Effects of Pectinase on Bacterial Succession during Hemp Retting
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Testing
2.2. Microbial Community Analysis by Illumina MiSeq Sequencing
2.3. Bioinformatics Analysis
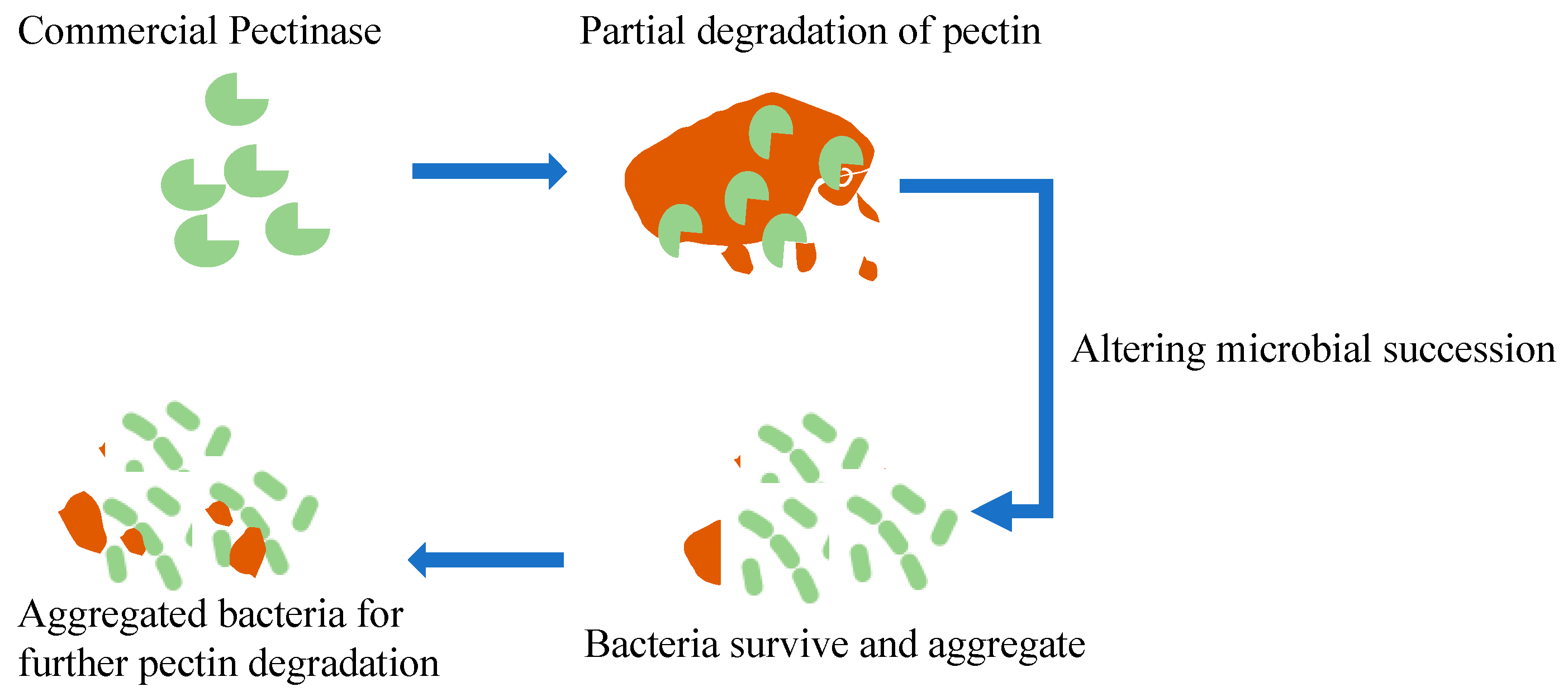
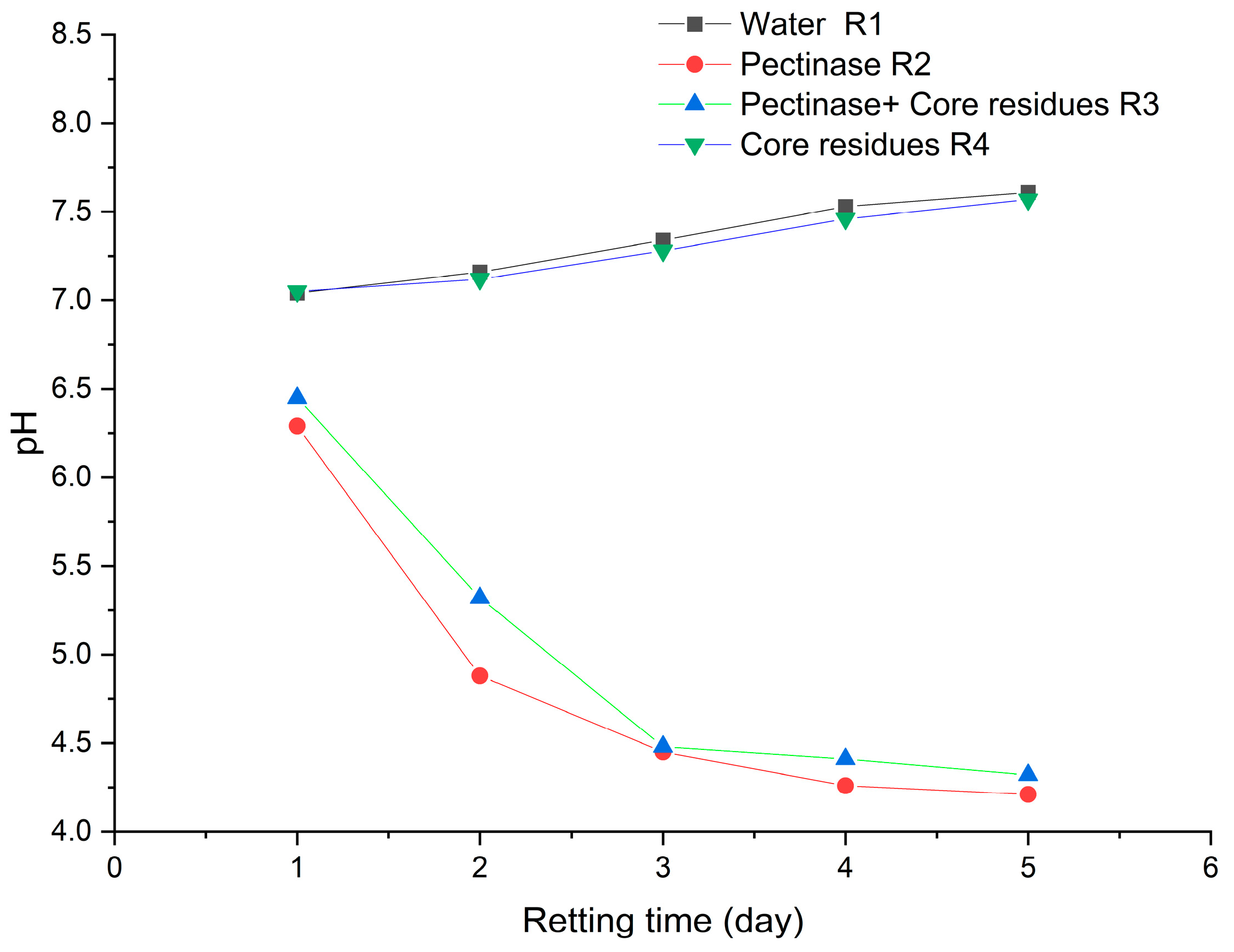
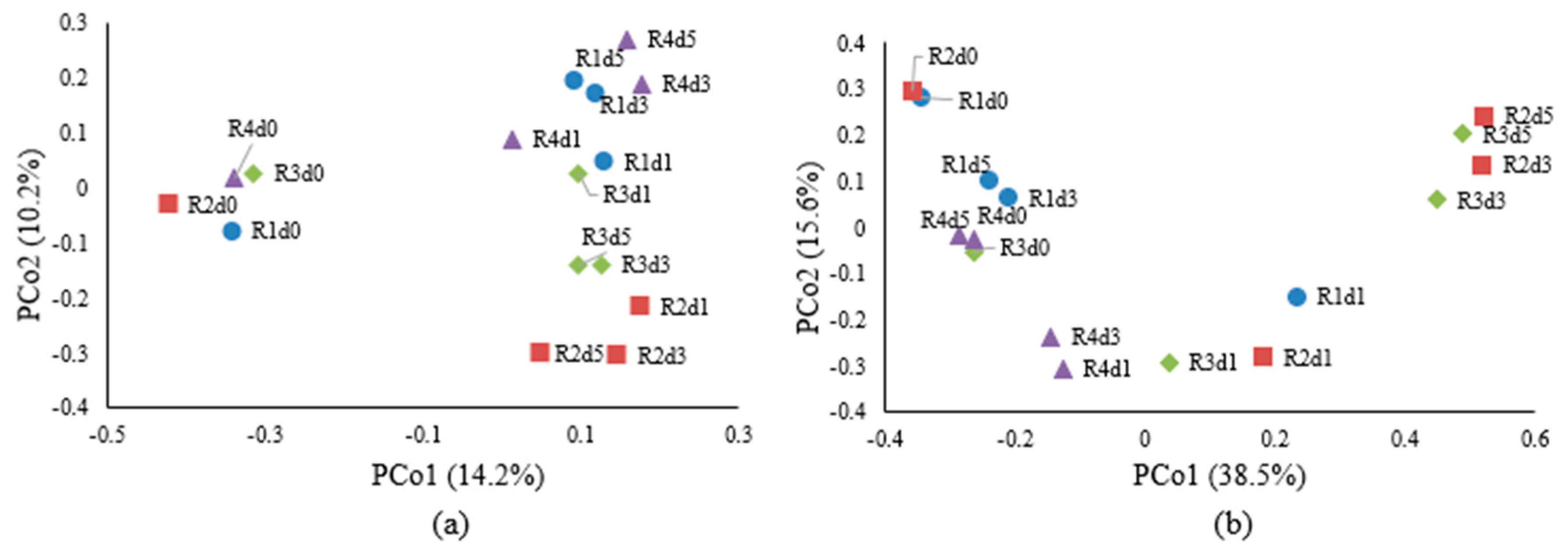
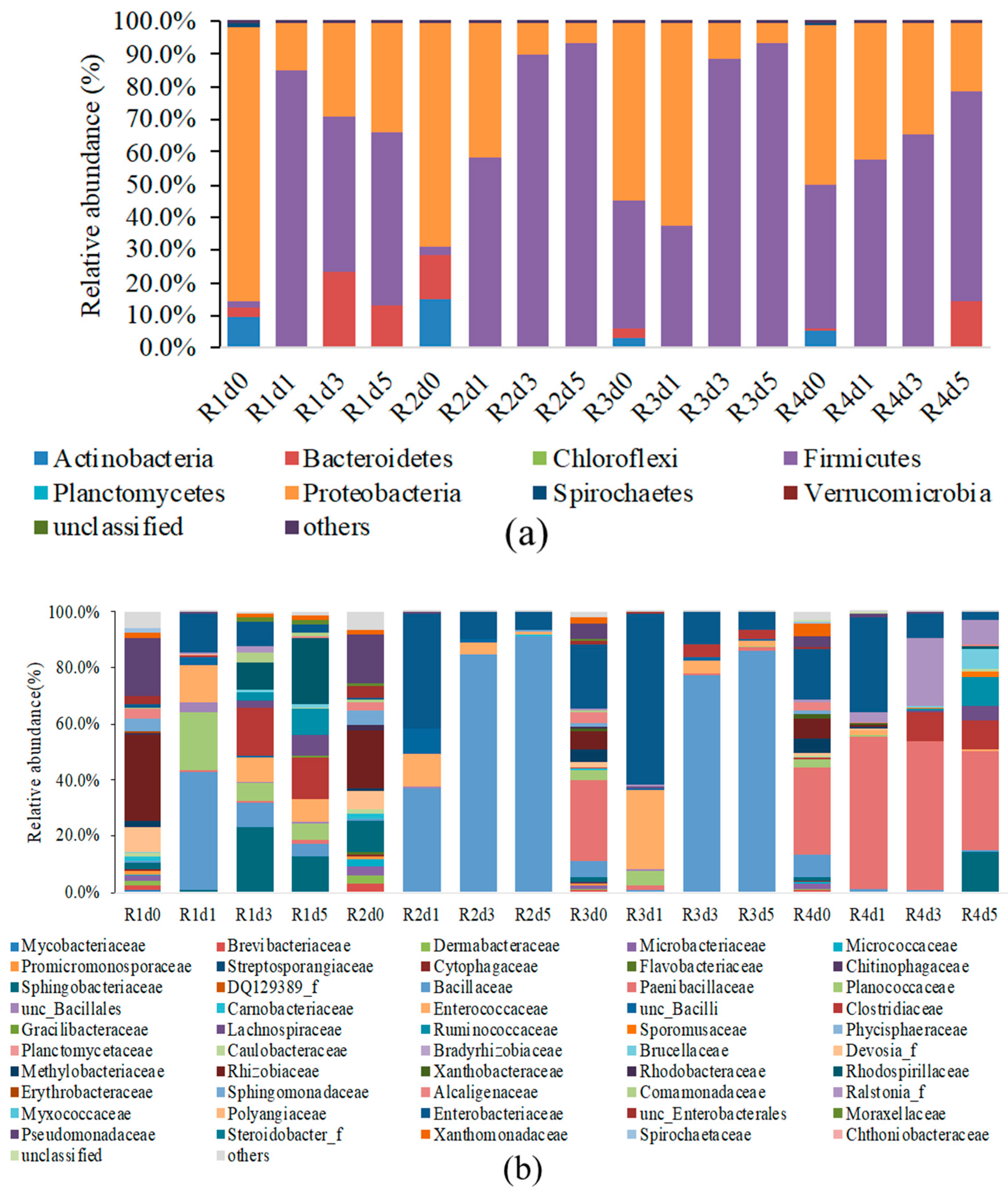
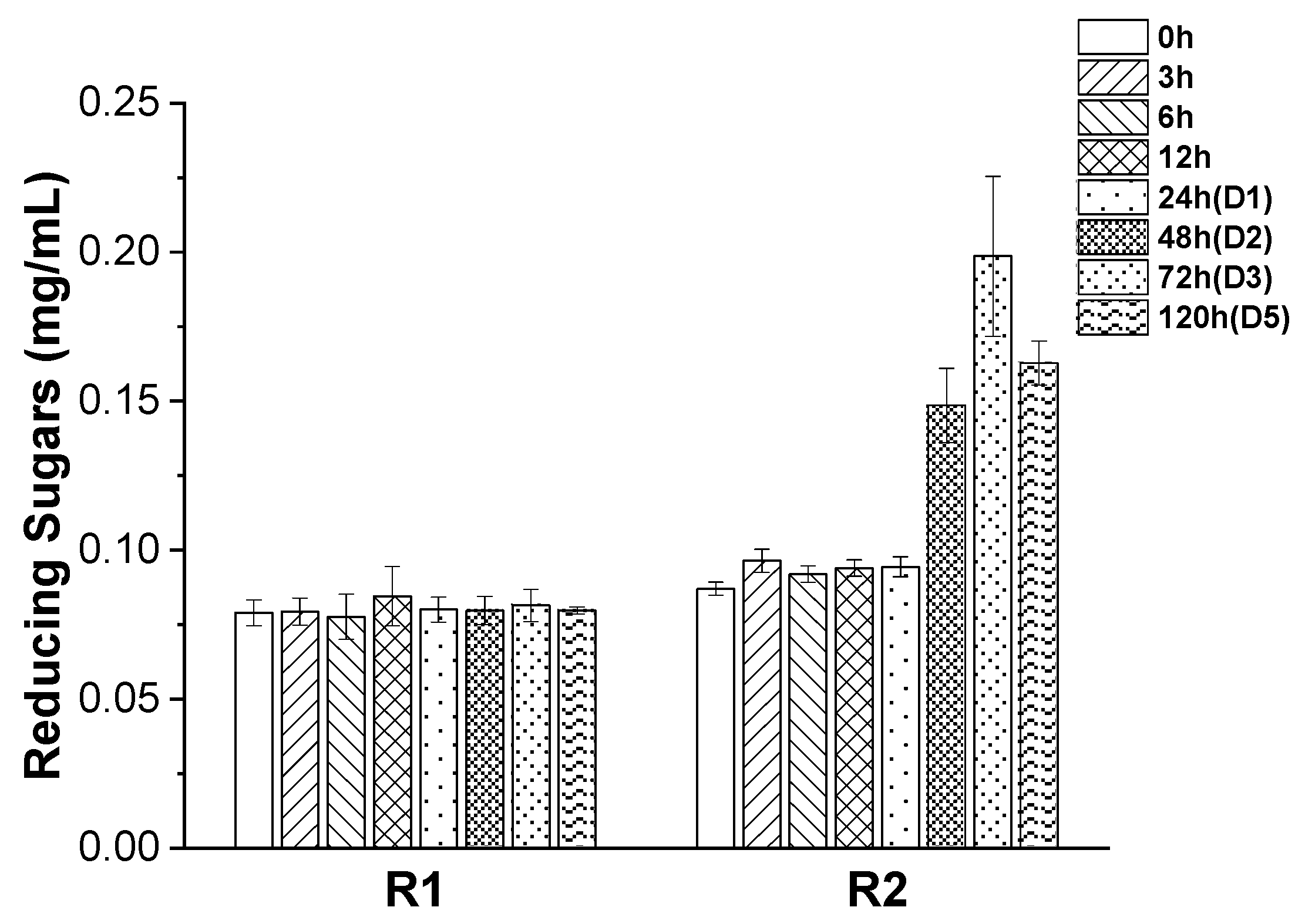
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- United States Department of Agriculture. National Hemp Report. 2022. Available online: https://downloads.usda.library.cornell.edu/usda-esmis/files/gf06h2430/xd07hw825/v692v917t/hempan22.pdf (accessed on 17 February 2022).
- Liang, K.; Shi, S.Q.; Wang, G. Effect of impregnated inorganic nanoparticles on the properties of the kenaf bast fibers. Fibers 2014, 2, 242–254. [Google Scholar] [CrossRef]
- Xia, C.; Zhang, S.; Shi, S.Q.; Cai, L.; Huang, J. Property enhancement of kenaf fiber reinforced composites by in situ aluminum hydroxide impregnation. Ind. Crops Prod. 2016, 79, 131–136. [Google Scholar] [CrossRef]
- Tulaphol, S.; Sun, Z.; Sathitsuksanoh, N. Chapter six—Biofuels and bioproducts from industrial hemp. Adv. Bioenergy 2021, 6, 301–338. [Google Scholar] [CrossRef]
- Paridah, M.T.; Basher, A.B.; SaifulAzry, S.; Ahmed, Z. Retting process of some bast plant fibres and its effect on fibre quality: A review. BioResources 2011, 6, 5260–5281. [Google Scholar]
- Zimniewska, M. Hemp fibre properties and processing target textile: A review. Materials 2022, 15, 1901. [Google Scholar] [CrossRef] [PubMed]
- Hurren, C.J.; Wang, X.; Dennis, H.G.; Clarke, A.F.K. Evaluation of bast fibre retting systems on hemp. In Proceedings of the 82nd Textile Institute World Conference, Cairo, Egypt; 2002. Available online: https://hdl.handle.net/10536/DRO/DU:30013875 (accessed on 23 March 2002).
- Hoondal, G.; Tiwari, R.; Tewari, R.; Dahiya, N.; Beg, Q. Microbial alkaline pectinases and their industrial applications: A review. Appl. Microbiol. Biotechnol. 2002, 59, 409–418. [Google Scholar] [CrossRef]
- Foulk, J.A.; Rho, D.; Alcock, M.M.; Ulven, C.A.; Huo, S. Modifications caused by enzyme-retting and their effect on composite performance. Adv. Mater. Sci. Eng. 2011, 2011, 179023. [Google Scholar] [CrossRef]
- Di Candilo, M.; Ranalli, P.; Bozzi, C.; Focher, B.; Mastromei, G. Preliminary results of tests facing with the controlled retting of hemp. Ind. Crops Prod. 2000, 11, 197–203. [Google Scholar] [CrossRef]
- Hossain, M.M.; Siddiquee, S.; Kumar, V. Critical factors for optimum biodegradation of bast fiber’s gums in bacterial retting. Fibers 2021, 9, 52. [Google Scholar] [CrossRef]
- Di Candilo, M.; Bonatti, P.M.; Guidetti, C.; Focher, B.; Grippo, C.; Tamburini, E.; Mastromei, G. Effects of selected pectinolytic bacterial strains on water-retting of hemp and fibre properties. J. Appl. Microbiol. 2010, 108, 194–203. [Google Scholar] [CrossRef]
- Tamburini, E.; León, A.G.; Perito, B.; Di Candilo, M.; Mastromei, G. Exploitation of bacterial pectinolytic strains for improvement of hemp water retting. Euphytica 2004, 140, 47–54. [Google Scholar] [CrossRef]
- Donaghy, J.A.; Levett, P.N.; Haylock, R.W. Changes in microbial populations during anaerobic flax retting. J. Appl. Bacteriol. 1990, 69, 634–641. [Google Scholar] [CrossRef]
- Visi, D.K.; D’Souza, N.; Ayre, B.G.; Webber III, C.L.; Allen, M.S. Investigation of the bacterial retting community of kenaf (Hibiscus cannabinus) under different conditions using next-generation semiconductor sequencing. J. Ind. Microbiol. Biotechnol. 2013, 40, 465–475. [Google Scholar] [CrossRef]
- Zhao, D.; Liu, P.; Pan, C.; Du, R.; Ping, W.; Ge, J. Bacterial succession and metabolite changes during flax (Linum usitatissimum L.) retting with bacillus cereus HDYM-02. Sci. Rep. 2016, 6, 31812. [Google Scholar] [CrossRef]
- De Prez, J.; Van Vuure, A.W.; Ivens, J.; Aerts, G.; de Van Voorde, I. Enzymatic treatment of flax for use in composites. Biotechnol. Rep. 2018, 20, e00294. [Google Scholar] [CrossRef] [PubMed]
- Akin, D.E.; Dodd, R.B.; Perkins, W.; Henriksson, G.; Eriksson, K.L. Spray enzymatic retting: A new method for processing flax fibers. Text. Res. J. 2000, 70, 486–494. [Google Scholar] [CrossRef]
- Akin, D.E.; Foulk, J.A.; Dodd, R.B.; McAlister, D.D., III. Enzyme-retting of flax and characterization of processed fibers. J. Biotechnol. 2001, 89, 193–203. [Google Scholar] [CrossRef]
- Bernava, A.; Reihmane, S.; Strazds, G. Influence of pectinase enzyme beisol PRO on hemp fibres retting. Pap. Present. Proc. Est. Acad. Sci. 2015, 64, 77–81. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Sandri, I.G.; Silveira, M.M.d. Production and application of pectinases from aspergillus niger obtained in solid state cultivation. Beverages 2018, 4, 48. [Google Scholar] [CrossRef]
- Ajayi, A.A.; Lawal, B.; Salubi, A.E.; Onibokun, A.E.; Oniha, M.I.; Ajayi, O.M. Pectinase production by aspergillus niger using pineapple peel pectin and its application in coconut oil extraction. Pap. Present. IOP Conf. Ser. Earth Environ. Sci. 2021, 655, 012014. [Google Scholar] [CrossRef]
- Jalil, M.T.M.; Ibrahim, D. Partial purification and characterization of pectinase produced by aspergillus niger LFP-1 grown on pomelo peels as a substrate. Trop. Life Sci. Res. 2021, 32, 1. [Google Scholar] [CrossRef] [PubMed]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Westram, R. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Ha, S.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Lee, C.H.; Khalina, A.; Lee, S.; Liu, M. A comprehensive review on bast fibre retting process for optimal performance in fibre-reinforced polymer composites. Adv. Mater. Sci. Eng. 2020, 2020, 6074063. [Google Scholar] [CrossRef]
- Müller, M.J.; Stachurski, S.; Stoffels, P.; Schipper, K.; Feldbrügge, M.; Büchs, J. Online evaluation of the metabolic activity of Ustilago maydis on (poly) galacturonic acid. J. Biol. Eng. 2018, 12, 34. [Google Scholar] [CrossRef]
- Sharma, S.; Mandhan, R.P.; Sharma, J. Pseudozyma sp. SPJ: An economic and eco-friendly approach for degumming of flax fibers. World J. Microbiol. Biotechnol. 2011, 27, 2697–2701. [Google Scholar] [CrossRef]
- Zhao, D.; Pan, C.; Ping, W.; Ge, J. Degumming crude enzyme produced by bacillus cereus HDYM-02 and its application in flax retting. BioResources 2018, 13, 5213–5224. [Google Scholar] [CrossRef]
- Garg, G.; Singh, A.; Kaur, A.; Singh, R.; Kaur, J.; Mahajan, R. Microbial pectinases: An ecofriendly tool of nature for industries. 3 Biotech 2016, 6, 47. [Google Scholar] [CrossRef]
- Zhang, G.; Li, S.; Xu, Y.; Wang, J.; Wang, F.; Xin, Y.; Liu, H. Production of alkaline pectinase: A case study investigating the use of tobacco stalk with the newly isolated strain bacillus tequilensis CAS-MEI-2-33. BMC Biotechnol. 2019, 19, 45. [Google Scholar] [CrossRef]
- Alqahtani, Y.S.; More, S.S.; Shaikh, I.A.; KJ, A.; More, V.S.; Niyonzima, F.N.; Khan, A.A. Production and purification of pectinase from Bacillus subtilis 15a-b92 and its biotechnological applications. Molecules 2022, 27, 4195. [Google Scholar] [CrossRef] [PubMed]
- Arotupin, D.J.; Akinyosoye, F.A.; Onifade, A.K. Purification and characterization of pectinmethylesterase from Aspergillus repens isolated from cultivated soil. Afr. J. Biotechnol. 2008, 7, 1991–1998. [Google Scholar] [CrossRef]
- Fahmy, A.S.; El-Beih, F.M.; Mohamed, S.A.; Abdel-Gany, S.S.; Abd-Elbaky, E.A. Characterization of an exopolygalacturonase from Aspergillus niger. Appl. Biochem. Biotechnol. 2008, 149, 205–217. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, R. Production of alkaline pectinase by bacteria (cocci sps.) isolated from decomposing fruit materials. J. Phytol. 2012, 4, 1–5. [Google Scholar]
- Ahlawat, S.; Mandhan, R.P.; Dhiman, S.S.; Kumar, R.; Sharma, J. Potential application of alkaline pectinase from Bacillus subtilis SS in pulp and paper industry. Appl. Biochem. Biotechnol. 2008, 149, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Pakarinen, A.; Zhang, J.; Brock, T.; Maijala, P.; Viikari, L. Enzymatic accessibility of fiber hemp is enhanced by enzymatic or chemical removal of pectin. Bioresour. Technol. 2012, 107, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.; Macfarlane, G.T. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 2003, 62, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Sar, A.; Misra, A.; Pal, S.; Chakraborty, A.; Dam, B. Increased productivity in poultry birds by sub-lethal dose of antibiotics is arbitrated by selective enrichment of gut microbiota, particularly short-chain fatty acid producers. Microbiology 2018, 164, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.; Pochart, P.; Day, A.; Mennuni, S.; Bono, P.; Baret, J.; Mangin, I. Microbial diversity observed during hemp retting. Appl. Microbiol. Biotechnol. 2015, 99, 4471–4484. [Google Scholar] [CrossRef] [PubMed]
- Law, A.D.; McNees, C.R.; Moe, L.A. The microbiology of hemp retting in a controlled environment: Steering the hemp microbiome towards more consistent fiber production. Agronomy 2020, 10, 492. [Google Scholar] [CrossRef]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [PubMed]
- Bajpai, P. Chapter 8—Industrial applications of xylanases. In Xylanolytic enzymes; Bajpai, P., Ed.; Academic Press: Amsterdam, Netherlands, 2014; pp. 69–104. [Google Scholar] [CrossRef]
- Barnett, S.E.; Cala, A.R.; Hansen, J.L.; Crawford, J.; Viands, D.R.; Smart, L.B.; Buckley, D.H. Evaluating the microbiome of hemp. Phytobiomes J. 2020, 4, 351–363. [Google Scholar] [CrossRef]
- Djemiel, C.; Grec, S.; Hawkins, S. Characterization of bacterial and fungal community dynamics by high-throughput sequencing (HTS) metabarcoding during flax dew-retting. Front. Microbiol. 2017, 8, 2052. [Google Scholar] [CrossRef]
- Djemiel, C.; Goulas, E.; Badalato, N.; Chabbert, B.; Hawkins, S.; Grec, S. Targeted metagenomics of retting in flax: The beginning of the quest to harness the secret powers of the microbiota. Front. Genet. 2020, 11, 581664. [Google Scholar] [CrossRef]
- Munshi, T.K.; Chattoo, B.B. Bacterial population structure of the jute-retting environment. Microb. Ecol. 2008, 56, 270–282. [Google Scholar] [CrossRef]
- Das, B.; Chakrabarti, K.; Tripathi, S.; Chakraborty, A. Review of some factors influencing jute fiber quality. J. Nat. Fibers 2014, 11, 268–281. [Google Scholar] [CrossRef]
- Mandic-Mulec, I.; Stefanic, P.; van Elsas, J.D. Ecology of bacillaceae. In The Bacterial Spore: From Molecules to Systems; John Wiley & Sons: Hoboken, NJ, USA, 2016; pp. 59–85. [Google Scholar] [CrossRef]
- Demir, N.; Nadaroglu, H.; Demir, Y.; Isik, C.; Taskin, E.; Adiguzel, A.; Gulluce, M. Purification and characterization of an alkaline pectin lyase produced by a newly isolated Bbrevibacillus borstelensis (P35) and its applications in fruit juice and oil extraction. Eur. Food Res. Technol. 2014, 239, 127–135. [Google Scholar] [CrossRef]
- Tamburini, E.; León, A.G.; Perito, B.; Mastromei, G. Characterization of bacterial pectinolytic strains involved in the water retting process. Environ. Microbiol. 2003, 5, 730–736. [Google Scholar] [CrossRef]
- Bowman, J.P. 14—Protein-based analysis and other new and emerging non-nucleic acid based methods for tracing and investigating foodborne pathogens. In Tracing Pathogens in the Food Chain; Brul, S., Fratamico, P.M., McMeekin, T.A., Eds.; Woodhead Publishing: Cambridge, UK, 2011; pp. 292–341. [Google Scholar] [CrossRef]
- Park, G.W.; Seo, C.; Jung, K.; Chang, H.N.; Kim, W.; Kim, Y. A comprehensive study on volatile fatty acids production from rice straw coupled with microbial community analysis. Bioprocess. Biosyst. Eng. 2015, 38, 1157–1166. [Google Scholar] [CrossRef] [PubMed]


| Schedule | Description of the Retting Schedule | Sample Labeling | |||
|---|---|---|---|---|---|
| Day 0 | Day 1 | Day 3 | Day 5 | ||
| R1 | Hemp fiber (5 g) was retted with tap water (200 mL) only | R1d0 | R1d1 | R1d3 | R1d5 |
| R2 | Hemp bast fiber (5 g) was retted with 1% (m/v) pectinase in tap water (200 mL) | R2d0 | R2d1 | R2d3 | R2d5 |
| R3 | Hemp bast fiber (5 g) was retted with 1% (m/v) pectinase in tap water (200 mL) and 1 g solid residues from hemp core retting (with cultured bacteria) | R3d0 | R3d1 | R3d3 | R3d5 |
| R4 | Hemp bast fiber (5 g) was retted with tap water (200 mL) and 1 g of solid residues from hemp core retting (with cultured bacteria) | R4d0 | R4d1 | R4d3 | R4d5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Y.; Zhang, Y.; Allen, M.S.; Shi, S.Q. Effects of Pectinase on Bacterial Succession during Hemp Retting. Processes 2024, 12, 1725. https://doi.org/10.3390/pr12081725
Fu Y, Zhang Y, Allen MS, Shi SQ. Effects of Pectinase on Bacterial Succession during Hemp Retting. Processes. 2024; 12(8):1725. https://doi.org/10.3390/pr12081725
Chicago/Turabian StyleFu, Yu, Yan Zhang, Michael S. Allen, and Sheldon Q. Shi. 2024. "Effects of Pectinase on Bacterial Succession during Hemp Retting" Processes 12, no. 8: 1725. https://doi.org/10.3390/pr12081725
APA StyleFu, Y., Zhang, Y., Allen, M. S., & Shi, S. Q. (2024). Effects of Pectinase on Bacterial Succession during Hemp Retting. Processes, 12(8), 1725. https://doi.org/10.3390/pr12081725







