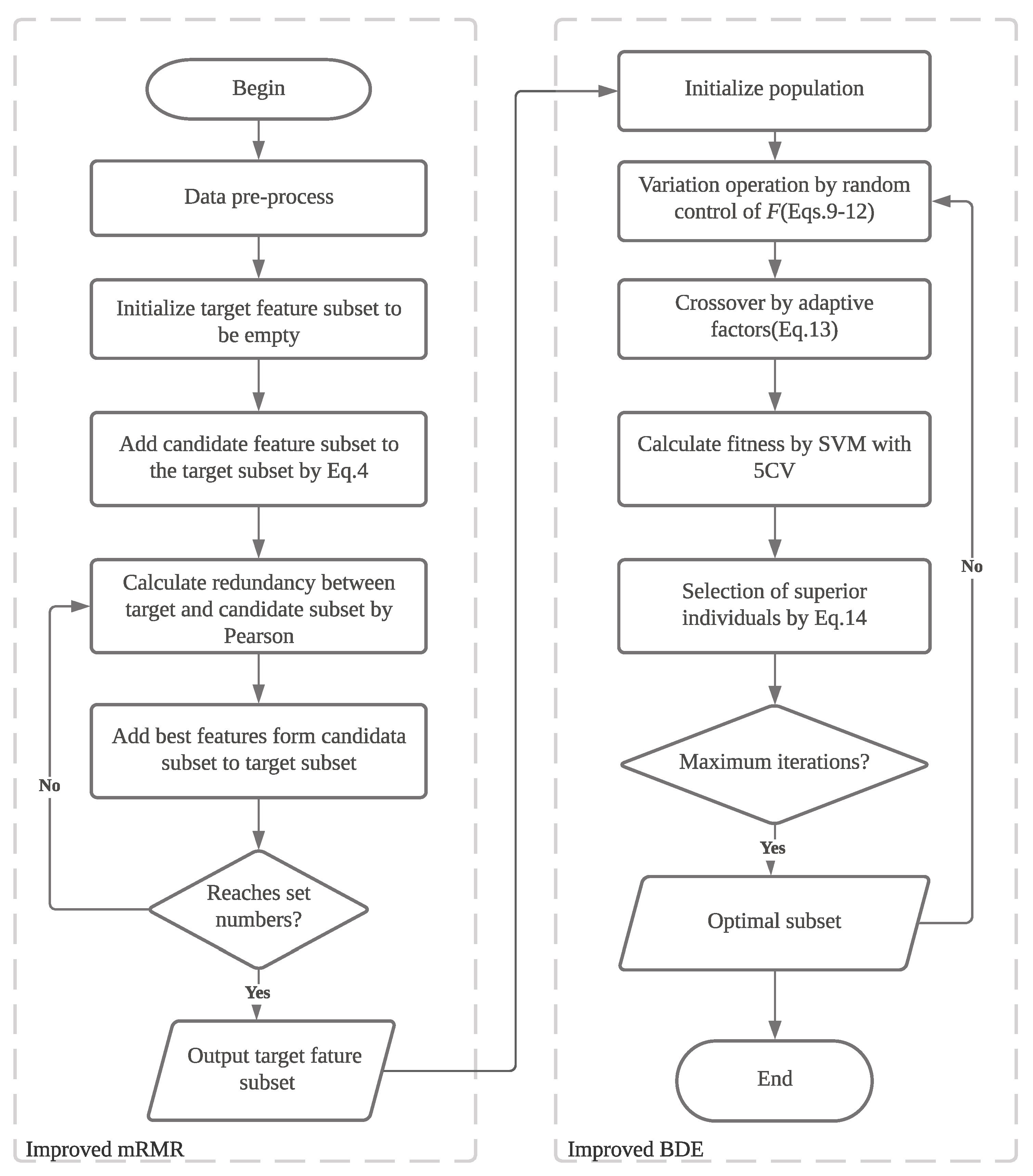
3.1. Overall Framework of the Proposed Method
Most of the previous research on feature selection in microarray data analysis has overlooked the continuity of attributes, leading to the need for discretization methods such as information entropy-based approaches to obtain satisfactory results. Additionally, the interdependence among genes, which is a crucial aspect of microarray data, has often been neglected.
To address these limitations, we propose the MBDE algorithm. This approach utilizes the mRMR method to capture the correlations between features and updates the quantization function to handle continuous attributes. Furthermore, the BDE method is employed for fine-scale feature selection. The general workflow of the proposed method is illustrated in
Figure 1. It can be divided into three stages.
In the first stage, the data are preprocessed, which includes handling missing values and performing data normalization. In the second stage, an improved mRMR algorithm is employed for initial feature filtering, resulting in the retention of 500 features in our experimental setup. Finally, in the third stage, the improved BDE algorithm is utilized for further feature selection, ultimately outputting the best subset of features. By integrating these stages, the proposed method aims to effectively address the challenges of feature selection in microarray data analysis, considering both the attribute continuity and the interdependence among genes.
3.2. Stage One: Preprocessing Method
The original dataset contains outliers and missing values, which can negatively impact the data quality and subsequent analysis. To address this issue, we apply the principle to identify outliers in the data. Specifically, we consider data points outside the range of as outliers, where represents the mean and represents the standard deviation.
To handle both outliers and missing values, we employ the K-nearest neighbors (KNN) imputation method. This approach fills in the missing values and replaces outliers with values derived from neighboring data points. By using KNN, we can ensure that the imputed values are representative of the local data distribution. Furthermore, we apply a logarithmic transformation to all expressed data. This transformation helps to reveal data relationships more effectively and facilitates better statistical inference.
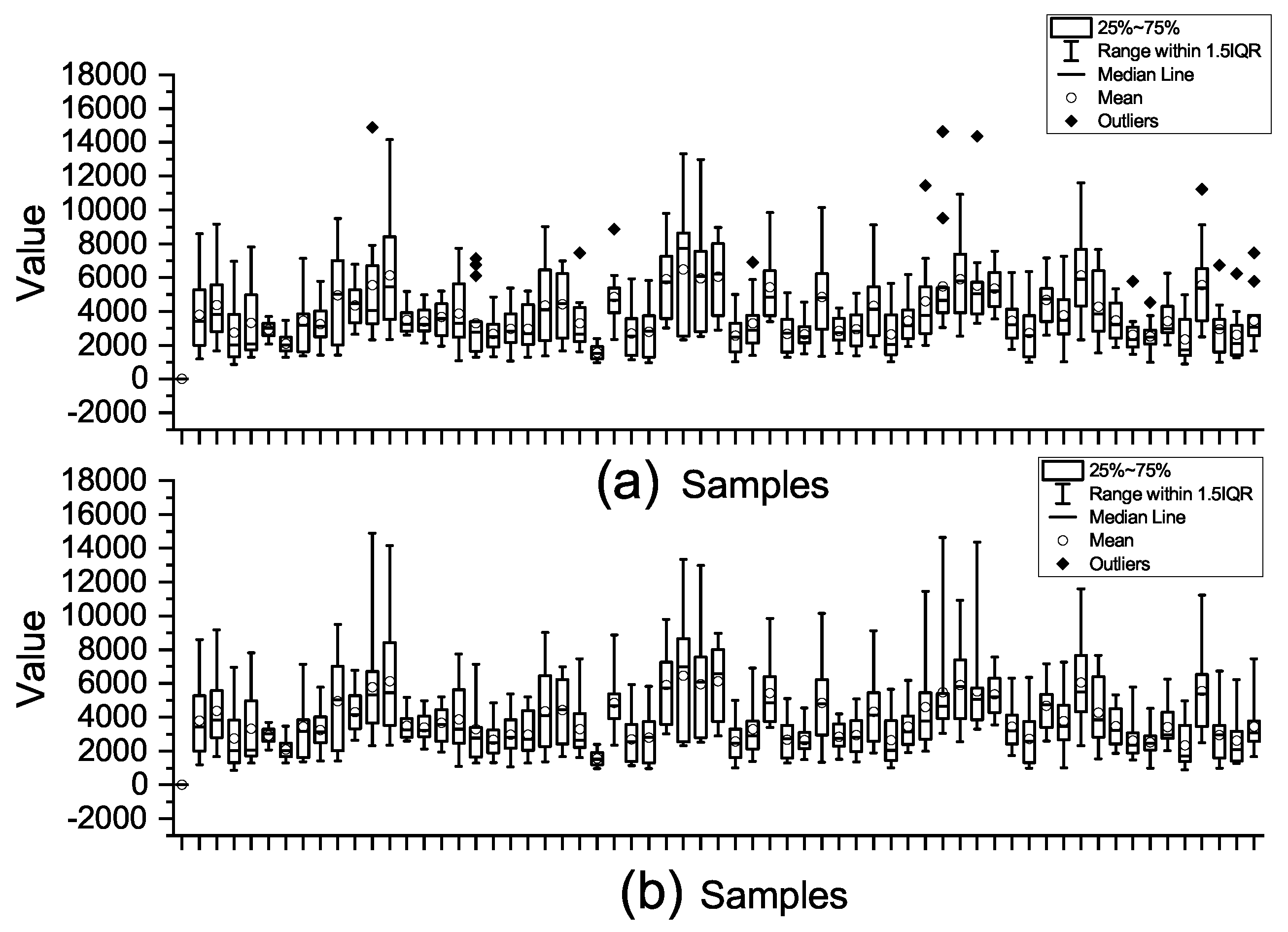
To illustrate the effect of the preprocessing steps,
Figure 2 shows the impact of outlier and missing value processing by using the Colon dataset as an example. As observed, the preprocessing steps effectively remove outliers and prepare the dataset for subsequent analysis and tasks.
3.3. Stage Two: Improved mRMR Algorithm
In the context of microarray data analysis, the dimensionality of the data is typically high. Applying the wrapper method directly to select features would result in a significant increase in the algorithm complexity. Therefore, it is common to use a filter method for coarse-scale feature selection initially. One effective filtering feature-selection method is the Minimum Redundancy Maximum Relevance (mRMR) algorithm. The mRMR algorithm is an incremental search algorithm that aims to select features with the highest correlation to the target variable while minimizing redundancy with the already-selected features.
Traditionally, the mRMR algorithm employs two objectives for feature selection: maximizing the relevance between the features and the target variable, and minimizing the redundancy among the selected features. These objectives are mathematically described by Equation (
1) and Equation (
2), respectively:
In Equations (
1) and (
2),
S represents the feature subset,
C represents the label variable, and
and
represent features within the feature subset
S. The term
denotes the correlation between the target feature subset
S and the label
C, while
represents the redundancy within the feature subset
S.
In the traditional mRMR algorithm, the correlation and redundancy are quantitatively calculated by using mutual information. The mutual information between two variables
X and
Y is mathematically represented by Equation (
3):
In Equation (
3),
represents the joint probability distribution of the random variables
X and
Y, while
and
represent their respective marginal probability distributions.
The traditional mRMR algorithm incorporates the two objective functions, Equations (
1) and (
2), into the feature-selection process. There are two common methods of integration: subtractive integration and divisive integration. This paper adopts the subtractive integration approach.
While the traditional mRMR algorithm utilizes mutual information to quantify the relationships between features and between features and labels, applying mutual information to microarray data poses challenges, as it is more suitable for data with continuous attributes and may require expert guidance for the binning operation. In contrast, the
t-test and Pearson correlation coefficient are not subject to such limitations and have demonstrated superior performance in feature-selection tasks for microarray data [
23]. The equations for the
t-test and Pearson correlation coefficient are described in Equation (
4) and Equation (
5), respectively:
In Equation (
4),
represents the mean value of feature
in the positive samples, while
represents the mean value of feature
in the negative samples.
and
denote the variances of feature
in the positive and negative samples, respectively.
and
represent the number of samples in the positive and negative classes, respectively.
Considering a dataset with all features denoted as
F and the subset of features to be selected as
, the search process of mRMR involves iteratively selecting the optimal features from the candidate feature subset
and adding them to
. The selection conditions for the optimal features can be obtained by using Equation (
6):
The original mRMR algorithm utilizes a predetermined number of features as the stopping criterion for the feature subset search. However, this criterion is determined empirically and may not guarantee optimal performance for the final feature subset selection. In this paper, we propose a modified stopping criterion for mRMR based on the average classification accuracy (Acc) of the classification model by using five-fold cross-validation on the current feature subset. The calculation of Acc is given by Equation (
7), where
represents the number of true positive samples that are correctly predicted;
represents the number of true negative samples that are correctly predicted; and
and
represent the number of false negative and false positive predictions, respectively:
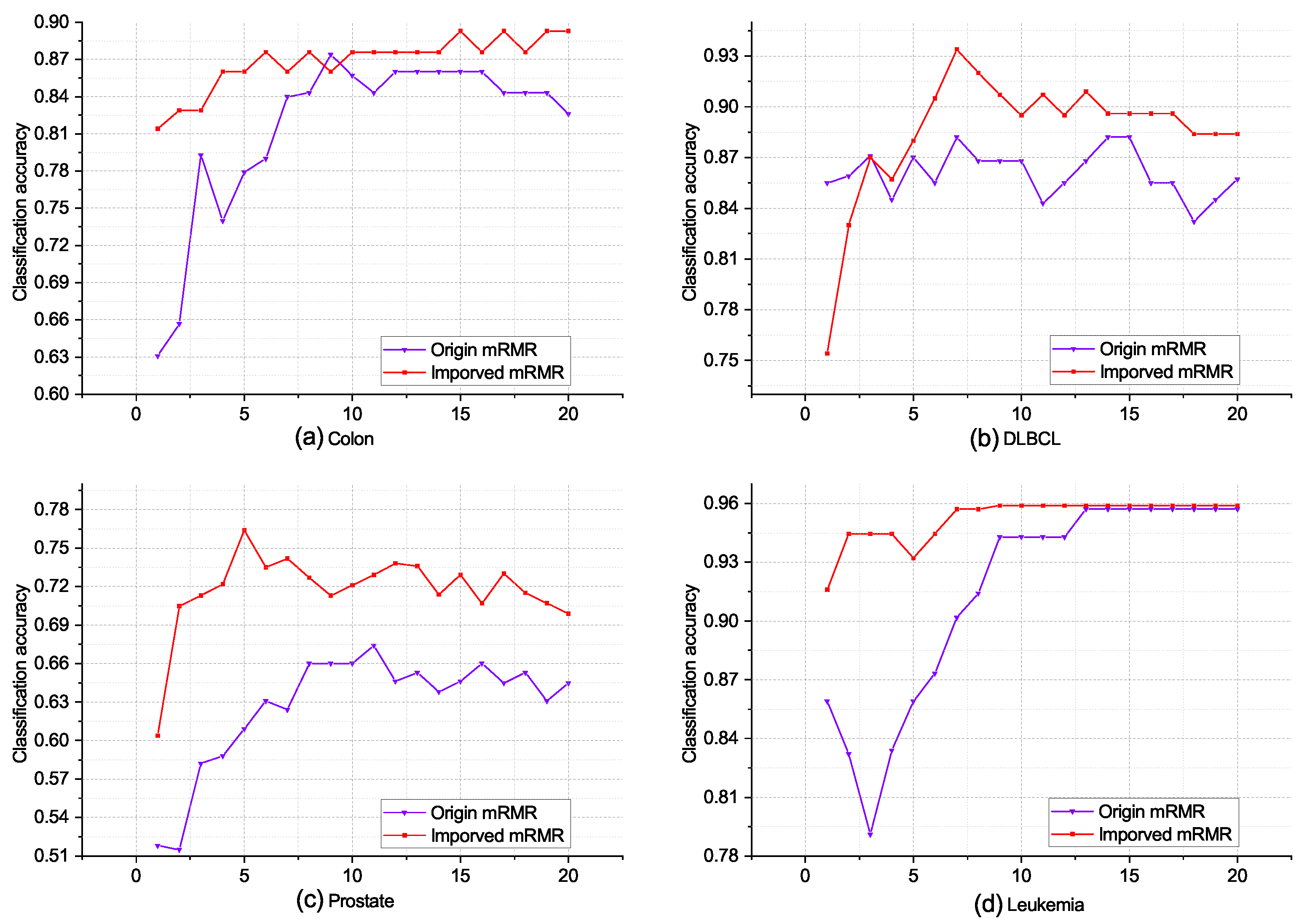
In this paper, we introduce a modified stopping criterion for the incremental search process in the mRMR algorithm. The criterion is considered to be met in the current case if either the classification accuracy of the classification model reaches 100% during the total search process or if there is no improvement observed for k consecutive iterations. This criterion ensures that the search process is terminated when the algorithm achieves an optimal classification performance or when further iterations do not yield significant improvements.
Moreover, the improved mRMR algorithm incorporates a quantization function suitable for continuous data to evaluate the correlation and redundancy between features. By applying this algorithm to microarray data, we can effectively filter out redundant, irrelevant, or weakly correlated genes. As a result, we obtain a subset of candidate biomarkers from the complete set of genes, which enhances the efficiency and relevance of the feature-selection process for microarray data analysis.
3.4. Stage Three: Improved BDE Algorithm
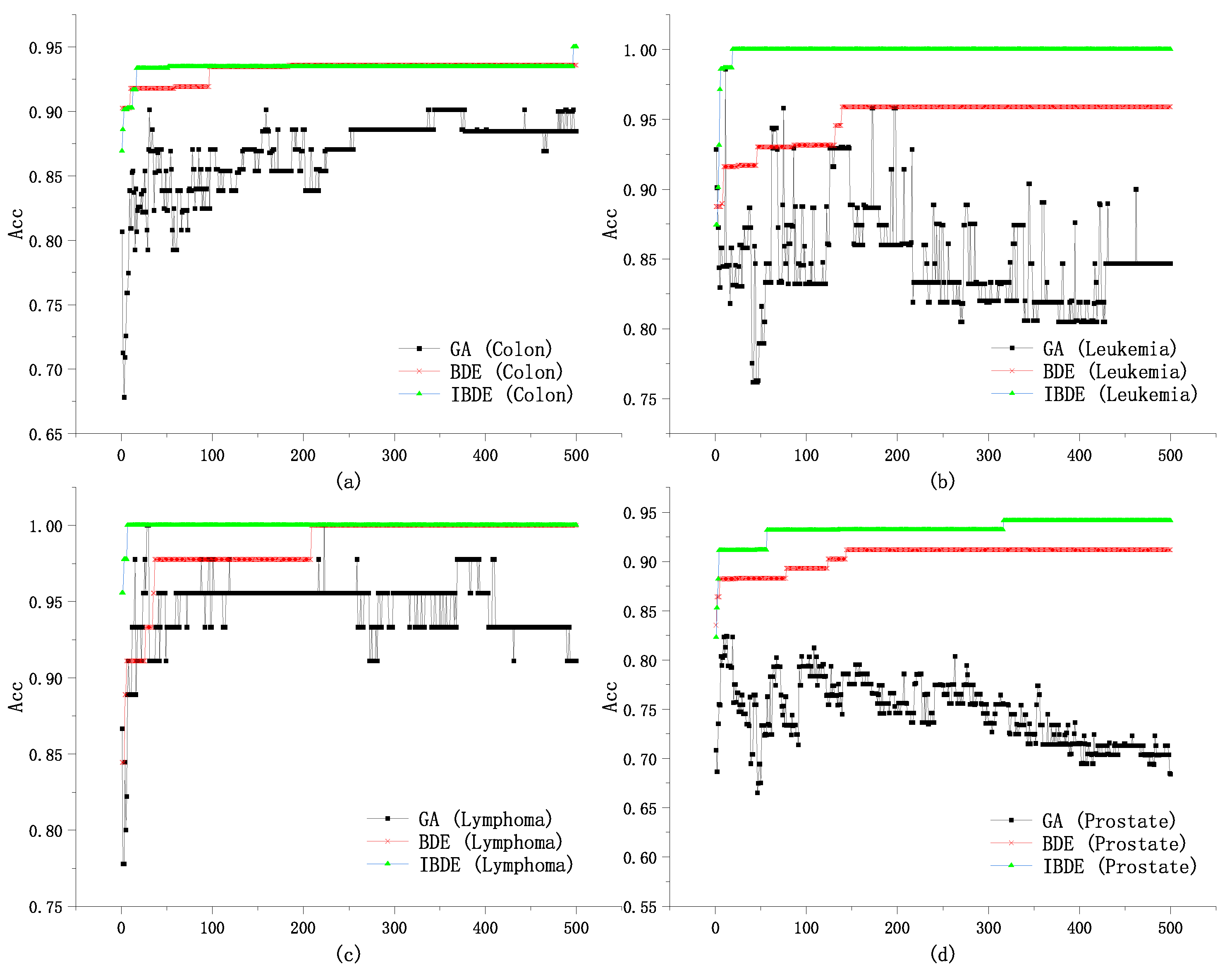
The proposed approach in this subsection introduces an improved binary differential evolution (DE) algorithm specifically designed for feature selection in microarray data analysis. DE is a widely utilized adaptive global optimization algorithm known for its simplicity, ease of implementation, rapid convergence, and robustness. It has found applications in various domains including data mining, pattern recognition, and artificial neural networks.
In our work, we enhance the DE algorithm by incorporating a new binary quantization method and scaling factor. This extension aims to enhance the algorithm’s exploration capability in the initial stage, thus improving population diversity. Additionally, we ensure the algorithm’s exploitation capability in the subsequent stage to exploit local advantages. To achieve this, we introduce an adaptive crossover operator, which not only accelerates the convergence speed in the initial stage but also maintains the algorithm’s exploitation capability in later stages.
The improved binary differential evolution algorithm presented in this paper addresses the challenges of feature-selection in microarray data by effectively balancing exploration and exploitation. This enhancement allows for the more efficient and accurate identification of relevant features for microarray analysis, contributing to the overall performance and effectiveness of the feature-selection process.
The traditional binary differential evolution algorithm calculates the variance vector
through the evolution process, as described in Equation (
8). In this equation, three individuals
,
, and
are randomly chosen from the population, with the condition
:
However, this direct manipulation of the binary strings in the traditional approach does not effectively emulate the behavior of the continuous differential evolution algorithm. Consequently, it exhibits suboptimal performance, particularly in scenarios involving high-dimensional data [
24].
Therefore, in the improved binary difference evolution algorithm, we use the vector
to represent the
j-th binary code of the final mutation vector, which is shown as Equation (
9):
In Equation (
9), the value of
is calculated according to Equation (
10) to ensure that the vector approximation after binary quantization falls within the range of 0 to 1. The proposed binary quantization method was inspired by [
25]. In this method, when
F is set to 0.5,
is approximately 0.462, and when
F is set to 1,
is approximately 0.762. This implies that in Equation (
9), even if
F is 1, it will not cause
to exceed the value of rand (0, 1), thereby reducing the likelihood of selecting the
j-th feature. As a result, the number of features is effectively reduced:
The
represents the
j-th dimension binary code of the difference vector, which is calculated in Equation (
11):
The scaling factor F plays a crucial role in balancing exploration and exploitation in the improved BDE algorithm. Increasing the value of F helps expand the search range and enhance population diversity, thereby promoting exploration. On the other hand, decreasing the value of F improves the exploitation ability and accelerates convergence, but may lead to premature convergence.
In the improved BDE algorithm, the value of
F is determined based on Equation (
12), where
g represents the current iteration number and
G represents the total number of iterations. By incorporating the iteration information, the scaling factor
F dynamically adjusts over the course of the algorithm. Moreover, in the selection of
, we ensure that
and
are not equal, preserving the element of randomness. This improved scaling factor strategy effectively strikes a balance between exploration and exploitation in the algorithm:
The crossover process plays a crucial role in maintaining population diversity in the improved BDE algorithm. The improved adaptive crossover operator is computed according to Equation (
13), where
represents a parameter that will be further discussed in the experimental section. The final selection operator is determined based on Equation (
14). In our method, we utilize the support vector machine (SVM) as the model to calculate the fitness function.
The adaptive crossover operator, as described in Equation (
13), adjusts the crossover probability based on the fitness value of the individual. This allows individuals with higher fitness values to have a higher probability of undergoing crossover, while individuals with lower fitness values have a lower probability. By adaptively adjusting the crossover probability, the algorithm can effectively balance exploration and exploitation, promoting the convergence of the population toward better solutions.
In our method, the fitness function is evaluated by using the support vector machine (SVM) model. The SVM is a popular and effective classifier that can distinguish between positive and negative samples based on the selected features. The fitness function quantifies the classification accuracy of the SVM model, guiding the search process toward selecting features that contribute to a better classification performance:
where
is a new individual and
is the average classification accuracy of SVM five-fold cross-validation.