyrosine | 80.32 | 77.93 | 26.83 |
| 306.64 | 118.08 | Valine | 23.85 | 4.40 | 3.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, L.; Zhou, Q.; Liang, J. Efficient Bioreduction of Cr(VI) by a Halotolerant Acinetobacter sp. ZQ-1 in High-Salt Environments: Performance and Metabolomic Mechanism. Processes 2025, 13, 3423. https://doi.org/10.3390/pr13113423
Yu L, Zhou Q, Liang J. Efficient Bioreduction of Cr(VI) by a Halotolerant Acinetobacter sp. ZQ-1 in High-Salt Environments: Performance and Metabolomic Mechanism. Processes. 2025; 13(11):3423. https://doi.org/10.3390/pr13113423
Chicago/Turabian StyleYu, Lei, Qi Zhou, and Jing Liang. 2025. "Efficient Bioreduction of Cr(VI) by a Halotolerant Acinetobacter sp. ZQ-1 in High-Salt Environments: Performance and Metabolomic Mechanism" Processes 13, no. 11: 3423. https://doi.org/10.3390/pr13113423
APA StyleYu, L., Zhou, Q., & Liang, J. (2025). Efficient Bioreduction of Cr(VI) by a Halotolerant Acinetobacter sp. ZQ-1 in High-Salt Environments: Performance and Metabolomic Mechanism. Processes, 13(11), 3423. https://doi.org/10.3390/pr13113423





