Abstract
Grape dehydration is practiced widely in the food industry with large yields of sultanas produced globally. This paper proposes an investigation into the microstructure changes of grapes as they are dried by imaging specimens at intervals during dehydration at two temperatures using scanning electron microscopy. Two main methods were developed to obtain the complex boundaries of cells present in grape tissue in over 36 SEM images. Segmentation of the binary image using an adapted watershed function obtained the most consistent and accurate morphological shape. This was compared to a secondary method which used Canny’s edge detection function, morphological closing and skeletonizing to outline the cellular microstructure. MATLAB was utilised to convert these boundaries into measurable areas so that quantitative data on average cell area, perimeter and cell axis lengths were acquired. It was found that over the drying time, the cell area and perimeter were reduced as expected. Some variability in the data was clear due to only single samples being analysed for each temperature and time combination. Trends in cell perimeter, diameter and shape will be used to demonstrate relationships between morphological structure, drying temperature, and duration. Detailed images of the microstructure were obtained, and a unique image processing algorithm was developed to quantitatively analyse the properties of this microstructure. The development of automatic image processing techniques and algorithms will enable quantitative data to be extracted from any image and extend to any plant/food material.
1. Introduction
The food industry develops many products worldwide, and there is a demand for improving technologies, which could benefit from advances in food engineering [1]. There is a large amount of energy consumed during these processes, and there is a need to investigate the science behind this to potentially enable more efficient technologies [2]. By examining the microstructure of food materials throughout the drying process, trends in changes to structure and moisture transfer can be found (Figure 1). During drying, moisture is transferred through the inner cells to the plant surface before transforming into vapour and evaporating into the surrounding environment [3]. As the cells lose their water content (which makes up 70–80% of the cell cytoplasm), the cells shrink and loose shape whilst retaining their continuous cell wall. In some cases, where equal water concentrations inside and outside cell walls exist, cells do not shrink at the same rate as adjoining cells, and there could be separation between neighbouring cells. If the dehydration process is rapid or not isotropic the cells could rupture from the high stresses placed on the cell membrane and cause a cavity to form [4].
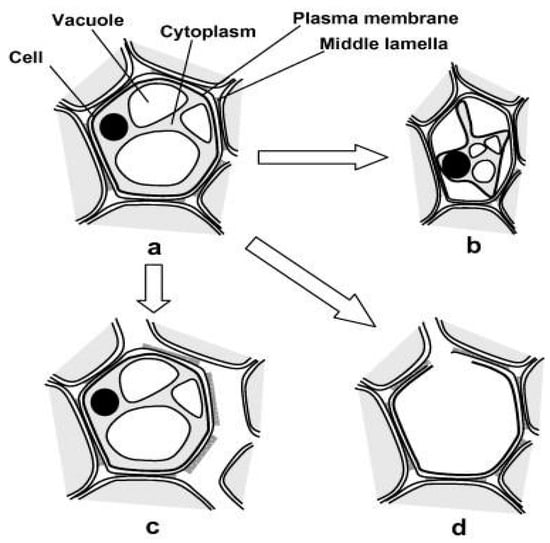
Figure 1.
Changes to plant microstructure during dehydration. (a) Fresh cell, (b) shrinkage and plasmolysis, (c) cell-to-cell debonding and (d) cell rupture and cavity formation [4].
It has been suggested by [2] and [5] that this process of cavity formation is very slow because the cells attempt to protect themselves from the dehydration process by adapting their cellular structure, depending on time and temperature changes. Previous research by Ramosa et al. [6] found that the grape microstructure retained its general shape whilst reducing uniformly by perimeter and area.
The kinematics of the drying process has been investigated in the current literature using a wide range of food products. The consensus appears to be that water content is proportional to cell area and appears to decrease uniformly as the drying time progresses [4,7]. Furthermore, a higher dying temperature increased the rate of shrinkage in the cell microstructure [6,8,9]. Several studies on the drying curves of grapes have been undertaken, all of which obtained similar drying curves to those carried out on Thompson seedless grapes [10]. Research conducted [11] into the effect of the particle size of drying times found that halved grape berries dry in considerably less time than whole grape berries while still exhibiting drying and moisture behaviour like other dried fruits.
There were comments made in several articles that during the final stages of drying the cell area seems to increase slightly, with varying hypotheses made that the cells either “reopened” after longer drying times [12] or created a rigid crust (casehardening) at the surface of the food sample during the very low moisture content stages [3]. The subsequent rehydration of the dried food tissues was also extensively covered by several sources [13,14,15].
Food microstructure controls water and nutrient transport by determining the pathways travelled between cells [16]. Thus, the relationship between the rate of drying and the transfer of moisture through not only the cells but the substructures within these cells should be examined [2,4].
As the food industry can control the external heat and mass transfer during drying but not internal cellular changes, there is a need for image processing techniques to uncover what these internal changes are to improve the drying process [2].
1.1. Scanning Electron Imaging
Scanning electron microscopes (SEMs) can be used to magnify an object up to 500,000 times to obtain images at the micro and nano levels [17]. The device focuses a concentrated beam of electrons onto the sample and scans the area side-to-side to get an overall reading of the structure the electrons come into contact with. The signals collected by the SEM are the result of interactions between the electron beam and atoms close to and at the surface of the sample. These reactions are recorded by collecting the secondary electrons produced from this interaction. As the electrons have the ability to contact atoms to a certain depth within the sample, it is sometimes feasible to produce an image with a characteristic three-dimensional appearance [17].
Scanning electron microscopy has been found to produce better images than past approaches which used light microscopes [16]. Furthermore, to obtain clear images, the sample must be conductive (to allow electron reactions with the surface atoms) and thus a thin coating of metal is often applied to organic tissue. In most cases, gold is sputter-coated across the mounted sample prior to imaging [18].
There are many cases in the literature where dried food microstructures have been examined using a scanning electron microscope. Carrot [18], potato [7,12,19], banana [20] and apple [16,21] are commonly chosen for investigations. Some researchers developed images of grape tissue for an investigation into the microscopic changes during dehydration [6,22]. However, there are few studies which quantitatively examine the grape microstructure using scanning electron microscopy.
1.2. Image Analysis
A similar investigation analysing SEM images which showed the microstructural changes of potatoes during drying employed image processing methods similar to this project [7,12]. This analysis utilised MATLAB algorithms to detect the edge of cells in a potato and measure the cell wall perimeter and area. The process initially detected cell boundaries by using the Canny’s edge detector algorithm, which specified two thresholds for analysis [23]. Pixels identified beneath the lower threshold were set to appear black and those above the upper threshold white. Pixels which sat between the two thresholds were set to white if they had a neighbouring pixel above this threshold, or black if they did not [24]. Morphological closing was performed which linked previously detected edges that were close to touching to create an enclosed cell [7]. The use of a thinning algorithm developed by [25] transformed these thick outlines and reduced the solid boundaries into one-pixel-wide outlines [26]. Pruning was also employed to remove small branches of the cell boundary that did not fully separate cells [12]. Furthermore, small regions within the image were removed by imposing a minimum number of connected pixels required of shapes (Figure 2).
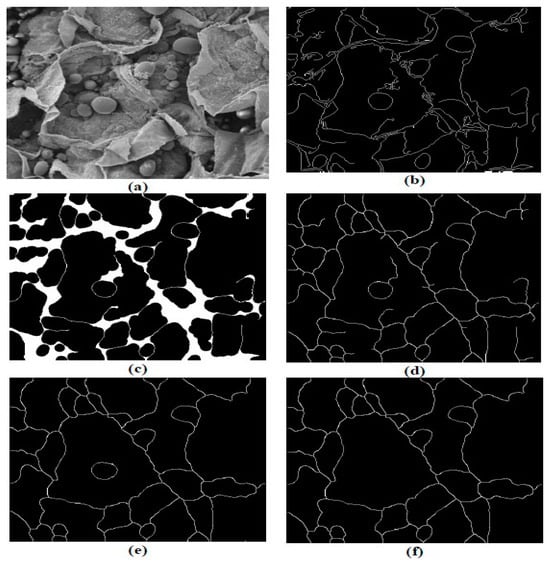
Figure 2.
Cell boundary detection process. (a) Original greyscale image, (b) after edge detection step, (c) following closing, (d) thinning, (e) pruning and (f) removal of small regions (Banks & Senadeera, 2012).
There are several other methods of cell segmentation image processing that have been identified in the literature on other types of specimens [27]). Medical and biological studies into cell growth have also utilized many image processing techniques which are relevant to this investigation [28]. The pre-processing stage shown in Figure 3 incorporates histogram equalisation and morphological closing techniques. Further analysis is undertaken by applying Gaussian kernel operations to the image. The final cell segmentation provided images that were easily analysed to obtain the dimensional properties of the cells [27,28] and to discuss methods of processing images obtained using confocal microscopy. The use of Gaussian filters to process the image further supported the segmentation methods by separating the background and foreground imaging using double thresholds like those employed in the Canny edge algorithm [28]. The method of segmentation using watershed functions is discussed thoroughly, with an emphasis on using distance functions to separate cells or objects [29].
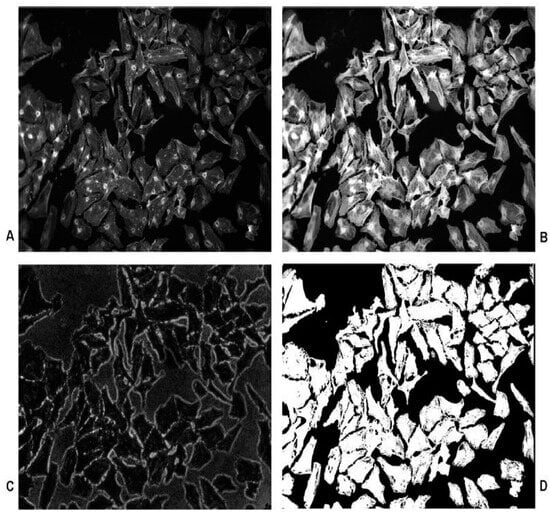
Figure 3.
Image segmentation: (A) the original microscopy image, (B) the effects of pre-processing, (C) Gaussian kernel applications and (D) final segmented image [29].
Although there were many studies found looking at the microstructure of food during dehydration, the current literature seems to be lacking a thorough analysis linking dehydration and microstructure during drying. It was found that grape specimens were often not a focus of the current literature, with the use of apple, potato, carrot, and banana favoured [1,8]. Image analysis methods were not covered in detail in many of the studies, and thus the need for the development of a custom SEM image analysis algorithm for the grape samples was confirmed. Image processing techniques such as the Canny edge detector, watershed function combined with distance measurements and closing techniques positively mentioned in the literature will be included in the development process of the custom algorithm.
The aim of the study was to find the relationship between cell size and the drying time of the grapes and also the rehydration characteristics of the dried grapes at different time intervals. To determine and develop an algorithm, different techniques of MATLAB image processing were used.
2. Material and Methods
2.1. Material
Thompson seedless grapes were used as the material for the experimentation. The internal structure of the grape is separated into fleshy tissue, skin, stalk and seeds [29]. The focus of this investigation was the internal flesh of the grape, and thus the skin, stalk, endocarp and pips were not important features. The skin (epicarp) is comprised of one layer of epidermis and three of four layers of subepidemic collenchymatous (exocarp). After these layers, the pulp (mesocarp) begins [22]. This is evident in the images of grapes produced using a light microscope, in which the epicarp and mesocarp layers can be identified (Figure 4).
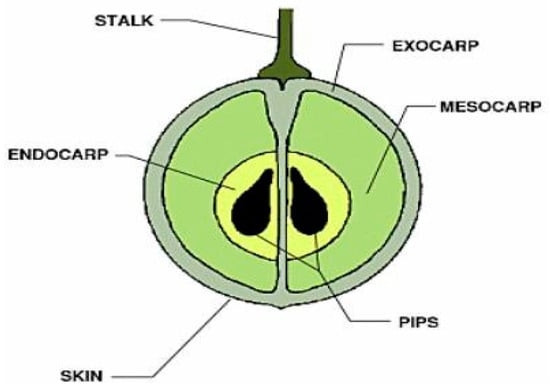
Figure 4.
Macrostructure of a typical grape [30].
2.2. Drying Procedure
The purpose of the drying procedure was to obtain specimens used in the food industry, namely raisins (dried grapes), which are usually dried until they reach a moisture content of 0.17 to 0.18 kg/kg, which is considered safe for long term storage [10]. The grapes were dried using a forced convection drying chamber (Model PA 00075773, Across International, Bayswater, Australia). In order to greatly reduce drying time, as shown in previous investigations [11], the grape specimens were first cut in two on a longitudinal line through the centre of the grape. The first group of grapes were heated at a constant temperature of 70 °C throughout the testing. A set of four grapes were placed facing upwards in the dehydrator and taken out after the first time interval (two hours). This was repeated for increasing durations a subsequent four times to obtain samples dried for 4, 6, 8 and 10 h. This method of drying the grapes in batches ensured that the amount of heat and air distributed to each grape was equal throughout the process instead of having 20 grapes in the dehydrator at the start reduce gradually to 4 grapes. A second group of grapes was dried at 55 °C in the same manner. Due to the availability of funds, only two temperatures were considered; the main idea was the development of the MATLAB algorithm. For each batch of four grapes, the weight of the sample prior to and after drying was recorded. Three grapes were refrigerated at approximately 8 °C for later rehydration and one was preserved in a dry and airtight container for imaging.
2.3. Rehydration Process
Prior to rehydration, the grape samples were weighed again to allow any changes in mass from storage in the refrigerator to be accounted for. This was used as the initial grape weight for the rehydration data. The grapes (three from each temperature and time duration set) were then submerged in a water bath for nearly seven hours. The temperature of water was consistent with ambient conditions. Within the first 100 min the grapes were removed and weighed every 20 min and then hourly until 400 min had passed. This was to record data frequently at the start of the rehydration process as it was expected that water absorption would slow after this point.
2.4. Sample Preparation for SEM Imaging
The specimens were placed in an osmotic dehydrating fluid (sucrose 60–80%) which removed any remaining fluid within the grape specimens without affecting the microstructure of the grape. Each sample was then mounted on an aluminium dish to support and transport the specimen. To ensure the surface was electrically and thermally conductive, and thus could be imaged by the scanning electron microscope, each piece of grape was sputter-coated with a thin layer of gold 5–10 nm. This layer enabled strong secondary electron generation, which improved the signal obtained by the SEM.
2.5. Imaging Process
QUT’s scanning electron microscope (SEM) was used to obtain high quality images of the grape microstructure at several stages of the drying process. Each sample was placed inside the SEM and the chamber was placed under vacuum. The electron beam was focussed on the sample and the image created by secondary electron generation was recorded. The image taken of each sample was at a region of the grape which appeared not physically damaged and was away from the edge of the grape to ensure only the mesocarp (large, inside grape cells) of the microstructure was captured. Each grape sample was imaged at 400× and 250× magnifications in the same region (NB, the grape sample from the 55 °C group after ten hours was imaged at 300× magnification instead of 400×). These images were saved for processing and labelled according to their drying conditions.
For example, sample H10T1_400 signifies the halved grape specimen that was dried for ten hours (H10) at Temperature 1 (T1) and imaged at 400× magnification (_400).
2.6. Image Processing Techniques
Image processing techniques were employed to automatically quantitate the microstructure found in each SEM image. Characteristics of the cell shape, including average area and perimeter of each sample, were obtained to enable relationships to be drawn between changes in the microstructure and drying times and temperatures. The first stage of the image processing was to identify regions of the image which signified cell walls and create an outline or boundary of these cells. Once this was established, analysis of the segmented/boundary image could be analysed to determine the mean dimensional characteristics of each sample. The image processing algorithm was required to omit holes and non-typical microstructures from the analysis without affecting the quality of the output. MATLAB was used in conjunction with the image processing toolbox to create the algorithm to analyse the images. Throughout the development of the image processing algorithms, the SEM image taken at 400 times magnification of the grape specimen was used as an example, as the 400× image, upon visual inspection of all the SEM images, provided a clearer and easier to analyse microstructure. Techniques were drawn from examples of image processing of cells in the literature and by expanding upon processing techniques outlined by Gonzalez and Woods in their 2009 text “Digital Image Processing using MATLAB” [24].
2.7. Procedure Followed for Determination of Outline of Cells
The first step after importing the chosen image into MATLAB was to convert it to a binary greyscale image. The size of this image was found and used to crop off the information panel at the bottom of the image.
2.8. Pre-Processing
A range of pre-processing measures or filters were applied to several SEM images with different appearances to note the effect. The three most effective of those attempted were the histogram equalisation, top hat and bottom hat filtering [24,30]. The histogram equalisation improved the visibility of the cell boundaries effectively in both very different contrasting SEM images. The direct impact of the final segmentation from the top and bottom hat filters was also confirmed by processing the final algorithm using each one, to detrimental effect. Thus, the histogram equalisation method was used to emphasise the cell boundary prior to further analysis.
2.9. Pyramid Reduction
The pyramid function was used to reduce the above-processed image to enable smoother and easier identification of the cell boundaries. The process reduced the image in both the horizontal and vertical dimensions by using a Gaussian-shaped weighting function [25]. This reduced the size of the image by half and enhanced salient image features. Pyramid reduction was conducted twice on the histogram-equalised image to reduce it to a quarter of the original size.
2.10. Cell Boundary Modelling
Three techniques were employed for this procedure.
2.10.1. Technique 1: Watershed Segmentation Method
The first technique used to model the cell boundaries in the SEM images was segmentation using the watershed function. The watershed function utilises the behaviour of water catchments in nature, which shows that a basin will drain water into the lowest/deepest point [24]. Figure 5 shows how catchment basins are formed around large depths and that watershed or ridge lines appear on the boundaries of these catchments.
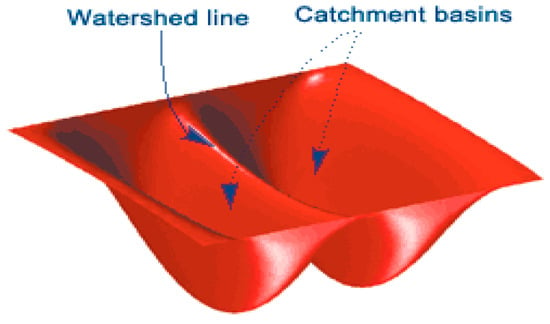
Figure 5.
Schematic of watershed theory [24].
This function works by interpreting the image gradient as heights. The image gradient is calculated by looking at the differential of the binary image. In this process, the minima of the gradient function are found and used as markers for the bottom of the basins. A threshold is used to exclude any ‘shallow’ basins which are not relevant to the image. Midway points between significant minima indicate ridge lines within the image, which segment the image into the important regions, in this case the grape cells.
There are several methods of using the watershed function, all of which were attempted in this process to determine the most effective process. Firstly, the gradient of the image was calculated by running a Sobel mask over the binary image and the derivative of the binary image and finding the squared average of the two. The watershed function of this gradient was then found. When this value was set to be logically equal to zero, the cell segments were shown [24].
To counteract this, some of the minima gradients used to segment the image were eliminated by using a threshold (h). This eliminated any ‘shallow’ basins which did not contribute strongly to the segmentation of the image. This was carried out by specifying a “height” which corresponded to the magnitude gradient, of which minima which fell below are not included in the analysis. Next, the distance each pixel is situated away from these minima (which meet the threshold requirements) was computed, and the watershed function was applied to it. When these values were logically equal to zero, the ridge lines, and thus the cell boundaries, were identified.
2.10.2. Technique 2: Edge Detection Methods
The second method of finding the cell boundaries incorporated edge detection procedures. Technique 2 comprised several steps to obtain a clear boundary of the grapes’ microstructure in the SEM images. These steps utilised the maxima found in the gradients in contrast to the minima, which was a focus of the watershed function. The Canny edge detection algorithm was chosen to base the cell outline on as it generates computer gradients in both the horizontal and vertical directions, which enabled the direction of the detected edge to be accounted for. It was the best method to optimise the signal-to-noise ratio of the SEM images as it is favoured over other edge detection methods in the literature [23].
Closing
The next step applied the closing function to the edges detected by the Canny function. The closing function used a structural element to dilate the existing lines and then erode the dilated image.
Skeleton
The skeleton function was used to link the closed image to boundary lines a single pixel width. The function found each point in the closed image and computed its closest neighbour. If there was more than one neighbour, then it belonged to the skeleton. Pixels belonging to the skeleton body are reduced until they are one pixel width, without breaking any objects apart. The pixels remaining make up the image skeleton. This option preserves Euler’s number.
Remove Small Regions
Spurs are the small regions of the skeleton which protrude from the boundary outline. If required, a function was used to remove any small regions of cells that were not fully connected to the main boundary/outline. This was to remove any accidental detection of lines that were too small to indicate cells. The function removes any group of pixels totally less than a set threshold (T).
2.10.3. Technique 3: Edge Detection Methods—Remove Inside Regions
An alternate to using the skeleton function and subsequent steps of Technique 2 was to remove the inside “white” regions of the closed image. This thinned the large cell boundaries from the inside of the white to the outside, the opposite of the skeleton function [24].
2.11. Determination of Morphological Properties
Boundaries obtained using Techniques 1 and 2 were used to quantify the microstructure of the SEM images. The first step in calculating the morphological properties (e.g., area, perimeter and axis length) was to invert the segmented or boundary image previously found using the two techniques. This was simply done by setting all zero-pixel values to a value of one and all one-value pixels to a zero value.
In order to remove any cells that were over segmented, the cell regions analysed were restricted by a minimum area. This value was adjusted for SEM images which by inspection were seen to obtain smaller cell sizes. In turn, cells with very large perimeters were also excluded as these were often cells that had been merged in the boundary detection stage due to unclear edges in the original SEM image. A computer code developed by the authors was used to perform the cell exclusion steps.
The mean microstructural characteristics were determined by the region props function, which calculated the properties of each cell included in the analysed boundary image. The mean of these outputs was calculated and scaled depending on what level magnification was used to capture the analysed image. The scale factor for each size was determined by determining the number of pixels for the scale bar shown in the origin image before it was cropped and the number of pixels for the original image. This was then divided by four to account for the multiple pyramid reductions.
3. Results and Discussion
3.1. Dehydration and Rehydration
Drying kinetics were calculated using the equation [31] (1961)
where Mi—moisture content of the sample (kg/kg db), Mo—initial moisture content (kg/kg db), Me—equilibrium moisture content (kg/kg db), k—drying constant (h−1), A—constant and t—time (h).
Me is the material equilibrium constant, which has been found to often have a negligible effect on the moisture ratio and was thus excluded from the calculation for simplicity [31].
The fitted curves in Figure 6 show the average of three replications with errors; there is a clear exponential relationship between the moisture ratio and the drying time of the grapes. This is consistent with the literature data, which confirmed that these grapes are suitable to be examined further to analyse the microstructural changes of dried grapes. Table 1 lists the drying coefficients of each temperature group modelled by the exponential curve and the goodness of fit indicator R-squared (signifies the perfect fit).
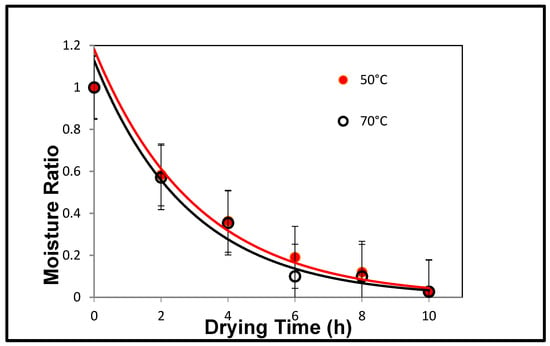
Figure 6.
Drying kinetics of grapes at two different temperatures.

Table 1.
Drying coefficients for both temperature ranges.
The initial stages of this investigation required measuring the moisture contents and producing the drying curves of the dehydrated grapes to confirm their suitability for further investigations. Figure 6 shows a plot of the exponential relationship between moisture ratio and drying time for both temperatures. This relationship was comparable to similar experiments found in literature searches [10]. In particular, the exponential curves fitted to the data had very high R–squared values (>0.9), indicating that the grapes could be modelled well to expected behaviours. Although it was expected that there was a larger difference between the drying curves of the two differing temperatures, the higher temperature did in general average higher drying rates than the lower range. The drying coefficients obtained, of 0.336 (Temp1) and 0.329 (Temp 2) measured in hours−1, are comparable to values in the literature when converted to similar units. It should be noted that although the drying constant for Temperature 1 is higher than Temperature 2 (as expected), it is only a difference of approximately 2% and thus not significant.
Following dehydration, the grape samples that were not preserved for image analysis were rehydrated. The percentage weight increase of the three grapes from each temperature and drying period group were averaged over the duration they were submerged in the hydrating solution. The results of this process are shown in Figure 7.
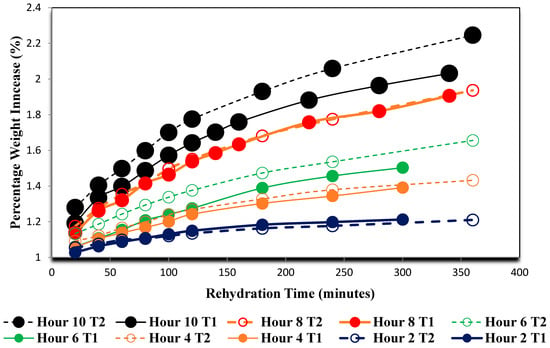
Figure 7.
Rehydration rate curves of grapes dried at 70 °C (T1) and 55 °C (T2).
The rehydration data shown in Figure 7 further demonstrate that there is a microstructural difference between the higher temperature grape samples and the lower temperature ones as grapes dried for the same duration at Temperature 1 (70 °C) almost always gained in weight percentage while submerged in the water bath at a slower rate than at Temperature 2 (55 °C).
3.2. Image Processing
Following dehydration, a halved grape sample from each drying period and temperature were imaged using the scanning electron microscope. The images taken at 70 °C drying at 2 h intervals are shown in Figure 8. Also, similar images were taken for 55 °C.

Figure 8.
Variation of microstructure at 70 °C drying in 2 h intervals.
Images were processed using the MATLAB algorithm previously developed to obtain quantitative values for mean area, perimeter, mean maximum and minimum axis length and a value of eccentricity. Two techniques for obtaining these values were used, where Technique 1 utilised the watershed segmentation method, as shown in Figure 9, for different thresholds for 70 °C with a magnification of ×400; Technique 2 used Canny edge detection with several trials to ensure the selection of the best threshold values. Figure 10 shows some of the results altering the thresholds using Canny algorithms and the selected final algorithm for further morphological transformations. Each sample was analysed individually, and the algorithm was altered slightly to negate the effect of over-segmentation (by removing areas that were below a minimum threshold) and to remove most of the cells which were combined and cut off on the edge of the image (by removing cells with a perimeter over the set threshold). Figure 11 shows the cell regions superimposed onto each SEM image by two methods: the thresholds used and the resulting cells, which were analysed to obtain the morphological properties.
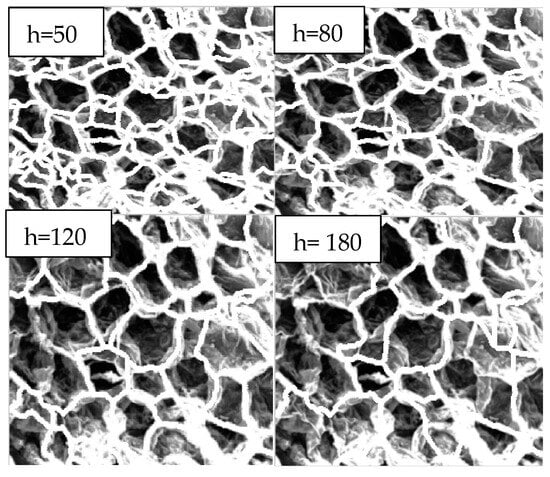
Figure 9.
Watershed segmentation at various thresholds (h) for 70 °C, 10_400. Chosen threshold h = 120.
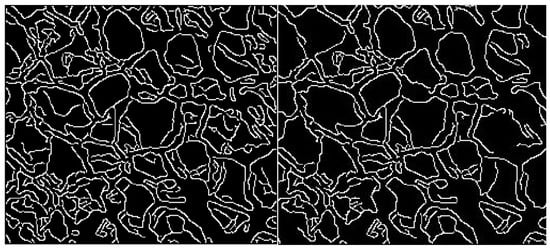
Figure 10.
Canny edge detector with varying thresholds applied.
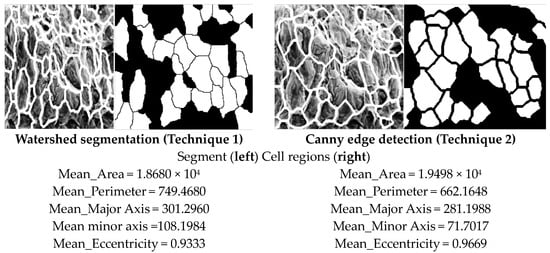
Figure 11.
Cell regions superimposed onto each SEM image by two methods.
The image processing stage also accounted for a lot of the unexpected results. Although the image processing techniques worked very well for most specimens, there were a few images where accurate cell boundaries were not able to be properly defined. This was due to a range of problems. The first issue was that the SEM image was too difficult to analyse. This was often due to the specimen itself. As the SEM captures an almost three-dimensional topographic image of the surface, the sample is very uneven; if physically damaged, it is difficult for the edges to stand out when compared to other parts of the cell (including the inside surface), which may be closer to the SEM focus point. In addition, if the gold sputter coating method did not successfully reach all areas of the specimen, then some key features would not have been captured properly. Although this did not seem to be an obvious problem, the effects may have been subtle. Although due care was taken when the SEM was positioned on each sample, some were small and damaged, and it was difficult to find a part of the grape mesocarp (centralized body cells) to entirely fill in the 250× magnification frame. Thus, some of the smaller cells on the edge of the grape berry might have been captured and it would not be a true reflection of the mean cell size of a typical grape specimen after being dried for the same period.
There are also clear problems with the image processing method. Firstly, it is difficult to use almost the same algorithm to analyse images with shapes that are different sizes and shapes. The contrast and highlights in the images also change dramatically between samples. Although the histogram equalisation step tried to account for this, it was not always successful. The biggest problem seen with obtaining the boundary outline was with grape samples with either very large or very small cell sizes. It was inherently difficult to find the outline of the some cells as they are very large and nearly every cell intersects the boundary of the image and cannot be modelled fully. In contrast, the very small cells were not able to be closely referenced by the algorithm at all.
It was, however, clear that the segmentation method using the watershed function was more successful than the edge detection method at mimicking the microstructure shapes, as shown when compared in Figure 12; all three techniques were also better at analysing the 400× magnification images, which was expected as it was designed to read those size images first.
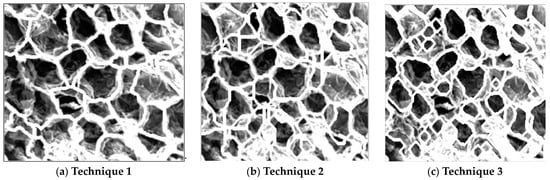
Figure 12.
Superimposed cell boundary outlines.
The method for measuring the properties of the cells, once they had been identified by the boundary finding techniques, was quite successful. Several sources found in the literature used very loose methods for averaging cell size by approximating the number of pixels which were allocated to the perimeter, and which were used to denote the area of the cell. In this case, each cell was identified and measured separately. The only inaccuracy of this stage of the process was from poor extraction of the segmented image. In particular, the constraints applied to minimise the cell area and limit the cell perimeter included in the analysis improved the boundary outline by the illuminated cells located in error. The parameters were changed as required, when the images of some cells were examined in addition to the cells selected comparing other images.
3.3. Microstructural Changes and Determination of Morphological Properties
Cell boundaries obtained by Techniques 1 and 2 were used to quantify the microstructure of the images. Before the calculation of the properties, it was necessary to invert the segmented or boundary images. Figure 13 shows the cell detection process for microstructural analysis.
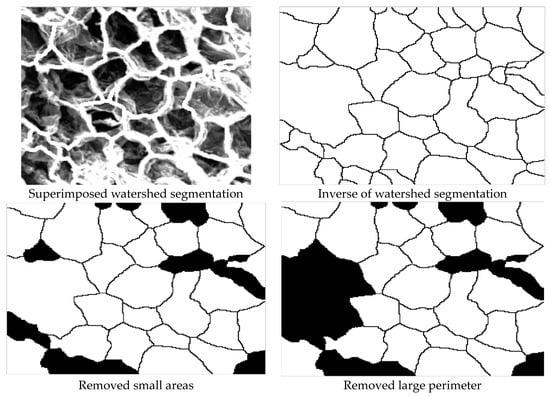
Figure 13.
Cell detection process for microstructural analysis.
Figure 14 shows the mean area that was calculated by summing the pixels within each separate region and multiplying them by a scaling factor to obtain metric units. Each of these values was averaged for each SEM image. The mean perimeter was calculated using a similar method where the number pixels along each boundary was converted to micrometres and graphed.
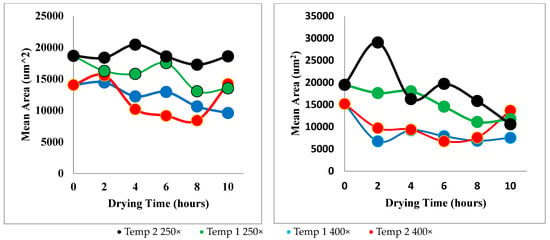
Figure 14.
Mean area versus drying time. Technique 1 (left), Technique 2 (right).
Unfortunately, the microstructural properties obtained from the SEM images did not clearly show many trends when compared with drying time and temperature. When the results of each property versus drying time are compared between the two techniques, it is clear that Technique 1 gives results with less unexpected or outlying data points. It is also noticeable that the outlying data points are usually part of the data set taken from images with a 250× magnification.
3.4. Area
In spite of this, there are some trends that are visible. Figure 14 reveals that the mean cell area did, in general, decrease with drying time. This is consistent with known trends, as shrinkage of the grape berry on the macro scale is expected to cause shrinkage in the microstructure. Also, samples dried at Temperature 1 (70 °C) seemed to have, on average, a lower average area than samples dried at Temperature 2 (55 °C). This confirms the theory that increased temperature affects the microstructure more quickly. The data obtained using Technique 2 appeared more variable than with Technique 1, further emphasising the superiority of the watershed segmentation method.
3.5. Perimeter
The data obtained from the image analysis for the perimeter was not very successful. It was clear that, on average, for both techniques used to analyse the images, the perimeter decreased with drying time. However, there does not seem to be any clear trend that the perimeter measurements for the Temperature 1 samples were on average less than those for the Temperature 2 samples, which was an expected outcome. In contrast to the other measured parameters, Technique 2 seemed better at modelling these relationships; both 400× magnification data followed similar trends, and both magnification levels of the Temperature 1 data seem to be aligned.
3.6. Axis Length
Mean axis length showed some very general trends, which were supported by findings in the literature. As the drying time increased, the mean axis length (in particular the minimum decreased). For the Temperature 1 samples, both the 400× and the 250× axis lengths seemed to follow a similar pattern, which positively indicated that the image processing correctly analysed those specimens. It was not always clear that the size of the axis length data for Temperature 1 was less than Temperature 2, although it did seem to maintain this relationship better in the maximum length data.
3.7. Eccentricity
Eccentricity versus time did not indicate any trends. It reinforced that the outlying values are more likely to appear in graphs recording data obtained using Technique 2. It was clear, though, that it is possible for the shape of the grape cells to vary greatly, as some eccentricities were recorded as low as 0.5, which when compared to the original image to confirm this value, it was clear the variance did truly exist (see H2T1_250 with e = 0.5540 and H0T1_250 with e = 0.9669).
When we look back at the dehydration and rehydration curve to see if there are any unexpected values, we see that H6T1, on average, did not fit within the fitted drying curve. When the raw data was examined, the moisture content of grape two (which was retained for imaging) was compared to the other three specimens to see if with any data group there was a clear variance. The only unusual value was H8T2, which had a significantly higher moisture content than the other three grapes, which may indicate that it was not dried sufficiently, which suggests the cells were more intact, and thus they had retained their larger size. These observations could account for some of the unexpected variance in the results.
The Results of Image Processing
Table 1 provide the morphological parameters of cells during drying periods using the watershed management technique. Canny edge detection data is shown in the In Table 2, A signifies threshold cell areas that were restricted by (pixels), P signifies threshold cell perimeters that were restricted by (pixels) at different magnifications. Table 3 provides the morphological parameters from the Canny edge detection technique.

Table 2.
Morphological properties from the watershed management technique.

Table 3.
Morphological properties from the Canny edge detection technique.
4. Conclusions
Major relationships between cell size and drying time and temperature were confirmed; the variance in the data did not allow any further in-depth analysis of the results or any new findings. The moisture ratio change to time during drying confirmed the shrinkage of the grape specimens over longer drying periods, and that a higher drying temperature generally increased moisture loss and the subsequent ability to rehydrate as successfully as their cooler counterparts. The images were analysed using image processing techniques established in this project. Two main techniques were established to outline the cells in the grapes’ microstructure recorded in each scanning electron microscope image. Segmentation of the cells using the watershed function was more successful and consistent at extracting the morphological shape than the use of the Canny edge detector, closing and skeletisation methods. From the obtained cell boundaries, quantitative data were extracted on mean grape cell area, perimeter and axis length for each sample. It was found that over the drying time, the cell area and perimeter were reduced as expected. Furthermore, in general, the higher temperature samples had smaller microstructural elements. Further work in the future could progress to develop a more accurate image processing algorithm and obtain further SEM images to analyse so that anomalies are removed from the results. It is clear from viewing the results that the techniques used modelled the cell microstructure very well. If more SEM images could be obtained, then the data could be averaged, or actual anomalies in the data could be excluded to create more reliable results.
The development of automatic image processing techniques will enable quantitative data to be extracted from these images.
Author Contributions
Conceptualization and methodology, J.B. and K.B.; Writing—original draft, W.S.; Writing—review & editing, W.S. and G.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data are contained within the article.
Acknowledgments
The authors acknowledge the Queensland University of Technology for their support in providing laboratory access to use their laboratory facilities to conduct this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Priyadarshini, A.; Rajauria, G.; O’Donnell, C.P.; Tiwari, B.K. Emerging food processing technologies and factors impacting their industrial adoption. Crit. Rev. Food Sci. Nutr. 2019, 59, 3082–3101. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, J.M. Drying and Dried Products under the Microscope. Food Sci. Technol. 2003, 9, 137–143. [Google Scholar] [CrossRef]
- Khan, M.I.H.; Nagy, S.A.; Karim, M.A. Transport of cellular water during drying: An understanding of cell rupturing mechanism in apple tissue. Food Res. Int. 2018, 105, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.K.; Singh, S.V. Osmotic dehydration of fruits and vegetables: A review. J. Food Sci. Technol. 2014, 51, 1654–1673. [Google Scholar] [CrossRef] [PubMed]
- García, A.H. Anhydrobiosis in bacteria: From physiology to applications. J. Biosci. 2011, 36, 939–950. [Google Scholar] [CrossRef] [PubMed]
- Ramos, I.N.; Silvaa, C.; Serenob, A.M.; Aguilera, J.M. Quantification of microstructural changes during first stage air drying of grape tissue. J. Food Eng. 2004, 62, 159–164. [Google Scholar] [CrossRef]
- Senadeera, W.; Banks, J. Analysis of Micro-Structural Changes and Measurement of their Parameters of a Food Material. In Proceedings of the 5th Nordic Drying Conference (NDC 2011), Helsinki, Finland, 19–21 June 2011. [Google Scholar]
- Niamnuy, C.; Devahastin, S.; Soponronnarit, S. Some recent advances in microstructural modification and monitoring of foods during drying: A review. J. Food Eng. 2014, 123, 148–156. [Google Scholar] [CrossRef]
- Nguyen, T.K.; Mondor, M.; Ratti, C. Shrinkage of cellular food during air drying. J. Food Eng. 2018, 230, 8–17. [Google Scholar] [CrossRef]
- Sawhney, R.; Pangavhane, D.; Sarsavadia, P. Drying Studies of Single Layer Thompson Seedless Grapes. In Proceedings of the International Solar Food Processing Conference, Indore, India, 14–16 January 2009. [Google Scholar]
- Khodaei, J.; Akhijahani, H. Some Physical Properties of Rasa Grape (Vitis vinifera L.). World Appl. Sci. J. 2012, 18, 818–825. [Google Scholar]
- Banks, J.; Senadeera, W. Measurement of Structural Changes to a Food Material during Dehydration. In Proceedings of the 4th International Conference on Computational Methods, Good Coast, Australia, 25–28 November 2012. [Google Scholar]
- Aguilera, J.M. Rational food design and food microstructure. Trends Food Sci. Technol. 2022, 122, 256–264. [Google Scholar] [CrossRef]
- Tepe, T.K.; Tepe, B. The comparison of drying and rehydration characteristics of intermittent-microwave and hot-air dried-apple slices. Heat Mass Transf. 2020, 56, 3047–3057. [Google Scholar] [CrossRef]
- Lopez-Quiroga, E.; Prosapio, V.; Fryer, P.J.; Norton, I.T.; Bakalis, S. Model discrimination for drying and rehydration kinetics of freeze-dried tomatoes. J. Food Process Eng. 2020, 43, e13192. [Google Scholar] [CrossRef]
- Tortoe, C.; Orchard, J. Microsturctual changes of osmotically dehydrated tissuies of apple, banana, and potato. Scanning 2006, 28, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Australian Microscopy and Microanalysis Research Facility. MyScope: SEM. 2013. Available online: http://www.ammrf.org.au/myscope/sem/background/ (accessed on 8 April 2013).
- Georget, D.M.R.; Smith, A.C.; Waldron, K.W. Thermal transitions in freeze-dried carrot and its cell wall components. Thermochim. Acta 1999, 332, 203–210. [Google Scholar] [CrossRef]
- Lewicki, P.P.; Pawlaka, G. Effect of mode of drying on microstructure of potato. Dry. Technol. 2005, 23, 847–869. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, M.; Mujumdar, A.; Lim, R. Analysis of Temperature Distribution and SEM Images of Microwave Freeze Drying Banana Chips. Food Bioprocess Technol. 2013, 6, 1144–1152. [Google Scholar] [CrossRef]
- Karunasena, H.C.P.; Hesami, P.; Senadeera, W.; Gu, Y.; Brown, R.J.; Oloyede, A. Scanning electron microscopic study of microstructure of gala apples during hot air drying. Dry. Technol. 2014, 32, 455–468. [Google Scholar] [CrossRef]
- Favaa, J.; Hodarac, K.; Nietob, A.; Guerrerob, S.; Alzamorab, S.M.; Castro, M.A. Structure (micro, ultra, nano), color and mechanical properties of Vitis labrusca L. (grape berry) fruitstreated by hydrogen peroxide, UV–C irradiation and ultrasound. Food Res. Int. 2011, 44, 2938–2948. [Google Scholar] [CrossRef]
- Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, 8, 679–698. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.; Meschino, G.; Ballarin, V. Solving the over segmentation problem in applications of Watershed Transform. J. Biomed. Graph. Comput. 2013, 3, 29–40. [Google Scholar] [CrossRef]
- Adellson, B. The Laplacian Pyramid as a Compact Image Code. IEEE Trans. Commun. 1983, 31, 532–540. [Google Scholar]
- Zhou, R.W.; Quek, Q.; Ng, G.S. A novel single-pass thinning algorithm and an effective set of performance criteria. Pattern Recognit. Lett. 1995, 16, 1267–1275. [Google Scholar] [CrossRef]
- Fisher, R.; Perkins, S.; Walker, A.; Wolfart, E. Thinning. 2003. Available online: http://homepages.inf.ed.ac.uk/rbf/HIPR2/thin.htm (accessed on 20 April 2023).
- Forero, M.; Hidalgo, A. Image Processing Methods for Automatic Cell Counting In Vivo or In Situ Using 3D Confocal Microscopy. In Advanced Biomedical Engineering; Gargiulo, G., Ed.; InTech: Croatia, Poland, 2011; pp. 183–204. Available online: http://www.intechopen.com/books/advanced-biomedicalengineering/image-processing-methods-for-automatic-cell-counting-in-vivo-or-in-situ-using-3d-confocalmicroscopy (accessed on 10 January 2024).
- Farhan, M.; Ruusuvuori, P.; Emmenlauer, M.; Ramo, P.; Dehio, C.; Yli-Harja, O. Multi-scale Gaussian representation and outline-learning based cell image segmentation. BMC Bioinform. 2013, 14, S6. Available online: http://www.biomedcentral.com/1471-2105/14/S10/S6 (accessed on 12 January 2023). [CrossRef] [PubMed]
- Margaritis, E.; Jones, M. Beyond cereals: Crop processing and Vitis vinifera L. Ethnography, experiment and charred grape remains from Hellenistic Greece. J. Archeol. Sci. 2006, 33, 784–805. [Google Scholar] [CrossRef]
- Adiletta, G.; Russo, P.; Senadeera, W.; Di Matteo, M. Drying characteristics and quality of grape under physical pretreatment. J. Food Eng. 2016, 172, 9–18. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).