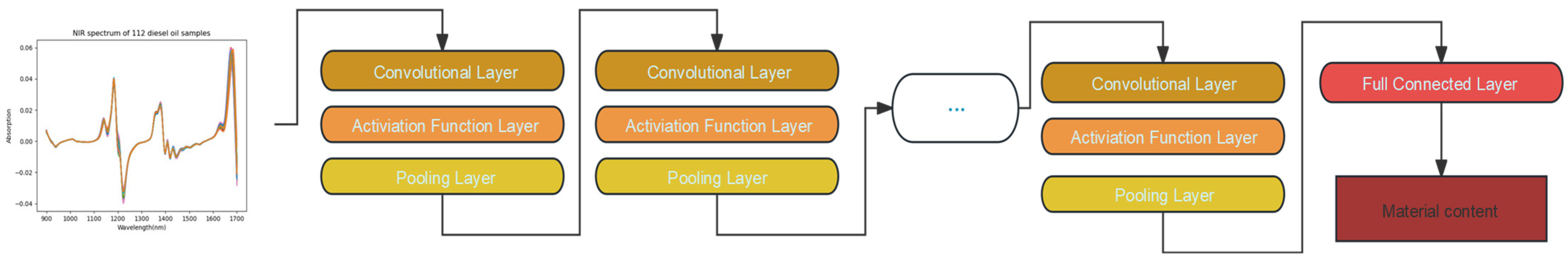
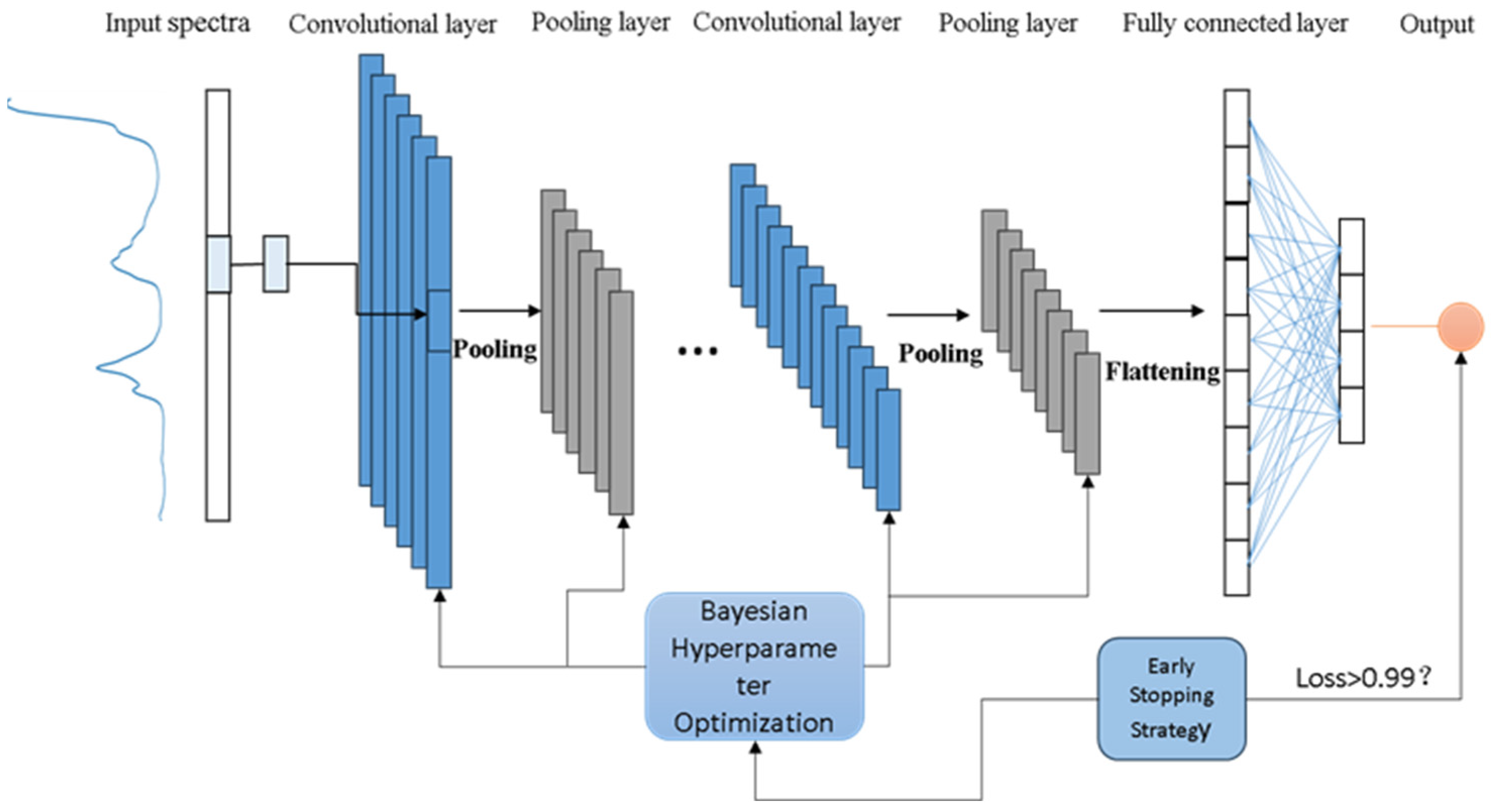
1. Introduction
Advancements in chemometrics and instrumental measurement techniques have increasingly highlighted the application of near-infrared (NIR) spectroscopy analysis in scientific research and industrial applications because of its nondestructive, rapid, and precise nature. Its widespread application spans various sectors, including food science [
1], environmental chemistry [
2], daily chemical industry [
3], traditional Chinese medicine [
4], polymer materials [
5], petroleum, and biochemistry [
6].
The primary objective of NIR spectroscopic analysis is to establish quantitative analytical models that describe the relationship between NIR spectra and target components [
7]. NIR spectroscopy (NIRS)—a nondestructive, swift, and accurate analytical technique—has made significant strides in recent decades, presenting a broad spectrum of future applications.
Traditional NIR spectroscopy analysis methods include principal component analysis (PCA), principal component regression (PCR), partial least squares (PLS), partial least squares regression (PLSR), and support vector machine regression (SVR). PCA, introduced in the 1930s, is a traditional spectral preprocessing method that transforms basic vectors to reduce data dimensionality. Aguiar et al. [
8] effectively differentiated soil samples from various regions of Brazil using PCA and demonstrated the value of NIRS in rapid soil analysis. However, PCA is limited in its ability to directly consider the relationship between independent and dependent variables. In the 1960s, Herman Wold introduced PCR, which transforms the original independent variables into a new set of principal components for model construction, effectively addressing the shortcomings of traditional PCA. Stacey et al. [
9] employed PCR and Fourier-transform infrared spectroscopy to measure the properties of respirable quartz, kaolinite, and coal in various mineral samples from different countries. Nonetheless, PCR considers only the variance of the explanatory variables without factoring in their relationships with the response variables.
In the 1970s, the introduction of PLS shifted the focus to not only the relationships between predictive variables but also their correlation with response variables. Wang et al. [
10] combined three distinct PLS algorithms with NIRS for rapid quantification of polyphenols in dark tea. However, in practical applications, PLS is limited to linear relationships, potentially failing to capture the complex nonlinear relationships present in the data. The 1990s saw the emergence of support vector machine (SVM)—a method extensively used in NIRS estimations [
11]—to address the inability of PLS to conduct nonlinear regression analyses. By employing kernel tricks to map data to a higher-dimensional space, SVM can linearly separate nonlinear problems, thereby managing complex nonlinear relationships that are challenging for PLS. Ding et al. [
11] proposed a machine learning approach combining NIRS and comprehensive learning particle swarm optimization to enhance tea-quality grade classification, demonstrating the effectiveness of the proposed algorithm in boosting SVM prediction accuracy. However, the sensitivity of SVMs to hyperparameter selection necessitates cross-validation and entails high computational complexity.
Recently, artificial neural networks, particularly convolutional neural networks (CNNs), have been extensively applied across various domains. Their advantage lies in eliminating the need for a manual network structure or kernel function design and avoiding the computational intensity of cross-validation. Given the complexity and structural characteristics of one-dimensional spectral data, the CNN approach is suitable for NIRS. Jernelv et al. [
12] investigated CNN’s performance of a CNN in spectral data classification and regression analysis and compared it with other chemometric methods such as SVM and PLSR. Their findings revealed CNN’s superiority of CNNs over traditional methods in unprocessed classification tasks. However, both CNN and traditional methods benefit from appropriate preprocessing and feature selection. Wang et al. [
13] demonstrated the application of deep convolutional neural networks in NIRS data analysis, effectively distinguishing different tobacco cultivation regions. This research not only confirmed CNN’s robust capability in extracting complex spectral features but also highlighted its crucial role in big data mining and analysis. However, these successful applications are underpinned by a key challenge: the performance of a CNN relies heavily on the setting of its parameters, particularly the configuration of the network layers. In practical applications, optimizing these parameters, especially the network depth, to enhance learning efficiency and analysis accuracy remains an unresolved issue. Moreover, CNN models are prone to overfitting, especially with limited sample sizes. Deep networks also require significant computational resources, including high-performance GPUs and substantial memory, which may limit their application in resource-constrained environments. As “black box” models, the internal decision-making processes of CNNs are often difficult to interpret, which could be a drawback in applications requiring model transparency. Additionally, CNN models may need to be adjusted and trained for different datasets, reducing their generality and flexibility.
In the realm of CNN performance optimization, parameter setting, especially the network layer configuration, is of paramount importance. These parameters, including the number of layers, number of neurons per layer, choice of activation functions, learning rate, and batch size, profoundly influence learning methods and network efficiency [
14]. Different parameter configurations can lead to significant variations in learning effectiveness and efficiency, thereby affecting the final analysis results. Manual adjustment of these parameters is not only time-consuming but also prone to suboptimal network performance. In this context, Mishra et al. [
15] explored the utilization of deep learning models to improve traditional PLSR methods by employing Bayesian hyperparameter tuning for some parameter adjustments. Additionally, Benmouna et al. [
16] focused on using CNNs to estimate the ripeness of Fuji apples and compared them with three alternative methods: artificial neural networks (ANN), SVMs, and k-nearest neighbors (KNN), underscoring the significance of parameter adjustment in CNNs and their superior performance compared to other methods. This raises a critical question: in the absence of sufficient automation tools, how can one effectively optimize CNN parameter configurations, particularly network layers and convolutional neuron numbers, to enhance the network learning efficiency and accuracy of the analysis results?
The core contribution of this study lies in the novel integration of Bayesian network hyperparameter optimization with early stopping strategies applied to one-dimensional CNNs, addressing the current parameter adjustment challenges. This innovative approach to neural network architecture searches not only maintains model generalizability but also automatically identifies the optimal model structure combinations. This method demonstrates significant advantages in processing complex spectral data to improve prediction accuracy and reliability. Moreover, it substantially reduces the need for manual parameter adjustments, thereby enhancing the efficiency of model development. Notably, this method exhibits exceptional generalizability across different datasets, eliminating the need to construct separate models for each data type. Using only the input data, the system can automatically determine the optimal parameter configuration, thereby achieving efficient and accurate predictions of the analysis results. This innovation not only optimizes the model performance but also showcases its unique value in practical applications, particularly in the precise analysis and prediction of complex data.
3. Results
3.1. Dataset Split
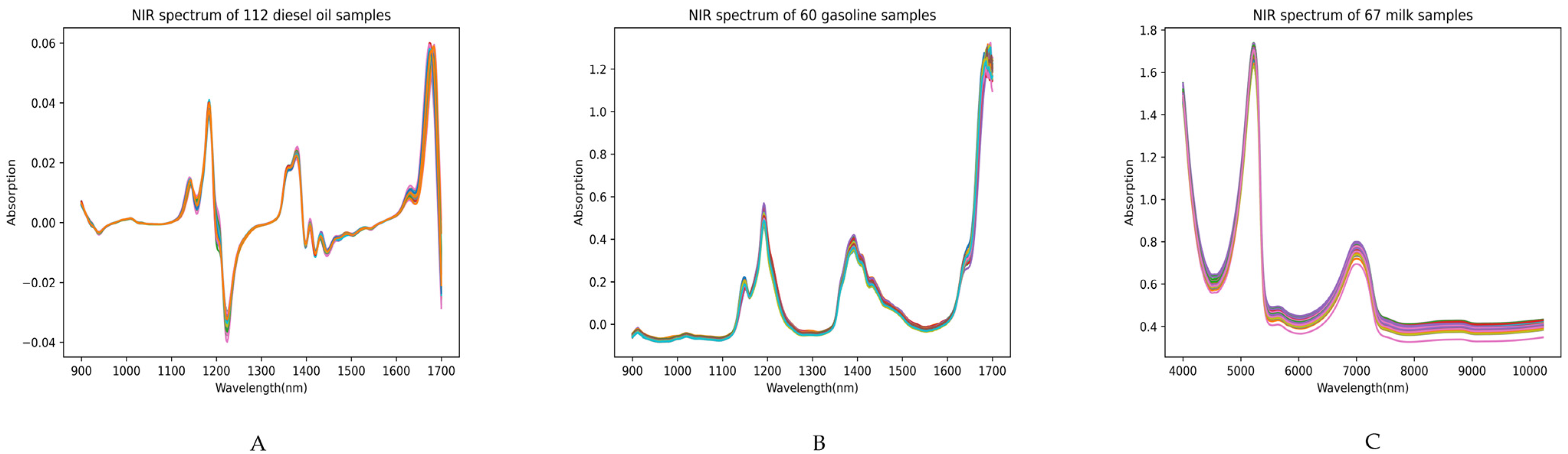
In this study, a 1D-CNN model is constructed using raw NIRS data. The dataset included 60 gasoline, 112 diesel, and 67 milk samples. These samples were divided into training and prediction sets in a 7:3 ratio. Specifically, for gasoline and diesel samples of varying concentrations, 70% were randomly selected for training the 1D-CNN model, whereas the remaining 30% were utilized for external prediction to evaluate the performance of the developed 1D-CNN model.
3.2. Training Results of SVM Modeling with Different Preprocessing Methods
A traditional machine learning technique, SVM, was employed for modeling. This approach was used to compare the post-preprocessing of the SVM and BEST-1D-CNN models. The SVM model utilizes the radial basis function as its kernel, with gamma set to ‘auto’ and the penalty parameter (C) set to 10. The performance metrics of the SVM model with and without preprocessing are listed in
Table 2. The results indicate significant variability in the impact of preprocessing on the different samples. Not all preprocessing combinations enhance the accuracy of predictions compared to those made with individual preprocessing techniques. For instance, in the case of the diesel sample analysis, predictions made post-MSC preprocessing alone exhibited a 12.44% higher accuracy than those made after combined preprocessing. Therefore, it is imperative to select preprocessing methods that are tailored to the specific characteristics of each sample.
Table 2 reveals that applying a combination of the three preprocessing methods prior to SVM modeling significantly enhances the predictive accuracy for the cetane number in diesel, octane number in petroleum, and protein content in milk compared to using each preprocessing method in isolation. The R
2 values for the combined MSC + SNV + 1D + SVM approach outperformed those of the standalone SVM, MSC + SVM, SNV + SVM, and 1D + SVM approaches, achieving values of 0.651380, 0.875780, and 0.883699, respectively. These results indicate performance improvements of 7.96%, 12.48%, and 14.32%, respectively, over the conventional SVM method, underscoring the efficacy of the combined preprocessing approach in regression training.
Figure 6 illustrates the training outcomes of the predictive models for the three analytes.
Figure 6 displays the test results of a SVM model with preprocessing applied to three distinct types of samples. The figure is subdivided into six panels, each illustrating different aspects of model performance.
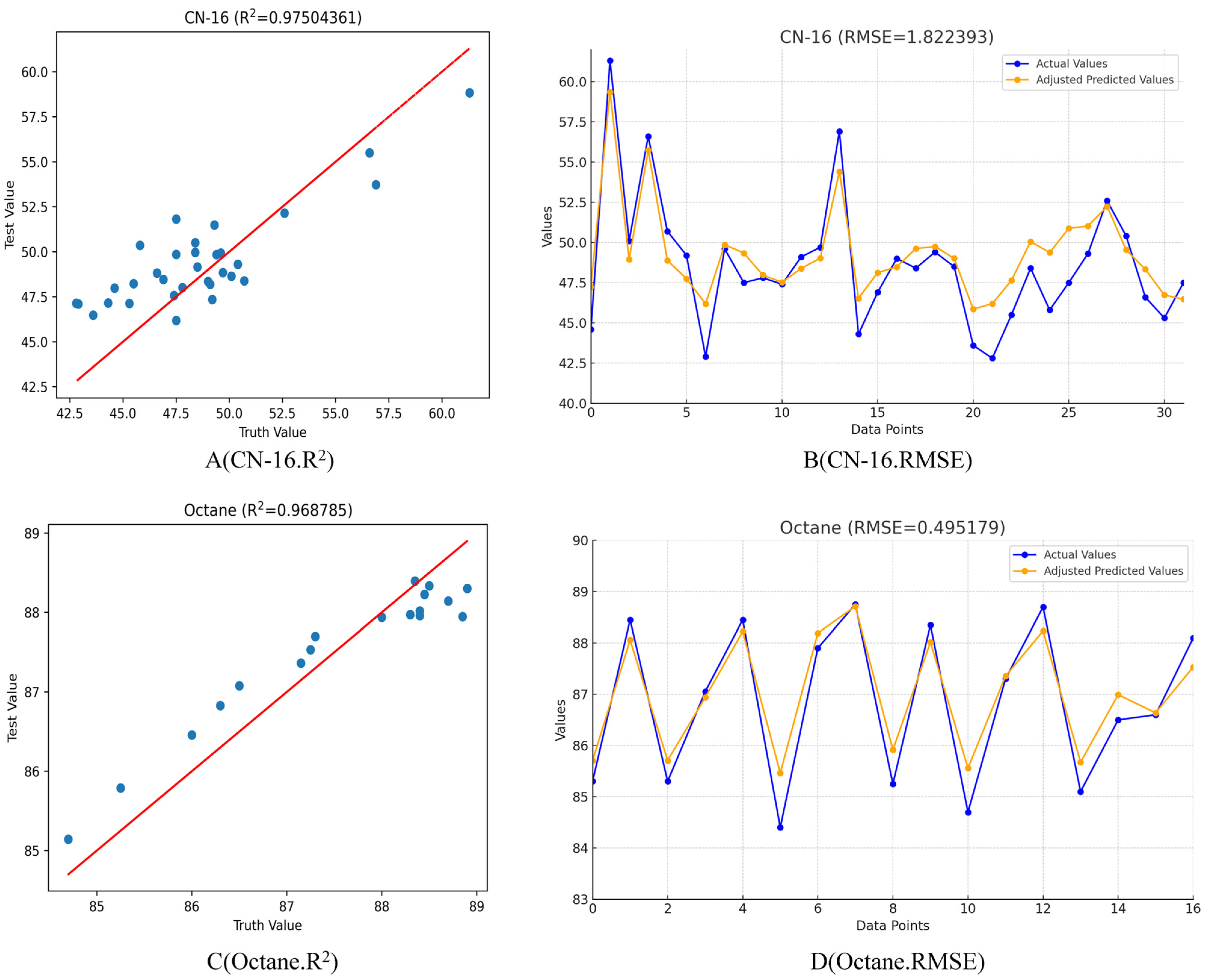
Figure 6A,B focus on the analyte CN-16. In
Figure 6A, a scatter plot showcases the relationship between the actual and predicted values, with an R
2 value of 0.636611, indicating a moderate positive correlation. The line of best fit emphasizes the trend in the data points.
Figure 6B presents a line graph detailing both the actual and the adjusted predicted values over a sequence of data points, with an RMSE of 1.839059, providing insight into the model’s prediction accuracy across the dataset. For the analyte octane, panels 6C-D depict similar trends.
Figure 6C’s scatter plot, with an R
2 value of 0.875788, suggests a strong positive correlation between the true and predicted values, with a closely adhering line of best fit. In panel 6D, the line graph compares the actual and adjusted predicted values with an RMSE of 0.512449, which is lower than that of CN-16, indicating a higher prediction accuracy for octane. Lastly, panels 6E-F represent the analyte protein. The scatter plot in panel 6E, with an R
2 value of 0.836699, demonstrates a strong correlation between the predicted and actual values. The line of best fit shows the model’s effectiveness at capturing the relationship.
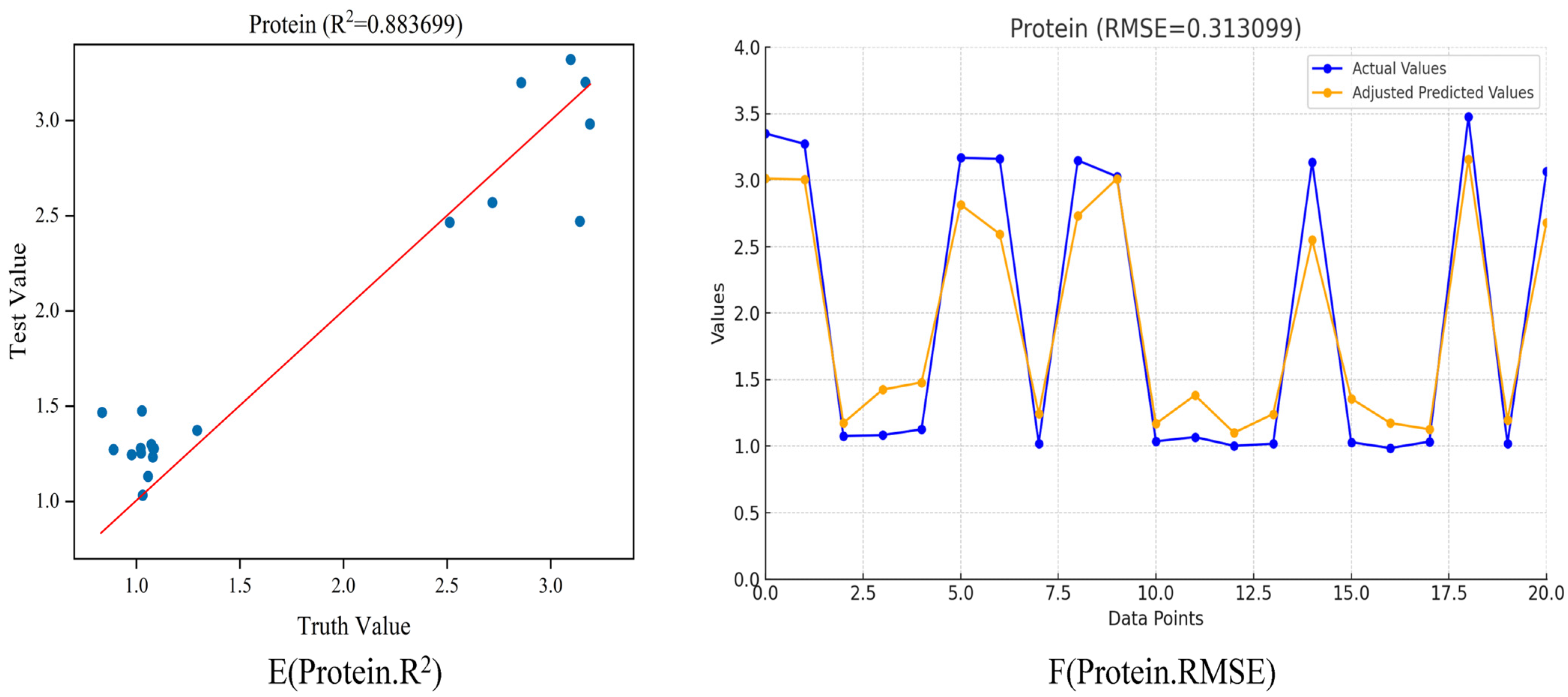
Figure 6F’s line graph, with an RMSE of 0.313099, suggests that the model’s predictions are quite precise, as indicated by the lowest RMSE among the three analytes.
Overall, these panels collectively indicate the effectiveness of the SVM model’s predictive capabilities, with varying degrees of accuracy and correlation strength across different analytes. The use of preprocessing techniques appears to enhance the model’s performance, as reflected in the relatively high R2 values and lower RMSE scores.
3.3. Training Results of the BEST-1DConvNet Model
The evaluation of the BEST-1DConvNet model, tailored for near-infrared spectroscopy data analysis, revealed significant findings. The model’s architectural optimization, achieved through Bayesian optimization techniques, focused on the fine-tuning of several key hyperparameters, notably enhancing its performance and generalization capabilities. Essential hyperparameters such as the number of convolution layers, the count of convolution kernels, their size, and stride each crucially contribute to the model’s efficacy. The convolution layers’ quantity directly influences the model’s depth and complexity, with a larger number potentially boosting feature extraction capabilities, albeit with an increased overfitting risk. The convolution kernels’ quantity and dimensions affect the range and types of features identifiable by the model at certain layers, thus impacting its recognition and generalization skills. Stride selection affects the spatial resolution of the feature maps, a factor critical to the model’s precision in processing input data details.
In addressing overfitting risks during training, the adoption of an early-stopping strategy was observed—if the loss on the validation set failed to decline significantly over a predetermined number of iterations, training was concluded prematurely. This method not only conserved time and computational resources but also curbed overfitting. The RMSE was deployed as the loss function for gauging the model’s predictive error, while the coefficient of determination (R
2) was utilized as the performance metric.
Table 3,
Table 4,
Table 5 and
Table 6 elucidate the parameters of the convolutional layers for the 1D-CNN and for the diesel, gasoline, and milk datasets, respectively. These tables display the optimal model parameter configurations for the respective datasets, evidencing the BEST-1DConvNet model’s efficacy in NIRS data analysis.
Table 4 and
Table 7 reveal that after conducting Bayesian hyperparameter search permutations, the CN value prediction model for gasoline comprises a single convolutional layer with a kernel size of 355, 156 filters, a stride of 494, and a learning rate of 0.02289. Training was halted at epoch 160, resulting in an R
2 value of 0.96878574419 and RMSE of 1.8223934 for the diesel CN value model, as illustrated in
Figure 7A, B. Similarly,
Table 5 and
Table 7 indicate that the cetane-number model for gasoline, derived from a Bayesian hyperparameter search, consists of a single convolutional layer with a kernel size of 201, 16 filters, a stride of 100, and a learning rate of 0.01. Training ceased at epoch 140, yielding an R
2 value of 0.97504361 and RMSE value of 0.49517905 for the gasoline octane model, as depicted in
Figure 7C, D. Finally,
Table 6 and
Table 7 show that the protein-value model for milk, based on the Bayesian hyperparameter search, consists of a single convolutional layer with a kernel size of 67, 14 filters, a stride of 316, and a learning rate of 0.02289. The training concluded at epoch 200, with the milk protein value model achieving an R
2 value of 0.96098518661 and RMSE of 0.301546, as shown in
Figure 7E, F.
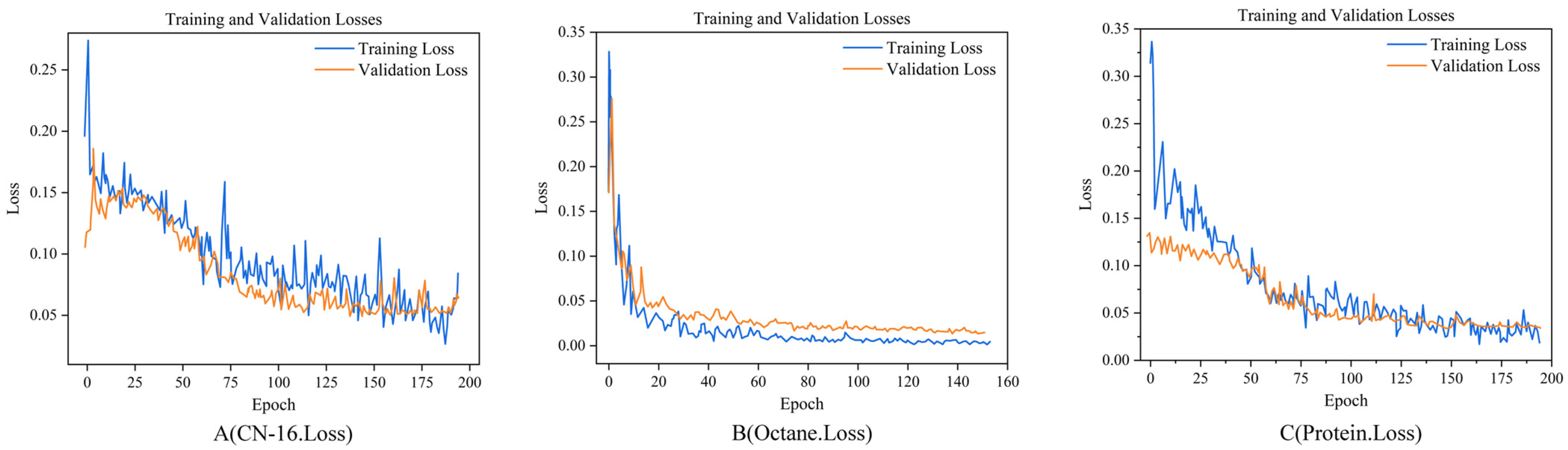
The loss curve graphs show variations in the loss function values of the model during the training process. Each graph comprises two lines: one representing the training loss (blue) and the other representing the validation loss (orange). Ideally, the loss curve should continuously decrease with the progress of training and stabilize towards the end of the training process. In
Figure 8A–C, the loss curves demonstrate a reduction in the loss function values as the training progressed, which stabilized after a certain number of iterations. This indicated that the models learned and progressively fitted the data. The close similarity of the training and validation loss curves suggests that the models do not suffer from overfitting and exhibit good generalization capabilities for unseen data. In all three datasets, the rapid decline in the loss curves reached a plateau phase, particularly in
Figure 8A, B, where the loss curves stabilized after approximately 50 training epochs. In
Figure 8B, this plateau phase appeared earlier, with the loss becoming extremely low and stable after approximately 20 epochs. These indicate that the model quickly captured the key features of the data for the gasoline dataset, whereas, for the other two datasets, more training time might be required to achieve similar levels of performance.
3.4. Model Comparison
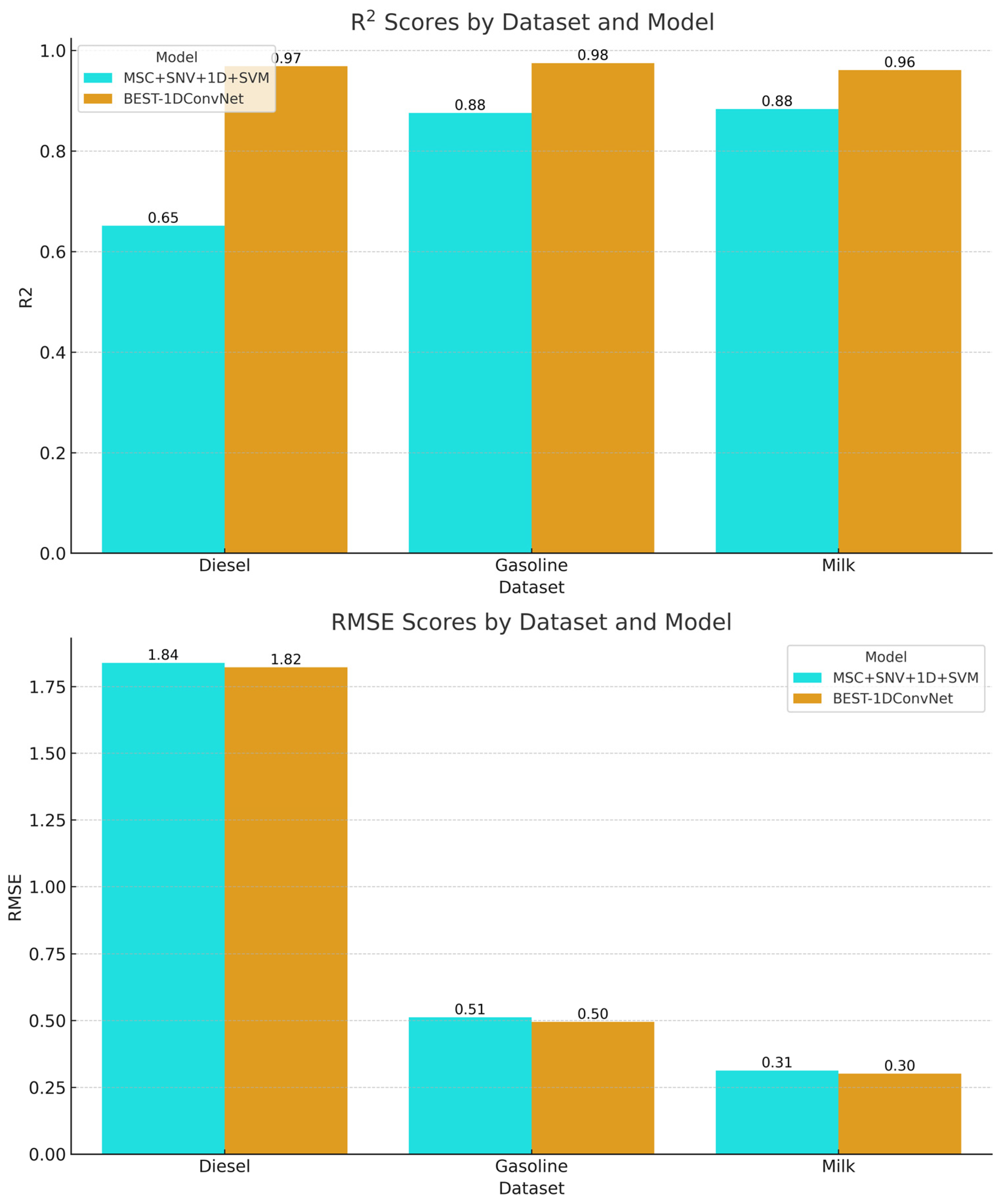
The R2 scores, which indicate the explanatory power of the models, were higher for the BEST-1DConvNet model across all datasets, signifying a stronger correlation between the predicted and actual values. For diesel, the R2 score was 0.969 for BEST-1DConvNet and 0.651 for MSC + SNV + 1D + SVM. For the gasoline dataset, BEST-1DConvNet achieved an R2 of 0.975, whereas the combined preprocessing SVM approach achieved a value of 0.876. The milk dataset results exhibited a similar trend to that of BEST-1DConvNet at 0.961 R2, outperforming the SVM combination by 0.884.
In terms of the RMSE, which measures the prediction error magnitude, lower scores indicate better performance. The BEST-1DConvNet model consistently exhibited lower RMSE values, demonstrating superior predictive accuracy. Specifically, for the diesel dataset, the RMSE values were 1.839 for MSC + SNV + 1D + SVM and 1.822 for BEST-1DConvNet. Gasoline showed a more notable difference, with an RMSE of 0.512 for the combined SVM and 0.495 for BEST-1DConvNet. The milk dataset reflected the closest RMSE values, with values of 0.313 for the SVM and 0.302 for the BEST-1DConvNet.
Overall, the bar chart in
Figure 9 elucidates the superior performance of the BEST-1DConvNet model over the SVM with preprocessing in both the R
2 and RMSE metrics, emphasizing its efficacy and precision in predictive tasks within the scope of the analyzed datasets.