Targeting GPR55 with Cannabidiol Derivatives: A Molecular Docking Approach Toward Novel Neurotherapeutics
Abstract
1. Introduction
2. Materials and Methods
2.1. Compounds Identification and Selection
2.2. Screening of Biologically Active Molecules
2.3. Pharmacokinetics (ADMET) Using Online Database
2.4. Potential Therapeutic Targets
2.5. Molecular Docking Protocol
2.6. Molecular Dynamics
3. Results
3.1. Molecular Properties and DrugLike Rules
3.2. Pharmacokinetics
3.2.1. Absorption
3.2.2. Distribution
3.2.3. Metabolism
3.2.4. Excretion
3.2.5. Toxicity
3.3. Pharmacodynamics
3.4. Molecular Docking
3.5. MD Simulations
3.5.1. Receptor Dynamics in the Protein—Ligand Complexes
3.5.2. Ligand Dynamics in the Protein—Ligand Complexes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CBD | Cannabidiol |
| THC | Tetrahydrocannabinol |
| CBC | Cannabichromene |
| CBDV | Cannabidivarin |
| CBL | Cannabicyclol |
| CBDP | Cannabidiphorol |
| THCV | Tetrahydrocannabivarin |
| 1″-HOCBD | 1″-Hydroxycannabidiol |
| 4″-HOCBD | 4″-Hydroxycannabidiol |
| 5″-HOCBD | 5″-Hydroxycannabidiol |
| 2″-HOCBD | 2″-Hydroxycannabidiol |
| 3″-HOCBD | 3″-Hydroxycannabidiol |
| CBDE | 5-ethyl-2-[(1R,6R)-3-methyl-6-(1-methylethenyl)-2-cyclohexen-1-yl]-1,3-benzenediol |
| GPR55 | G-protein coupled receptor 55 |
| CNR1 | Cannabinoid receptor 1 |
| CNR2 | Cannabinoid receptor 2 |
| GLRA1 | Glycine receptor subunit alpha-1 |
| MW | Molecular weight |
| TPSA | Topological polar surface area |
| LOG P | Coefficient of partition |
| CNS | Central nervous system |
| RMSD | Root mean squared deviation |
| RMSF | Root mean squared fluctuation |
| COM | Center of mass |
References
- Bonini, S.A.; Premoli, M.; Tambaro, S.; Kumar, A.; Maccarinelli, G.; Memo, M.; Mastinu, A. Cannabis Sativa: A Comprehensive Ethnopharmacological Review of a Medicinal Plant with a Long History. J. Ethnopharmacol. 2018, 227, 300–315. [Google Scholar] [CrossRef]
- Atalay, S.; Jarocka-Karpowicz, I.; Skrzydlewska, E. Antioxidative and Anti-Inflammatory Properties of Cannabidiol. Antioxidants 2019, 9, 21. [Google Scholar] [CrossRef]
- Aran, A.; Harel, M.; Cassuto, H.; Polyansky, L.; Schnapp, A.; Wattad, N.; Shmueli, D.; Golan, D.; Castellanos, F.X. Cannabinoid Treatment for Autism: A Proof-of-Concept Randomized Trial. Mol. Autism 2021, 12, 6. [Google Scholar] [CrossRef]
- McGuire, P.; Robson, P.; Cubala, W.J.; Vasile, D.; Morrison, P.D.; Barron, R.; Taylor, A.; Wright, S. Cannabidiol (CBD) as an Adjunctive Therapy in Schizophrenia: A Multicenter Randomized Controlled Trial. Am. J. Psychiatry 2018, 175, 225–231. [Google Scholar] [CrossRef]
- Wright, M.; Di Ciano, P.; Brands, B. Use of Cannabidiol for the Treatment of Anxiety: A Short Synthesis of Pre-Clinical and Clinical Evidence. Cannabis Cannabinoid Res. 2020, 5, 191–196. [Google Scholar] [CrossRef]
- Schiavon, A.P.; Bonato, J.M.; Milani, H.; Guimarães, F.S.; Weffort de Oliveira, R.M. Influence of Single and Repeated Cannabidiol Administration on Emotional Behavior and Markers of Cell Proliferation and Neurogenesis in Non-Stressed Mice. Prog. Neuropsychopharmacol. Biol. Psychiatry 2016, 64, 27–34. [Google Scholar] [CrossRef]
- Mooko, T.; Bala, A.; Tripathy, S.; Kumar, C.S.; Mahadevappa, C.P.; Chaudhary, S.K.; Matsabisa, M.G.; Cannabis Sativa, L. Flower and Bud Extracts Inhibited In Vitro Cholinesterases and β-Secretase Enzymes Activities: Possible Mechanisms of Cannabis Use in Alzheimer Disease. Endocr. Metab. Immune Disord. Drug Targets 2022, 22, 297–309. [Google Scholar] [CrossRef]
- Pisanti, S.; Malfitano, A.M.; Ciaglia, E.; Lamberti, A.; Ranieri, R.; Cuomo, G.; Abate, M.; Faggiana, G.; Proto, M.C.; Fiore, D.; et al. Cannabidiol: State of the Art and New Challenges for Therapeutic Applications. Pharmacol. Ther. 2017, 175, 133–150. [Google Scholar] [CrossRef]
- Elsaid, S.; Le Foll, B. The Complexity of Pharmacology of Cannabidiol (CBD) and Its Implications in the Treatment of Brain Disorders. Neuropsychopharmacology 2020, 45, 229–230. [Google Scholar] [CrossRef]
- Castillo-Arellano, J.; Canseco-Alba, A.; Cutler, S.J.; León, F. The Polypharmacological Effects of Cannabidiol. Molecules 2023, 28, 3271. [Google Scholar] [CrossRef]
- Millar, S.A.; Stone, N.L.; Yates, A.S.; O’Sullivan, S.E. A Systematic Review on the Pharmacokinetics of Cannabidiol in Humans. Front. Pharmacol. 2018, 9, 1365. [Google Scholar] [CrossRef]
- Lim, S.Y.; Sharan, S.; Woo, S. Model-Based Analysis of Cannabidiol Dose-Exposure Relationship and Bioavailability. Pharmacother. J. Hum. Human. Pharmacol. Drug Ther. 2020, 40, 291–300. [Google Scholar] [CrossRef]
- Taylor, L.; Gidal, B.; Blakey, G.; Tayo, B.; Morrison, G. A Phase I, Randomized, Double-Blind, Placebo-Controlled, Single Ascending Dose, Multiple Dose, and Food Effect Trial of the Safety, Tolerability and Pharmacokinetics of Highly Purified Cannabidiol in Healthy Subjects. CNS Drugs 2018, 32, 1053–1067. [Google Scholar] [CrossRef]
- Nadulski, T.; Pragst, F.; Weinberg, G.; Roser, P.; Schnelle, M.; Fronk, E.-M.; Stadelmann, A.M. Randomized, Double-Blind, Placebo-Controlled Study about the Effects of Cannabidiol (CBD) on the Pharmacokinetics of Delta9-Tetrahydrocannabinol (THC) after Oral Application of THC Verses Standardized Cannabis Extract. Ther. Drug Monit. 2005, 27, 799–810. [Google Scholar] [CrossRef]
- Elsohly, M.A.; Slade, D. Chemical Constituents of Marijuana: The Complex Mixture of Natural Cannabinoids. Life Sci. 2005, 78, 539–548. [Google Scholar] [CrossRef]
- Nader, D.A.; Sanchez, Z.M. Effects of Regular Cannabis Use on Neurocognition, Brain Structure, and Function: A Systematic Review of Findings in Adults. Am. J. Drug Alcohol. Abus. 2018, 44, 4–18. [Google Scholar] [CrossRef]
- Patil, N.; Chandel, V.; Rana, A.; Jain, M.; Kaushik, P. Investigation of Cannabis Sativa Phytochemicals as Anti-Alzheimer’s Agents: An In Silico Study. Plants 2023, 12, 510. [Google Scholar] [CrossRef]
- Patil, N.; Patil, K.; Jain, M.; Mohammed, A.; Yadav, A.; Dhanda, P.S.; Kole, C.; Dave, K.; Kaushik, P.; Azhar Abdul Razab, M.K.; et al. A Systematic Study of Molecular Targets of Cannabidiol in Alzheimer’s Disease. J. Alzheimers Dis. Rep. 2024, 8, 1339–1360. [Google Scholar] [CrossRef]
- Shen, S.-Y.; Yu, R.; Li, W.; Liang, L.-F.; Han, Q.-Q.; Huang, H.-J.; Li, B.; Xu, S.-F.; Wu, G.-C.; Zhang, Y.-Q.; et al. The Neuroprotective Effects of GPR55 against Hippocampal Neuroinflammation and Impaired Adult Neurogenesis in CSDS Mice. Neurobiol. Dis. 2022, 169, 105743. [Google Scholar] [CrossRef]
- Hill, J.D.; Zuluaga-Ramirez, V.; Gajghate, S.; Winfield, M.; Sriram, U.; Rom, S.; Persidsky, Y. Activation of GPR55 Induces Neuroprotection of Hippocampal Neurogenesis and Immune Responses of Neural Stem Cells Following Chronic, Systemic Inflammation. Brain Behav. Immun. 2019, 76, 165–181. [Google Scholar] [CrossRef]
- Liu, B.; Song, S.; Jones, P.M.; Persaud, S.J. GPR55: From Orphan to Metabolic Regulator? Pharmacol. Ther. 2015, 145, 35–42. [Google Scholar] [CrossRef]
- Marichal-Cancino, B.A.; Fajardo-Valdez, A.; Ruiz-Contreras, A.E.; Mendez-Díaz, M.; Prospero-García, O. Advances in the Physiology of GPR55 in the Central Nervous System. Curr. Neuropharmacol. 2017, 15, 771–778. [Google Scholar] [CrossRef]
- Xia, R.; Yuan, Q.; Wang, N.; Hou, L.; Abe, J.; Song, J.; Ito, Y.; Xu, H.E.; He, Y. Structural Insight into GPR55 Ligand Recognition and G-Protein Coupling. Cell Res. 2025, 35, 76–79. [Google Scholar] [CrossRef]
- Claff, T.; Ebenhoch, R.; Kley, J.T.; Magarkar, A.; Nar, H.; Weichert, D. Structural Basis for Lipid-Mediated Activation of G Protein-Coupled Receptor GPR55. Nat. Commun. 2025, 16, 1973. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New Data Content and Improved Web Interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Cherry, A.L.; Wheeler, M.J.; Mathisova, K.; Di Miceli, M. In Silico Analyses of the Involvement of GPR55, CB1R and TRPV1: Response to THC, Contribution to Temporal Lobe Epilepsy, Structural Modeling and Updated Evolution. Front. Neuroinform. 2024, 18, 1294939. [Google Scholar] [CrossRef]
- Putz, M.V.; Duda-Seiman, C.; Duda-Seiman, D.; Putz, A.-M.; Alexandrescu, I.; Mernea, M.; Avram, S. Chemical Structure-Biological Activity Models for Pharmacophores’ 3D-Interactions. Int. J. Mol. Sci. 2016, 17, 1087. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. In Chemical Biology: Methods and Protocols; Hempel, J.E., Williams, C.H., Hong, C.C., Eds.; Springer: New York, NY, USA, 2015; pp. 243–250. ISBN 978-1-4939-2269-7. [Google Scholar]
- Myung, Y.; de Sá, A.G.C.; Ascher, D.B. Deep-PK: Deep Learning for Small Molecule Pharmacokinetic and Toxicity Prediction. Nucleic Acids Res. 2024, 52, W469–W475. [Google Scholar] [CrossRef]
- Garisetti, V.; Varughese, R.E.; Anandamurthy, A.; Haribabu, J.; Dasararaju, G. Exploring Potential GPR55 Agonists Using Virtual Screening, Molecular Docking and Dynamics Simulation Studies. J. Biomol. Struct. Dyn. 2024, 1–13. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Avram, S.; Mernea, M.; Mihailescu, D.; Duda-Seiman, D.; Duda-Seiman, C. Advanced QSAR Methods Evaluated Polycyclic Aromatic Compounds Duality as Drugs and Inductors in Psychiatric Disorders. Curr. Org. Chem. 2013, 17, 2880–2890. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef] [PubMed]
- Updated Resources for Exploring Experimentally-Determined PDB Structures and Computed Structure Models at the RCSB Protein Data Bank|Nucleic Acids Research|Oxford Academic. Available online: https://academic.oup.com/nar/article/53/D1/D564/7912033?login=false (accessed on 23 July 2025).
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading-Trott-2010-Journal of Computational Chemistry-Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/10.1002/jcc.21334 (accessed on 23 July 2025).
- Citations—Autodock Vina 1.2.0 Documentation. Available online: https://autodock-vina.readthedocs.io/en/latest/citations.html (accessed on 23 November 2021).
- Dassault Systèmes BIOVIA. Discovery Studio Visualizer, Version 17.2.; Dassault Systèmes: San Diego, CA, USA, 2020.
- Elbegdorj, O.; Westkaemper, R.B.; Zhang, Y. A Homology Modeling Study toward the Understanding of Three-Dimensional Structure and Putative Pharmacological Profile of the G-Protein Coupled Receptor GPR55. J. Mol. Graph. Model. 2013, 39, 50–60. [Google Scholar] [CrossRef]
- Hetényi, C.; van der Spoel, D. Blind Docking of Drug-Sized Compounds to Proteins with up to a Thousand Residues. FEBS Lett. 2006, 580, 1447–1450. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable Molecular Dynamics on CPU and GPU Architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field (CGenFF): A Force Field for Drug-like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.; Jo, S.; Brooks, C.L.; Lee, H.S.; Im, W. CHARMM-GUI Ligand Reader & Modeler for CHARMM Force Field Generation of Small Molecules. J. Comput. Chem. 2017, 38, 1879–1886. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder Toward Realistic Biological Membrane Simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Avram, S.; Mernea, M.; Limban, C.; Borcan, F.; Chifiriuc, C. Potential Therapeutic Approaches to Alzheimer’s Disease By Bioinformatics, Cheminformatics And Predicted Adme-Tox Tools. Curr. Neuropharmacol. 2020, 18, 696–719. [Google Scholar] [CrossRef] [PubMed]
- Avram, S.; Stan, M.S.; Udrea, A.M.; Buiu, C.; Boboc, A.A.; Mernea, M. 3D-ALMOND-QSAR Models to Predict the Antidepressant Effect of Some Natural Compounds. Pharmaceutics 2021, 13, 1449. [Google Scholar] [CrossRef]
- Martínez-Pinilla, E.; Rico, A.J.; Rivas-Santisteban, R.; Lillo, J.; Roda, E.; Navarro, G.; Lanciego, J.L.; Franco, R. Expression of GPR55 and Either Cannabinoid CB1 or CB2 Heteroreceptor Complexes in the Caudate, Putamen, and Accumbens Nuclei of Control, Parkinsonian, and Dyskinetic Non-Human Primates. Brain Struct. Funct. 2020, 225, 2153–2164. [Google Scholar] [CrossRef]
- Miao, Y.; Zhao, F.; Guan, W. A Novel Insight into the Antidepressant Effect of Cannabidiol: Possible Involvement of the 5-HT1A, CB1, GPR55, and PPARγ Receptors. Int. J. Neuropsychopharmacol. 2025, 28, pyae064. [Google Scholar] [CrossRef]
- Oka, S.; Nakajima, K.; Yamashita, A.; Kishimoto, S.; Sugiura, T. Identification of GPR55 as a Lysophosphatidylinositol Receptor. Biochem. Biophys. Res. Commun. 2007, 362, 928–934. [Google Scholar] [CrossRef]
- Ryberg, E.; Larsson, N.; Sjögren, S.; Hjorth, S.; Hermansson, N.-O.; Leonova, J.; Elebring, T.; Nilsson, K.; Drmota, T.; Greasley, P.J. The Orphan Receptor GPR55 Is a Novel Cannabinoid Receptor. Br. J. Pharmacol. 2007, 152, 1092–1101. [Google Scholar] [CrossRef]
- Sun, L.; Apweiler, M.; Normann, C.; Grathwol, C.W.; Hurrle, T.; Gräßle, S.; Jung, N.; Bräse, S.; Fiebich, B.L. Anti-Inflammatory Effects of GPR55 Agonists and Antagonists in LPS-Treated BV2 Microglial Cells. Pharmaceuticals 2024, 17, 674. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Bao, N.; Chen, J.; Tang, M.; Yang, L.; Yang, Y.; Zhang, H.; Han, J.; Yu, P.; Zhang, S.; et al. Neuromolecular and Behavioral Effects of Cannabidiol on Depressive-Associated Behaviors and Neuropathic Pain Conditions in Mice. Neuropharmacology 2024, 261, 110153. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Yu, W.; Tan, Y. Activation of GPR55 Alleviates Neuropathic Pain and Chronic Inflammation. Biotechnol. Appl. Biochem. 2025, 72, 196–206. [Google Scholar] [CrossRef]
- Armin, S.; Muenster, S.; Abood, M.; Benamar, K. GPR55 in the Brain and Chronic Neuropathic Pain. Behav. Brain Res. 2021, 406, 113248. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-Y.; Tan, M.-S.; Yu, J.-T.; Tan, L. Role of Pro-Inflammatory Cytokines Released from Microglia in Alzheimer’s Disease. Ann. Transl. Med. 2015, 3, 136. [Google Scholar] [CrossRef]
- Solas, M.; Francis, P.T.; Franco, R.; Ramirez, M.J. CB2 Receptor and Amyloid Pathology in Frontal Cortex of Alzheimer’s Disease Patients. Neurobiol. Aging 2013, 34, 805–808. [Google Scholar] [CrossRef]
- Kurano, M.; Saito, Y.; Uranbileg, B.; Saigusa, D.; Kano, K.; Aoki, J.; Yatomi, Y. Modulations of Bioactive Lipids and Their Receptors in Postmortem Alzheimer’s Disease Brains. Front. Aging Neurosci. 2022, 14, 1066578. [Google Scholar] [CrossRef]
- Avram, S.; Milac, A.-L.; Mihailescu, D. 3D-QSAR Study Indicates an Enhancing Effect of Membrane Ions on Psychiatric Drugs Targeting Serotonin Receptor 5-HT1A. Mol. Biosyst. 2012, 8, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Avram, S.; Udrea, A.M.; Negrea, A.; Ciopec, M.; Duteanu, N.; Postolache, C.; Duda-Seiman, C.; Duda-Seiman, D.; Shaposhnikov, S. Prevention of Deficit in Neuropsychiatric Disorders through Monitoring of Arsenic and Its Derivatives as Well as Through Bioinformatics and Cheminformatics. Int. J. Mol. Sci. 2019, 20, 1804. [Google Scholar] [CrossRef] [PubMed]
- Geffrey, A.L.; Pollack, S.F.; Bruno, P.L.; Thiele, E.A. Drug-Drug Interaction between Clobazam and Cannabidiol in Children with Refractory Epilepsy. Epilepsia 2015, 56, 1246–1251. [Google Scholar] [CrossRef]
- Beers, J.L.; Fu, D.; Jackson, K.D. Cytochrome P450–Catalyzed Metabolism of Cannabidiol to the Active Metabolite 7-Hydroxy-Cannabidiol. Drug Metab. Dispos. 2021, 49, 882–891. [Google Scholar] [CrossRef]
- Ikezawa, N.; Iwasa, K.; Sato, F. Molecular Cloning and Characterization of CYP80G2, a Cytochrome P450 That Catalyzes an Intramolecular C-C Phenol Coupling of (S)-Reticuline in Magnoflorine Biosynthesis, from Cultured Coptis Japonica Cells. J. Biol. Chem. 2008, 283, 8810–8821. [Google Scholar] [CrossRef]
- Sholler, D.J.; Spindle, T.R.; Cone, E.J.; Goffi, E.; Kuntz, D.; Mitchell, J.M.; Winecker, R.E.; Bigelow, G.E.; Flegel, R.R.; Vandrey, R. Urinary Pharmacokinetic Profile of Cannabidiol (CBD), Δ9-Tetrahydrocannabinol (THC) and Their Metabolites Following Oral and Vaporized CBD and Vaporized CBD-Dominant Cannabis Administration. J. Anal. Toxicol. 2021, 46, 494–503. [Google Scholar] [CrossRef]
- Gunasekaran, S.; Purushothaman, A.; Anju, K. In Silico Assessment of Cannabidiol from Cannabis Sativa as an Antiviral Agent Against Key Shrimp Pathogens in Aquaculture. J. Fish Dis. 2025, e70015. [Google Scholar] [CrossRef]
- Dumitrascu, F.; Udrea, A.-M.; Caira, M.R.; Nuta, D.C.; Limban, C.; Chifiriuc, M.C.; Popa, M.; Bleotu, C.; Hanganu, A.; Dumitrescu, D.; et al. In Silico and Experimental Investigation of the Biological Potential of Some Recently Developed Carprofen Derivatives. Molecules 2022, 27, 2722. [Google Scholar] [CrossRef] [PubMed]
- Mareş, C.; Udrea, A.-M.; Şuţan, N.A.; Avram, S. Bioinformatics Tools for the Analysis of Active Compounds Identified in Ranunculaceae Species. Pharmaceuticals 2023, 16, 842. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Li, X.; Shen, L.; Ge, X.; Hao, S.; Wu, L.; Liu, S.; Liu, J.; Cherezov, V.; Hua, T. Structure Basis of Ligand Recognition and Activation of GPR55. Cell Res. 2025, 35, 80–83. [Google Scholar] [CrossRef]



| PubChem CID | Molecule Name | 2D Structure | Canonical SMILES |
|---|---|---|---|
| 644019 | CBD |  | CCCCCC1CC(O)C(C(C1)O)[C@@H]1C=C(C)CC[C@H]1C(=C)C |
| 16078 | THC |  | CCCCCC1=CC(=C2[C@@H]3C=C(CC[C@H]3C(OC2=C1)(C)C)C)O |
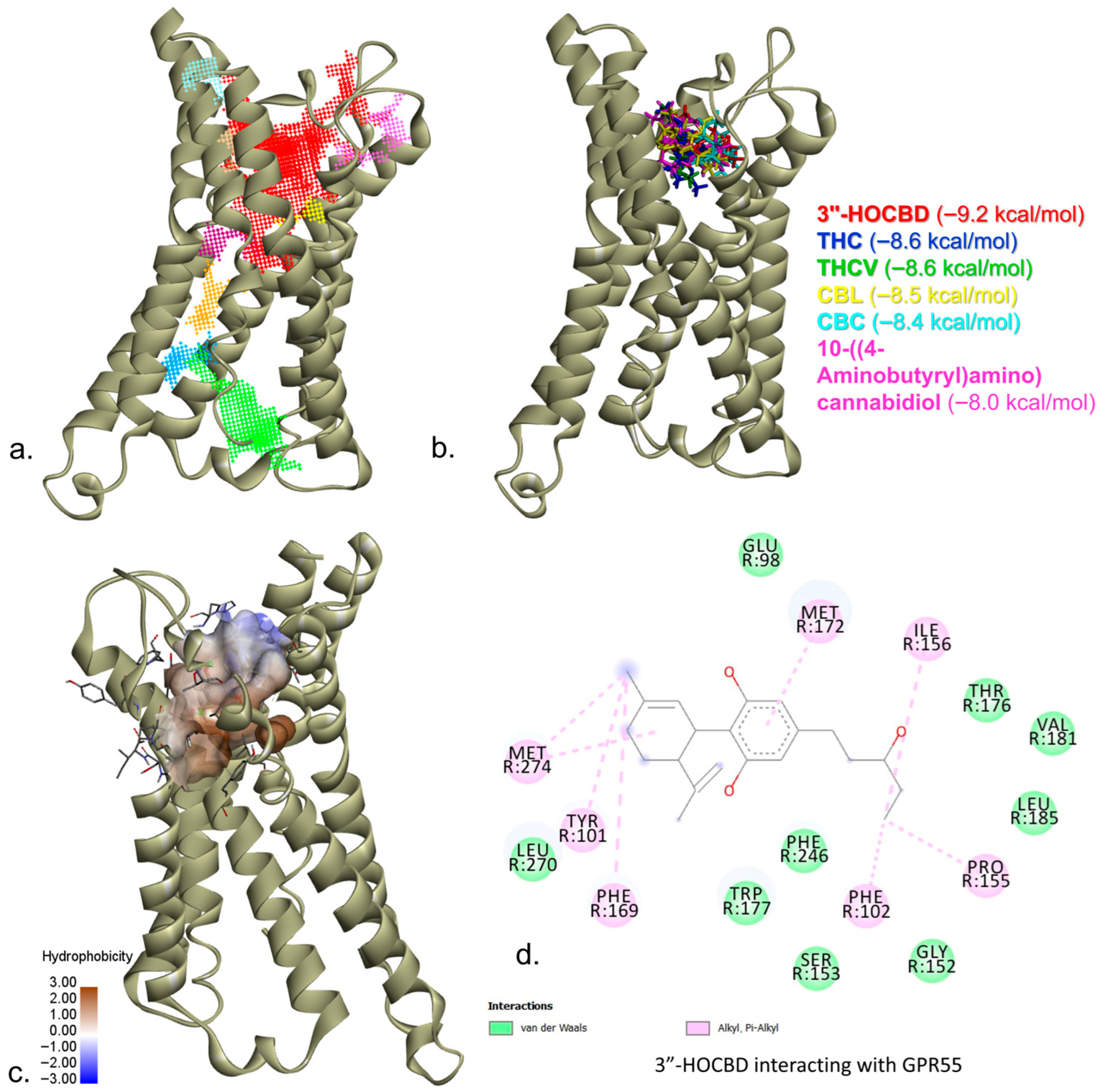
| 30219 | CBC | 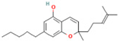 | CCCCCC1=CC(=C2C=CC(OC2=C1)(C)CCC=C(C)C)O |
| 11601669 | CBDV |  | CCCC1=CC(=C(C(=C1)O)[C@@H]2C=C(CC[C@H]2C(=C)C)C)O |
| 59444380 | CBL |  | CCCCCC1=CC(=C2[C@@H]3[C@H]4[C@@H](C3(C)C)CC[C@]4(OC2=C1)C)O |
| 49873141 | CBDP | 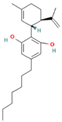 | CCCCCCCC1=CC(=C(C(=C1)O)[C@@H]2C=C(CC[C@H]2C(=C)C)C)O |
| 93147 | THCV |  | CCCC1=CC(=C2[C@@H]3C=C(CC[C@H]3C(OC2=C1)(C)C)C)O |
| 121596213 | 1″-HOCBD |  | CCCCC(C1=CC(=C(C(=C1)O)[C@@H]2C=C(CC[C@H]2C(=C)C)C)O)O |
| 53357352 | 4″-HOCBD | 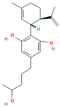 | CC1=C[C@H]([C@@H](CC1)C(=C)C)C2=C(C=C(C=C2O)CCCC(C)O)O |
| 101621543 | 5″-HOCBD | 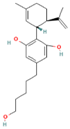 | CC1=C[C@H]([C@@H](CC1)C(=C)C)C2=C(C=C(C=C2O)CCCCCO)O |
| 121596214 | 2″-HOCBD | 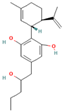 | CCCC(CC1=CC(=C(C(=C1)O)[C@@H]2C=C(CC[C@H]2C(=C)C)C)O)O |
| 121596215 | 3″-HOCBD | 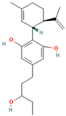 | CCC(CCC1=CC(=C(C(=C1)O)[C@@H]2C=C(CC[C@H]2C(=C)C)C)O)O |
| 129210056 | CBDE | 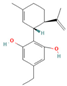 | CCC1=CC(=C(C(=C1)O)[C@@H]2C=C(CC[C@H]2C(=C)C)C)O |
| 44149593 | 10-((4-Aminobutyryl)amino)cannabidiol |  | CCCCCC1=CC(=C(C(=C1)O)[C@@H]2C=C(CC[C@H]2C(=C)CNC(=O)CCCN)C)O |
| Molecule | MW | #H-Bond Acceptors | #H-Bond Donors | TPSA | Consensus Log P |
|---|---|---|---|---|---|
| CBD | 314.46 | 2 | 2 | 40.46 | 5.2 |
| THC | 314.46 | 2 | 1 | 29.46 | 5.33 |
| CBC | 314.46 | 2 | 1 | 29.46 | 5.45 |
| CBDV | 286.41 | 2 | 2 | 40.46 | 4.5 |
| CBL | 314.46 | 2 | 1 | 29.46 | 5.08 |
| CBDP | 342.51 | 2 | 2 | 40.46 | 5.92 |
| THCV | 286.41 | 2 | 1 | 29.46 | 4.68 |
| 1″-HOCBD | 330.46 | 3 | 3 | 60.69 | 4.41 |
| 4″-HOCBD | 330.46 | 3 | 3 | 60.69 | 4.3 |
| 5″-HOCBD | 330.46 | 3 | 3 | 60.69 | 4.34 |
| 2″-HOCBD | 330.46 | 3 | 3 | 60.69 | 4.26 |
| 3″-HOCBD | 330.46 | 3 | 3 | 60.69 | 4.27 |
| CBDE | 272.38 | 2 | 2 | 40.46 | 4.17 |
| 10-((4-Aminobutyryl)amino)cannabidiol | 414.58 | 4 | 4 | 95.58 | 4.18 |
| Molecule | Lipinski #Violations | Ghose #Violations | Veber #Violations | Egan #Violations | Muegge #Violations | Bioavailability Score |
|---|---|---|---|---|---|---|
| CBD | 1 | 1 | 0 | 0 | 1 | 0.55 |
| THC | 1 | 1 | 0 | 0 | 1 | 0.55 |
| CBC | 1 | 1 | 0 | 1 | 1 | 0.55 |
| CBDV | 0 | 0 | 0 | 0 | 1 | 0.55 |
| CBL | 1 | 0 | 0 | 0 | 1 | 0.55 |
| CBDP | 1 | 1 | 0 | 1 | 1 | 0.55 |
| THCV | 0 | 0 | 0 | 0 | 1 | 0.55 |
| 1″-HOCBD | 0 | 0 | 0 | 0 | 1 | 0.55 |
| 4″-HOCBD | 0 | 0 | 0 | 0 | 0 | 0.55 |
| 5″-HOCBD | 0 | 0 | 0 | 0 | 0 | 0.55 |
| 2″-HOCBD | 0 | 0 | 0 | 0 | 0 | 0.55 |
| 3″-HOCBD | 0 | 0 | 0 | 0 | 0 | 0.55 |
| CBDE | 0 | 0 | 0 | 0 | 0 | 0.55 |
| 10-((4-Aminobutyryl)amino)cannabidiol | 0 | 0 | 1 | 0 | 0 | 0.55 |
| Compound Name | Human Oral Bioavailability 20% Probability | Human Oral Bioavailability 50% Probability | Human Intestinal Absorption Probability | Human Intestinal Absorption Interpretation |
|---|---|---|---|---|
| CBD | 0.57 | 0.337 | 0.985 | Absorbed (High Confidence) |
| THC | 0.674 | 0.41 | 0.994 | Absorbed (High Confidence) |
| CBC | 0.621 | 0.53 | 0.992 | Absorbed (High Confidence) |
| CBDV | 0.712 | 0.405 | 0.994 | Absorbed (High Confidence) |
| CBDV | 0.712 | 0.405 | 0.994 | Absorbed (High Confidence) |
| CBDP | 0.516 | 0.327 | 0.987 | Absorbed (High Confidence) |
| THCV | 0.757 | 0.461 | 0.997 | Absorbed (High Confidence) |
| 1″-HOCBD | 0.56 | 0.365 | 0.983 | Absorbed (High Confidence) |
| 4″-HOCBD | 0.676 | 0.337 | 0.989 | Absorbed (High Confidence) |
| 5″-HOCBD | 0.57 | 0.337 | 0.985 | Absorbed (High Confidence) |
| 2″-HOCBD | 0.607 | 0.38 | 0.984 | Absorbed (High Confidence) |
| 3″-HOCBD | 0.633 | 0.386 | 0.985 | Absorbed (High Confidence) |
| CBDE | 0.745 | 0.432 | 0.994 | Absorbed (High Confidence) |
| 10-((4-Aminobutyryl)amino)cannabidiol | 0.412 | 0.325 | 0.971 | Absorbed (High Confidence) |
| Compound Name | Central Nervous System Predictions | Blood–Brain Barrier Probability |
|---|---|---|
| CBD | −2.13 | 0.793 |
| THC | −2.53 | 0.998 |
| CBC | −2.29 | 0.981 |
| CBDV | −2.2 | 0.983 |
| CBDV | −2.2 | 0.983 |
| CBDP | −1.96 | 0.984 |
| THCV | −2.62 | 0.997 |
| 1″-HOCBD | −2.14 | 0.841 |
| 4″-HOCBD | −2.31 | 0.797 |
| 5″-HOCBD | −2.13 | 0.793 |
| 2″-HOCBD | −2.09 | 0.691 |
| 3″-HOCBD | −2.24 | 0.545 |
| CBDE | −2.2 | 0.989 |
| 10-((4-Aminobutyryl)amino)cannabidiol | −2.19 | 0.91 |
| Compound Name | CYP 1A2 Inhibitor | CYP 1A2 Substrate | CYP 2C19 Inhibitor | CYP 2C19 Substrate | CYP 2C9 Inhibitor | CYP 2C9 Substrate | CYP 2D6 Inhibitor | CYP 2D6 Substrate | CYP 3A4 Inhibitor | CYP 3A4 Substrate |
|---|---|---|---|---|---|---|---|---|---|---|
| CBD | 0.942 | 0.379 | 0.523 | 0.595 | 0.882 | 0.767 | 0.179 | 0.419 | 0.927 | 0.518 |
| THC | 0.995 | 0.599 | 0.963 | 0.634 | 0.986 | 0.974 | 0.32 | 0.567 | 0.732 | 0.756 |
| CBC | 0.931 | 0.539 | 0.883 | 0.604 | 0.659 | 0.975 | 0.917 | 0.578 | 0.637 | 0.808 |
| CBDV | 0.874 | 0.466 | 0.984 | 0.608 | 0.969 | 0.959 | 0.867 | 0.474 | 0.865 | 0.631 |
| CBDV | 0.874 | 0.466 | 0.984 | 0.608 | 0.969 | 0.959 | 0.867 | 0.474 | 0.865 | 0.631 |
| CBDP | 0.977 | 0.359 | 0.91 | 0.612 | 0.881 | 0.818 | 0.652 | 0.428 | 0.891 | 0.626 |
| THCV | 0.992 | 0.669 | 0.981 | 0.615 | 0.981 | 0.984 | 0.928 | 0.568 | 0.668 | 0.712 |
| 1″-HOCBD | 0.821 | 0.307 | 0.896 | 0.619 | 0.951 | 0.884 | 0.287 | 0.397 | 0.856 | 0.404 |
| 4″-HOCBD | 0.904 | 0.356 | 0.784 | 0.612 | 0.801 | 0.846 | 0.01 | 0.457 | 0.861 | 0.484 |
| 5″-HOCBD | 0.942 | 0.379 | 0.523 | 0.595 | 0.882 | 0.767 | 0.179 | 0.419 | 0.927 | 0.518 |
| 2″-HOCBD | 0.799 | 0.317 | 0.923 | 0.623 | 0.891 | 0.864 | 0.277 | 0.419 | 0.867 | 0.429 |
| 3″-HOCBD | 0.835 | 0.328 | 0.861 | 0.613 | 0.768 | 0.849 | 0.012 | 0.415 | 0.816 | 0.45 |
| CBDE | 0.722 | 0.485 | 0.985 | 0.604 | 0.947 | 0.964 | 0.689 | 0.483 | 0.792 | 0.624 |
| 10-((4-Aminobutyryl)amino)cannabidiol | 0.658 | 0.195 | 0.664 | 0.626 | 0.629 | 0.098 | 0.985 | 0.436 | 0.956 | 0.294 |
| Compound Name | Organic Cation Transporter 2 Probability | Organic Cation Transporter 2 Interpretation | Half-Life of Drug Predictions | Half-Life of Drug Probability | Half-Life of Drug Interpretation |
|---|---|---|---|---|---|
| CBD | 0.382 | Non-Inhibitor (Low Confidence) | Half-Life < 3 hs | 0.243 | Half-Life < 3 hs (Medium Confidence) |
| THC | 0.394 | Non-Inhibitor (Low Confidence) | Half-Life < 3 hs | 0.135 | Half-Life < 3 hs (High Confidence) |
| CBC | 0.322 | Non-Inhibitor (Medium Confidence) | Half-Life < 3 hs | 0.178 | Half-Life < 3 hs (Medium Confidence) |
| CBDV | 0.307 | Non-Inhibitor (Medium Confidence) | Half-Life < 3 hs | 0.243 | Half-Life < 3 hs (Medium Confidence) |
| CBDV | 0.307 | Non-Inhibitor (Medium Confidence) | Half-Life < 3 hs | 0.243 | Half-Life < 3 hs (Medium Confidence) |
| CBDP | 0.358 | Non-Inhibitor (Low Confidence) | Half-Life < 3 hs | 0.206 | Half-Life < 3 hs (Medium Confidence) |
| THCV | 0.425 | Non-Inhibitor (Low Confidence) | Half-Life < 3 hs | 0.157 | Half-Life < 3 hs (High Confidence) |
| 1″-HOCBD | 0.263 | Non-Inhibitor (Medium Confidence) | Half-Life < 3 hs | 0.319 | Half-Life < 3 hs (Medium Confidence) |
| 4″-HOCBD | 0.326 | Non-Inhibitor (Medium Confidence) | Half-Life < 3 hs | 0.234 | Half-Life < 3 hs (Medium Confidence) |
| 5″-HOCBD | 0.382 | Non-Inhibitor (Low Confidence) | Half-Life < 3 hs | 0.243 | Half-Life < 3 hs (Medium Confidence) |
| 2″-HOCBD | 0.269 | Non-Inhibitor (Medium Confidence) | Half-Life < 3 hs | 0.314 | Half-Life < 3 hs (Medium Confidence) |
| 3″-HOCBD | 0.313 | Non-Inhibitor (Medium Confidence) | Half-Life < 3 hs | 0.252 | Half-Life < 3 hs (Medium Confidence) |
| CBDE | 0.266 | Non-Inhibitor (Medium Confidence) | Half-Life < 3 hs | 0.255 | Half-Life < 3 hs (Medium Confidence) |
| 10-((4-Aminobutyryl)amino)cannabidiol | 0.407 | Non-Inhibitor (Low Confidence) | Half-Life < 3 hs | 0.297 | Half-Life < 3 hs (Medium Confidence) |
| Compound Name | AMES Mutagenesis | Carcinogenesis | Liver Injury I (DILI) | Liver Injury II | hERG Blockers |
|---|---|---|---|---|---|
| CBD | Safe | Toxic | Safe | Safe | Safe |
| THC | Safe | Safe | Safe | Toxic | Safe |
| CBC | Safe | Safe | Safe | Safe | Safe |
| CBDV | Safe | Safe | Safe | Safe | Safe |
| CBDV | Safe | Safe | Safe | Safe | Safe |
| CBDP | Safe | Toxic | Safe | Safe | Safe |
| THCV | Safe | Safe | Safe | Toxic | Safe |
| 1″-HOCBD | Safe | Safe | Safe | Safe | Safe |
| 4″-HOCBD | Safe | Toxic | Safe | Safe | Safe |
| 5″-HOCBD | Safe | Toxic | Safe | Safe | Safe |
| 2″-HOCBD | Safe | Toxic | Safe | Safe | Safe |
| 3″-HOCBD | Safe | Toxic | Safe | Safe | Safe |
| CBDE | Safe | Safe | Safe | Safe | Safe |
| 10-((4-Aminobutyryl)amino)cannabidiol | Safe | Toxic | Safe | Toxic | Toxic |
| Target | Common Name | Target Class | Probability |
|---|---|---|---|
| CBD | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.893165 |
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.893165 |
| G-protein coupled receptor 55 | GPR55 | Family A G protein-coupled receptor | 0.818184 |
| THC | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.959682 |
| N-arachidonyl glycine receptor | GPR18 | Family A G protein-coupled receptor | 0.959682 |
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.959682 |
| Glycine receptor subunit alpha-1 | GLRA1 | Ligand-gated ion channel | 0.959682 |
| CBC | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.959682 |
| N-arachidonyl glycine receptor | GPR18 | Family A G protein-coupled receptor | 0.959682 |
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.959682 |
| Glycine receptor subunit alpha-1 | GLRA1 | Ligand-gated ion channel | 0.959682 |
| CBDV | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.897728 |
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.897728 |
| G-protein coupled receptor 55 | GPR55 | Family A G protein-coupled receptor | 0.641391 |
| CBL | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.693215 |
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.693215 |
| CBDP | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.825425 |
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.825425 |
| G-protein coupled receptor 55 | GPR55 | Family A G protein-coupled receptor | 0.758299 |
| THCV | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.906315 |
| N-arachidonyl glycine receptor | GPR18 | Family A G protein-coupled receptor | 0.786927 |
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.786927 |
| Glycine receptor subunit alpha-1 | GLRA1 | Ligand-gated ion channel | 0.786927 |
| Vascular endothelial growth factor receptor 2 | KDR | Kinase | 0.542516 |
| 1″-HOCBD | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.371182 |
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.371182 |
| 4″-HOCBD | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.573001 |
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.573001 |
| 5″-Hydroxycannabidiol | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.613334 |
| 2″-HOCBD | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.459956 |
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.459956 |
| 3″-HOCBD | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.629642 |
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.629642 |
| CBDE | |||
| Cannabinoid receptor 1 | CNR1 | Family A G protein-coupled receptor | 0.608509 |
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.608509 |
| 10-((4-Aminobutyryl)amino)cannabidiol | |||
| Cannabinoid receptor 2 | CNR2 | Family A G protein-coupled receptor | 0.106166 |
| Molecule Name | Ligand | Beste Pose Binding Affinity (kcal/mol) | Interacting Aminoacids |
|---|---|---|---|
| 3”-HOCBD | 8zx5_121596213_uff_E = 233.98 | −9.2 | TYR101, PHE102, PRO 155, ILE 156,PHE 169, MET 172, MET274 |
| THC | 8zx5_16078_uff_E = 312.91 | −8.6 | HIS27, LEU77, LYS80, TYR 101, PHE 169, TRP177, MET172,LEU270, MET274 |
| THCV | 8zx5_93147_uff_E = 308.78 | −8.6 | HIS27, LYS80, TYR101, MET172, PHE169, TRP177, LEU 270, MET 274 |
| CBL | 8zx5_59444380_uff_E = 965.10 | −8.5 | TYR 101, PHE 169, LEU270, MET274 |
| CBC | 8zx5_30219_uff_E = 202.54 | −8.4 | PHE102, PRO155,ILE156, LEU185, PHE169, MET 172 |
| 10-((4-Aminobutyryl)amino)cannabidiol | 8zx5_44149593_uff_E = 278.31 | −8 | TYR101, PHE169, MET172, TRP 177, PHE 246, LEU 270 MET 274, GLN 271 |
| 4”-HOCBD | 8zx5_53357352_uff_E = 242.61 | −7.8 | PHE 102, ILE 156, PHE 169, LEU270, MET 274 |
| 5”-HOCBD | 8zx5_101621543_uff_E = 233.63 | −7.7 | LYS80, TYR101, PHE169, HIS170, MET 172, MET274 |
| CBDP | 8zx5_49873141_uff_E = 221.20 | −7.6 | TYR101, PHE102, ILE 156, PHE169, MET172, LEU185, PHE 246, LEU270, MET274 |
| CBDV | 8zx5_11601669_uff_E = 309.11 | −7.6 | HIS27, LYS80, TYR101, PHE169, MET172, PHE246, MET274 |
| 1”-HOCBD | 8zx5_121596213_uff_E = 214.48 | −7.5 | TYR101, PHE102, ILE156, MET172, TRP 177, LEU270 |
| 2”-HOCBD | 8zx5_121596214_uff_E = 242.42 | −7.5 | GLN23, TYR101, PHE102, ILE156, MET172, HIS170, PHE169, LEU270, GLN271, MET274 |
| CBD | 8zx5_644019_uff_E = 254.46 | −7.3 | TYR101, PHE 102, ILE156, PHE169, MET172, LEU270, MET274 |
| CBDE | 8zx5_129210056_uff_E = 205.71 | −7.2 | TYR101, PHE169 MET172, ILE156, TRP177, LEU270 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mares, C.; Paun, A.-M.; Mernea, M.; Matanie, C.; Avram, S. Targeting GPR55 with Cannabidiol Derivatives: A Molecular Docking Approach Toward Novel Neurotherapeutics. Processes 2025, 13, 3261. https://doi.org/10.3390/pr13103261
Mares C, Paun A-M, Mernea M, Matanie C, Avram S. Targeting GPR55 with Cannabidiol Derivatives: A Molecular Docking Approach Toward Novel Neurotherapeutics. Processes. 2025; 13(10):3261. https://doi.org/10.3390/pr13103261
Chicago/Turabian StyleMares, Catalina, Andra-Maria Paun, Maria Mernea, Cristina Matanie, and Speranta Avram. 2025. "Targeting GPR55 with Cannabidiol Derivatives: A Molecular Docking Approach Toward Novel Neurotherapeutics" Processes 13, no. 10: 3261. https://doi.org/10.3390/pr13103261
APA StyleMares, C., Paun, A.-M., Mernea, M., Matanie, C., & Avram, S. (2025). Targeting GPR55 with Cannabidiol Derivatives: A Molecular Docking Approach Toward Novel Neurotherapeutics. Processes, 13(10), 3261. https://doi.org/10.3390/pr13103261





