1. Introduction and Motivation
Alzheimer’s disease, also abbreviated as AD, is one of the most common forms of dementia, which is characterized by memory loss and cognitive decline. As of 2023, 6.7 million people are living with AD in United States [
1] and over 55 million worldwide [
2]. Nearly 60–80% of dementia cases comprise Alzheimer’s [
2,
3]. There are over 10 million new cases of dementia each year worldwide [
4], which means, on average, 7 million of them are suffering from Alzheimer’s. And it will only increase each year [
1] unless a satisfactory cure is invented. There are two main hypotheses from which the mathematical models have emerged. These hypotheses are used to describe the progression of AD. There are thousands of studies conducted using these hypotheses [
5,
6,
7,
8]. The first hypothesis is the amyloid hypothesis [
5,
6,
7,
9,
10,
11,
12,
13,
14,
15], which is a neuron-centric model, and the second is the Warbug hypothesis, which is the neuron-astrocytic model [
8,
15,
16,
17]. The neuron-centric model suggests that a mutation in the nuclear genome induces the overproduction of
and tau, which become toxic to neurons [
8,
16,
17]. The neuron-astrocytic model contends that the progression of AD is triggered by defects in the normal energy transduction process, a condition induced by mitochondrial dysregulation [
8,
15,
17].
According to the amyloid-beta hypothesis, Alzheimer’s is a neuro-degenerative disease, where the patient’s cognitive function progressively deteriorates, which is characterized by the progressive accumulation of amyloid beta called
peptides [
15,
18]. The major pathological hallmarks in the brain of AD patients are amyloid plaques [
5,
9,
10,
11,
12,
18], and neurofibrillary tangles (NFTs) formed by hyperphosphorylated tau protein [
5]. Neurofibrillary tangles (NFT) are pathological insoluble aggregates of hyperphosphorylated tau proteins, which are microtubule-associated proteins, observed within the neurons of AD patients [
12]. In healthy individuals, normal tau stabilizes the microtubules of the neuronal cytoskeleton; among their diverse functions, microtubules facilitate the transport of substances between nerve cell compartments. Aggregated tau filaments cannot be degraded by the host neurons and gradually accumulate and aggregate into insoluble filaments [
11]. Tau pathology is an early feature of AD, and the appearance of tangles correlates with neuronal loss [
19].
Amyloid beta is normally secreted from cells and degraded, and it is produced throughout one’s life, but in the case of AD patients, it is secreted and aggregated into insoluble plaques [
18,
20,
21].
is a proteolytic product of amyloid precursor protein (APP), and AD neuropathology is characterized by an abnormal metabolism of APP with an excessive accumulation of
peptides [
20,
22].
is believed to be the main reason for initiating the pathological cascade of the disease [
23]. Amyloid plaques are hard, insoluble accumulations of beta-amyloid proteins that clump together between the neurons in the brains of AD patients [
5,
9,
10,
11,
12,
24]. The lesions occur in brain regions involved in learning and memory, i.e, the hippocampus, the amygdala, and in the association cortices of the frontal, temporal and parietal lobes [
5].
deposits in senile and neuritic plaques and hyperphosphorylated tau proteins in neurofibrillary tangles (NFTs) are extracellular and intracellular expressions, respectively, of the AD neuropathological phenotype, together with selective neuronal loss in the hippocampal and neocortical regions [
25].
Figure 1 shows the complex interrelation of amyloid plaques and NFT and how they cause the loss of neurons. The presence of plaques around a neuron causes them to die, possibly by triggering an immune response in the immediate area. Tangles form inside of neurons and interfere with the cellular machinery used to create and recycle proteins, which ultimately kills the cell [
24].
One of the main characteristics of cells and tissues made of proteins is the self-assembly of proteins into filaments of a particular structure [
14,
26,
27]. Cells are sustained by cytoskeletal filaments, whereas tissues are sustained by collagen fibrils. Within all eukaryotic cells, there exists a network of filaments, which is a complex mesh of protein filaments and motor proteins that aid in cell movement and stabilize the cell [
28]. And cytoskeletal filaments are equally important in the correct formation of the nervous system and their various functions [
27,
28]. Given the importance of the cytoskeleton for neurons, its involvement in neurological disorders (taupathies), including AD, is not surprising, and it can be in the form of their mutation and changes in their dynamics and the stability of the cytoskeletal proteins [
27].
All of this self-assembly of the proteins, be it functional or dysfunctional, involves the assembly of elementary units to the ends of the growing structure [
9,
13,
14]. Hence, on that note, we are interested in studying the early aggregation of monomers forming the structure, like oligomers and filaments. These filaments are the most toxic structure [
13] and need attention for both understanding the disease and deriving the potential cure. Studies suggest that the aggregation of a primary structure, such as monomers, into the intermediate species, such as oligomers and unbreakable filaments, are correlated with cellular toxicity [
13], and such is the case of most amyloid-based diseases, including AD. In AD, the aggregation of soluble APP, into non-soluble amyloid fibrils is the major cause of cognitive loss, including NFT [
5,
9,
10,
11,
12].
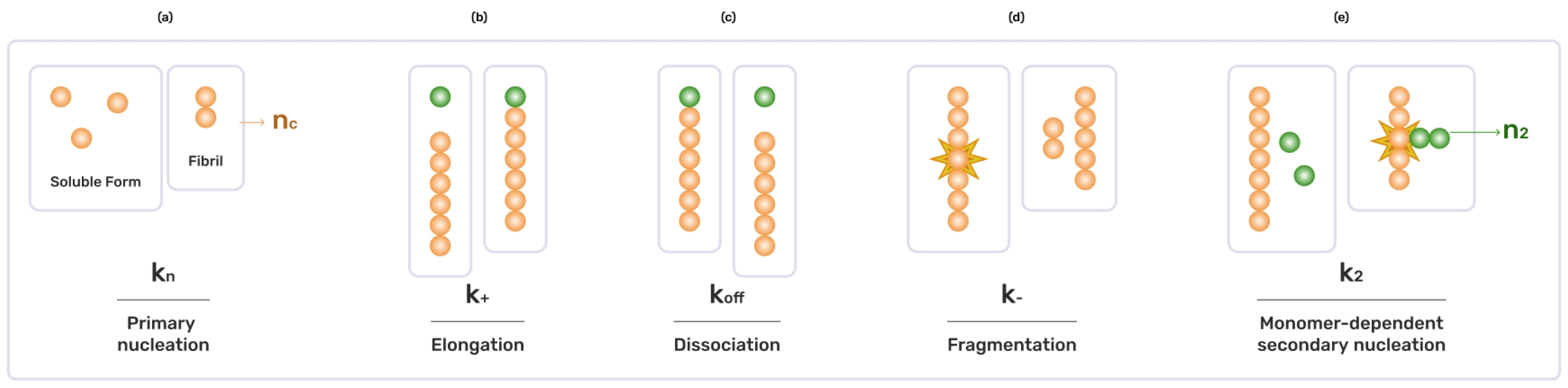
The law of mass action serves as the foundation for robust kinetic models that effectively define the chemical kinetics of intricate reaction networks controlling the growth of filamentous protein structures. These models also provide valuable insights into the complex processes underlying protein aggregation [
9]. In that respect, [
9,
14] established master equations to treat the aggregation of the amyloid precursor protein towards the formation of fibrils. They established the criterion for primary nucleation, elongation and other secondary events, such as secondary nucleation, fragmentation and monomer-dependent elongation [
9,
14]. Ref. [
13] established another master equation to capture the stochastic effects in early amyloid aggregation to describe the aggregation of monomers into oligomers. They used log-normal closure moment method rather than direct simulation. They also used the Gillespie algorithm to simulate the process of protein aggregation and compared it with the result from their master equation. Ref. [
6] introduced the method of second stochasticization by introducing random noises into already statistically averaged equations obtained from moment closure methods. They coupled their study with the stochastic simulation using the Gillespie algorithm. Ref. [
15] used a stochastic mathematical model to understand the various phases and transitions of AD.
Due to the complexity and the number of aspects involved in the protein aggregation, although the deterministic mathematical model has given some very good frameworks, they do not capture the inherent stochastic nature present in the bio-chemical reactions. From a physical point of view, the stochastic formulation of chemical kinetics is superior to the deterministic formulation: the stochastic approach is always valid whenever the deterministic approach is valid, and is sometimes valid when the deterministic approach is not [
29]. Hence, to unravel the complex dynamics preset in the amyloid-beta aggregation, we make use of the stochastic computational simulation to understand the dynamics of amyloid growth and the aggregation of monomers towards forming filaments. We study the formation of filaments from, at most, eight monomers. We treat all possible combinations of monomers forming the filaments of lengths up until eight. Our comprehensive simulation of reaction kinetics is the main contribution of our work, which sheds light on the aggregation and formation of no-soluble toxic filaments from soluble protein units. We experiment with the primary nucleation, followed by aggregation/elongation and fragmentation, which occur with different propensities. We also simulate the dynamics on different sets of propensities, using different propensities for different reaction events, which are not so evident in the studies that we reviewed. With that, we are also able to see how the individual filament of length
i evolved with time, and we also evaluate the dynamics by introducing different random propensities at different times. Our experiments show that the evolution of amyloid-beta aggregation can be modeled computationally by using stochastic modeling techniques of chemical reactions. We evaluate the convergence by plotting the phase diagram of the evolution of filaments of different lengths, and by also plotting the zeroth moment and first moment. We also find that the convergence is highly correlated with the propensities used for the reaction events, and some events dominate the overall reactions events that occur during the simulation. And there is a clear difference in convergence when we use fixed constants for each reaction and different constants for different reactions. We also evaluate that the initial conditions (the initial population of pre-existing filaments) affect the final convergence.
Although some 90,000+ research studies have been published since 1990 on AD [
25], it still remains an enigma largely due to the nature of the disease and its complexity [
25,
30]. Although the amyloid-beta hypothesis has been studied widely and believed to show the development of disease, it is still very hard to trace the root of the disease, or how, why and when it develops [
19,
25,
30]. And there is more and more speculation that
plaques are the result of AD, and not the other way around [
30]. But it is clear from all these studies that
and
are well-known markers of AD, and hence are worth studying further. On that note, we think that there is a space for contribution to be made in the stochastic nature of the
aggregation, and how they propagate and form the dysfunctional insoluble filaments. Hence, the main objective of this work is to utilize the mathematical and computational framework to unravel the stochastic nature of the elongation, propagation and possible fragmentation of
peptides. We treat this process as a chemical reaction process and use the chemical kinetics of the intricate reaction, which is responsible for forming the filamentous protein structure. Our main contribution lies in conducting detailed simulations and reaction kinetics analyses to understand the stochastic nature of amyloid-beta aggregation, which includes experimenting with various reaction propensities and their random switching over time, thus providing a comprehensive model of amyloid-beta aggregation as a stochastic chemical reaction process, an approach not extensively explored in previous studies.
The paper is organized as follows: This first section introduces the work, the motivation behind this study, and the objectives.
Section 3 introduces the mathematical theory behind the chemical reaction and how the stochastic modeling can be performed.
Section 4 shows the experiments that we conducted comprehensively. In the last three sections, we discuss the results, limitations and conclusions.
4. Computational Simulation Results
We use the Gillespie algorithm [
29], which is a seminal method for simulating the stochastic processes, and we use it to simulate the aggregation and fragmentation processes of amyloid beta. We use Monte Carlo simulation on top of the Gillespie algorithm and average out the results to reduce any anomalies that may be present in a single simulation. The general framework of the Gillespie algorithm lies in randomly picking up the next reaction time, and the next reaction event that can occur from the set of possible events. The pseudo-code for the Gillespie algorithm that we used is given in
Algorithm S2 of Supplementary Material. We wrap the Gillespie function with Monte Carlo simulation and derive the average simulation results over the number of simulations. The pseudo-code for the Monte Carlo simulation is provided in the additional materials. We utilize Python to simulate the reaction processes and conduct Monte Carlo simulations. The initial experiments were performed using the Jupyter Notebook on a standard notebook/desktop, with 16 GB of memory.
Different initial conditions are used for different simulations; similarly, we experimented with different reaction rates, e.g., different aggregation rates and fragmentation rates. Firstly, we only simulated the aggregation process, and later added the secondary events such as fragmentation. The monomers are labeled as , and when they aggregate they can take the form of filaments of length i, such as . , the population of free proteins, can be interpreted as amyloid precursor protein (APP), the primary nucleation which produces the oligomers, then the rest of the population can be interpreted as the aggregation of into toxic filaments.
4.1. Modeling the Aggregation Events
To model the monomer aggregation, we first define the kinetics of aggregation, listing all possible reactions that can occur and defining their aggregation rates/constants. The table of the kinetics of amyloid-beta aggregation in the appendix shows the possible reactions that we model in this study. represents the filament of length i, whereas the reaction rate represent the reaction rate for that particular reaction. For example, represents the aggregation rate of and , blending together to form . In the table of reaction kinetics, there are a total of 27 different types of aggregation reactions, producing filaments of different length i.
We use the stoichiometry matrix given in the
Supplementary Material Section S2.3.1, Equation (S1) derived for a total of 27 possible aggregation events for the filaments of length up until eight. The stoichiometry matrix is derived using the reaction kinetics given in
Table S2.1 of Supplementary Material. In this matrix, each column represents
, while each row describes the actual reactions. For example, two
make
; hence, in the first row of the matrix,
has a corresponding value
, and
has a value 1. This is essentially saying that from the total population of
, two
were lost to create one new
. For the reaction rates, we start the rate from a certain constant, for example,
, which is used for the result presented in
Figure 3.
starts from this initial value
;
is the exact half of
,
is the exact half of
and so on, and thus the last reaction constant
is half of
. The rationale behind choosing such a reaction rate is that, the greater the length of the filaments, the lower the likelihood of blending with others, and similarly, the propensity of, let us say,
is much higher than
, and the propensity of forming the filament of length eight (
) from the aggregation event
is the least likely. These complex reactions are assumed to be less likely than two monomers joining together.
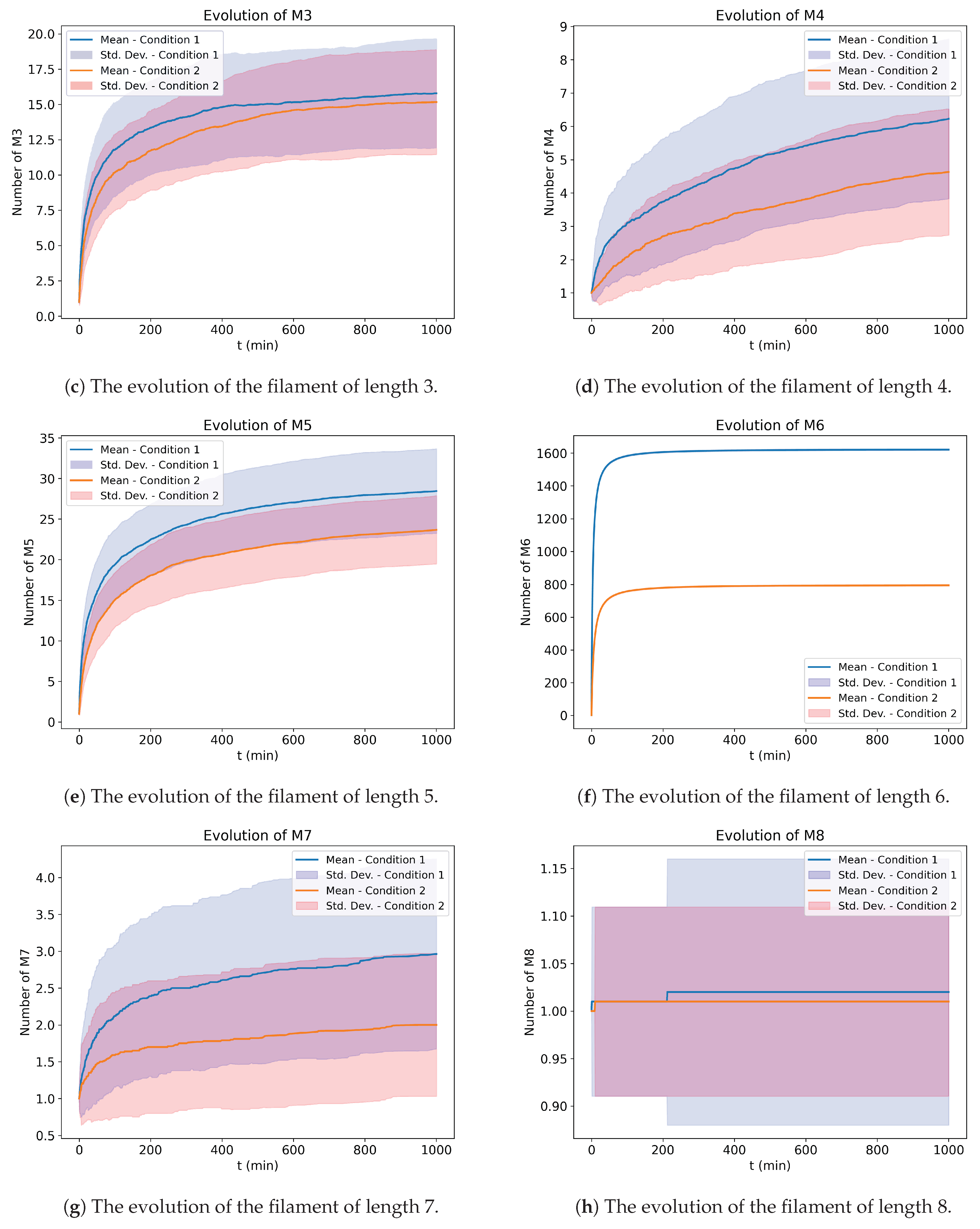
Figure 3 shows the time evolution of monomers and filaments of lengths up to eight, as we can see that there is smooth evolution of filaments of a length of six. We run 100 Monte Carlo simulations, and the average and standard deviation among the multiple simulations are shown in the shaded area. The simulation uses the matrix representation of the reaction as given in the extra materials. The essential difference in
Figure 3 and
Figure 4 and is the initial population of different lengths of the filaments. Our experiments for fixed constant rates with different initial conditions are in the extra material’s section, called “Aggregation of Filaments with Fixed Constant Rates”. It shows how keeping the constants fixed but changing just the initial conditions leads to different convergence.
4.2. Modeling the Aggregation and Fragmentation Events
In this section, we show the experiments conducted in different initial conditions and reaction rates, with both aggregation as well as secondary events, such as fragmentation. In
Section 4.1, we only considered the aggregation events; here, we add the fragmentation events and form a different kinetics and stoichiometry matrix. The kinetics and stoichiometry matrix are provided in the
Supplementary Material’s Table S2.2 the Equation (S2.3.2), respectively.
In
Figure 5, we take the initial population of
to be 100, and in
Figure 6, we keep it to be 1. We can notice the difference in convergence when we use the same reaction probabilities for the events while taking a different initial population.
Figure 7 shows how this changes when we keep the reaction rates constant (one for elongation, second for all aggregation events and third constant for fragmentation events). We denote the reduction in the overall population of filaments of length
. Also, they are converging slower than in
Figure 5, where we used different propensities for different events.
In
Figure 5, different aggregation and fragmentation rates are used. The starting reaction rate for aggregation starts from
, and the subsequent aggregation event, as listed in
Table S2 (aggregation and fragmentation kinetics) in the extra material, will be exactly half of the previous one and so on. And similarly for the fragmentation, the reaction rate starts from
, and each subsequent fragmentation reaction rate such as
and so on.
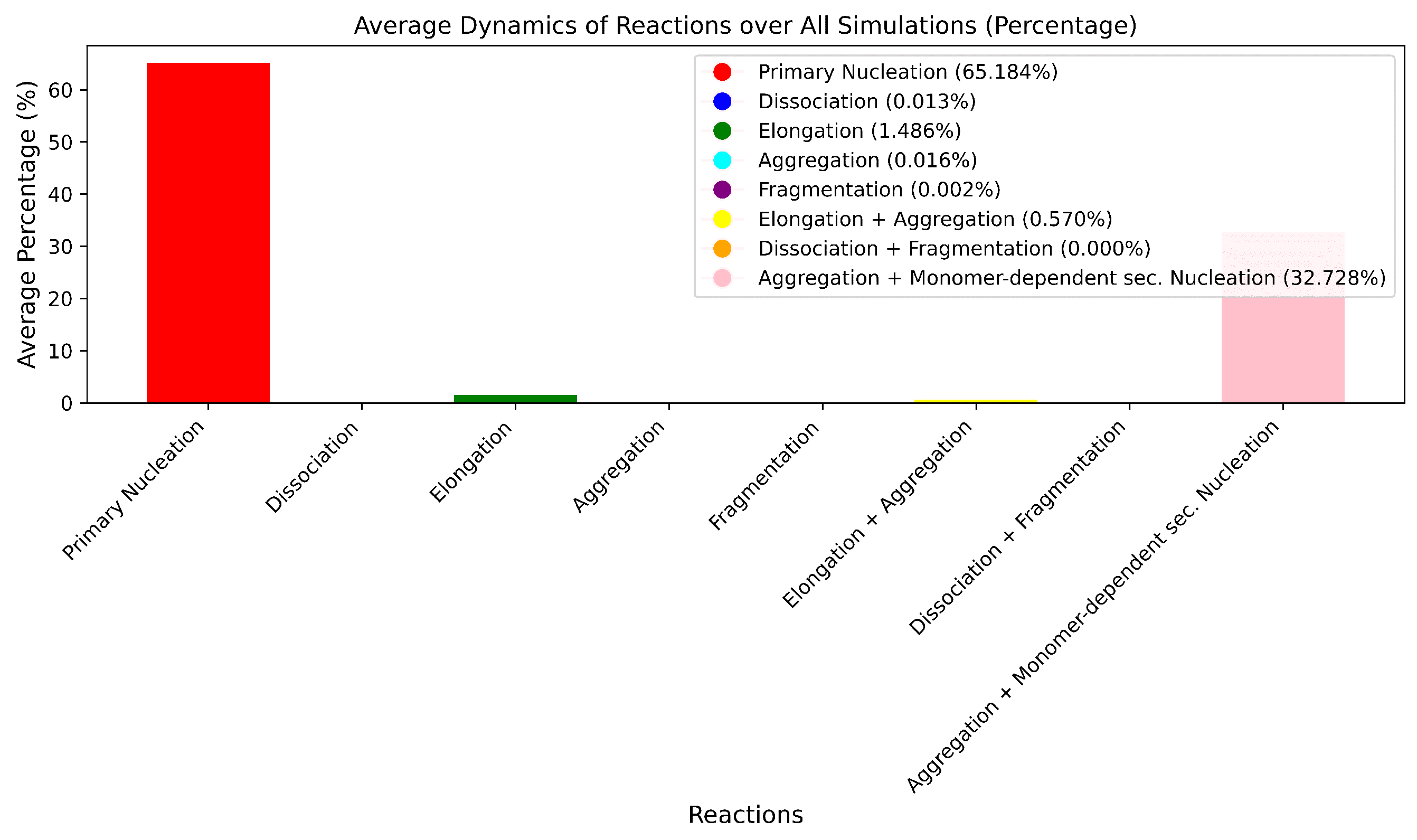
From
Figure 8, it is clear that certain reaction events dominate the overall dynamics of the process. This observation is critical in understanding the inherent behavior of the system under study. Subsequently, in
Figure 9, we introduce a modification to the Gillespie algorithm by incorporating random selection of reaction rates. This adaptation involves randomly choosing a reaction rate from the original set and applying it to a reaction event. We implement this change for
of the time, allowing for the possibility of any reaction propensity to be randomly selected. When we reduce this randomness to only
of the time, the results increasingly resemble those obtained without introducing randomness. Further reduction of the randomness leads to results that closely align with those from the original algorithm, demonstrating more uniform convergence. The impact of using random switching of reaction probabilities is evident in the results; specifically, introducing random propensities tends to decrease the converged population, thereby reducing the filament formation. This effect becomes even more apparent when we examine the evolution of individual populations.
Continuing this analysis,
Figure 10 shows that the introduction of random switching of probabilities does not significantly alter the dominance of certain reaction events in the overall dynamics. This observation indicates that, despite the random elements introduced in the reaction propensities, the core characteristics of the process are largely maintained.
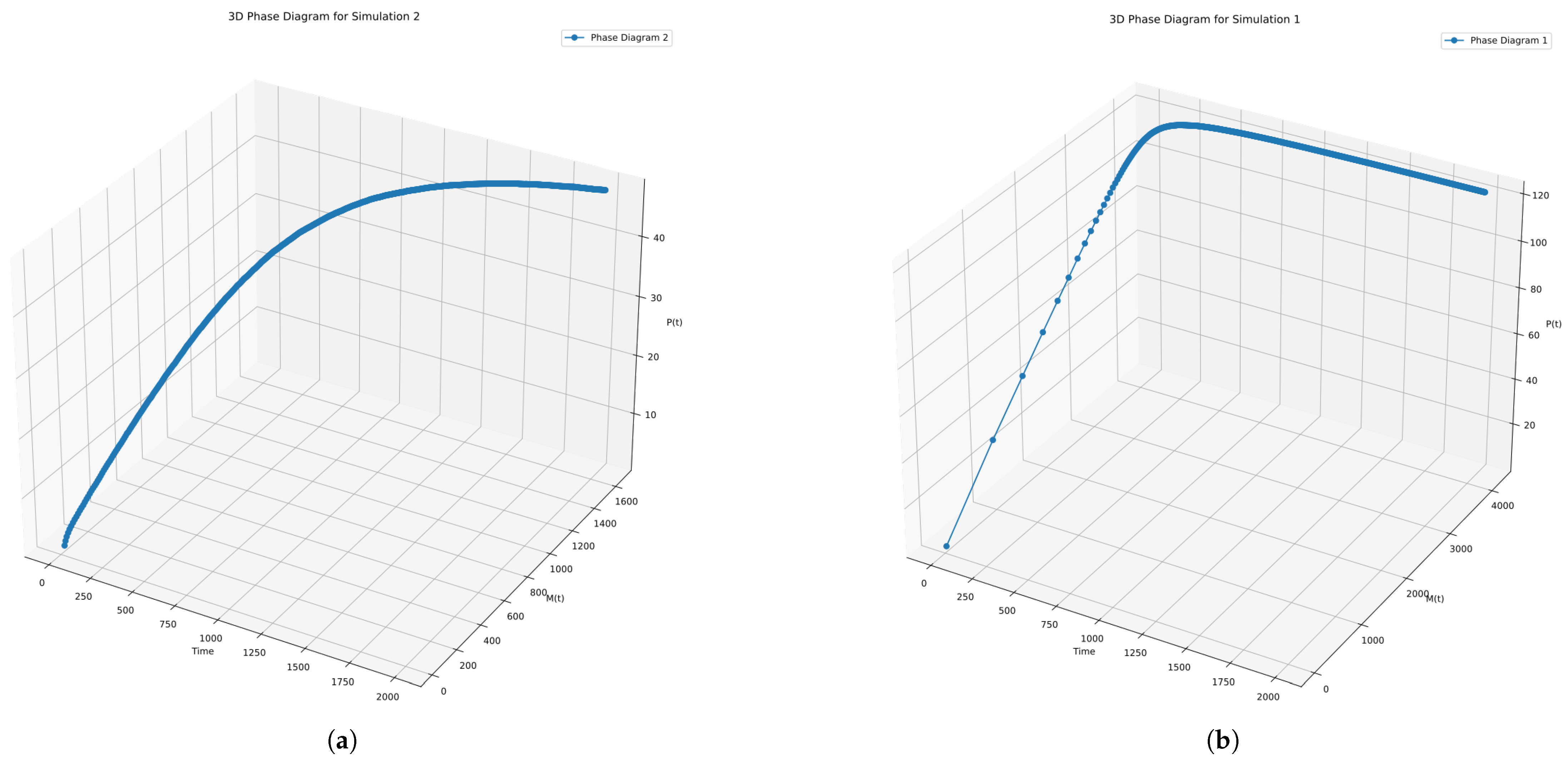
In
Figure 11, we plot the phase diagram, X-axis showing the time (t), Y-axis showing the first moment M(t) and Z-axis showing the zeroth moment (P(t)). It is evident that, without the random switching of the probabilities, the dynamics tend to aggregate more filaments of different lengths than in the case of using the random switching of probabilities.
4.3. Comparison with Other Studies
In this section, we compare our results with that of [
6,
13]. Both of these methods implement the moment-closure method for the stochastic modeling of the aggregation of proteins in amyloid disease. First we try to model the work of [
13] using their own stoichiometry but with different probabilities for the reaction events than were originally used in their work.
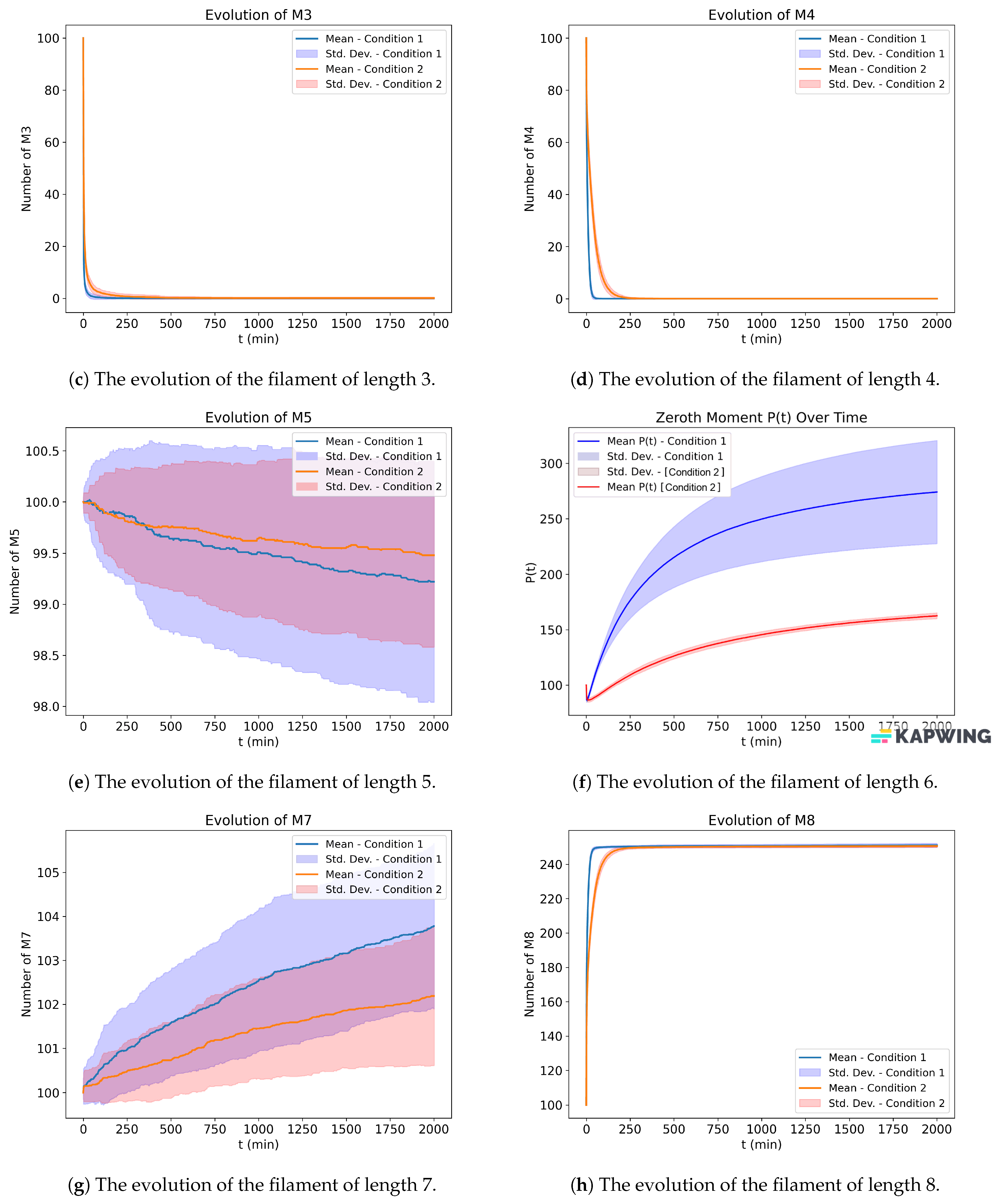
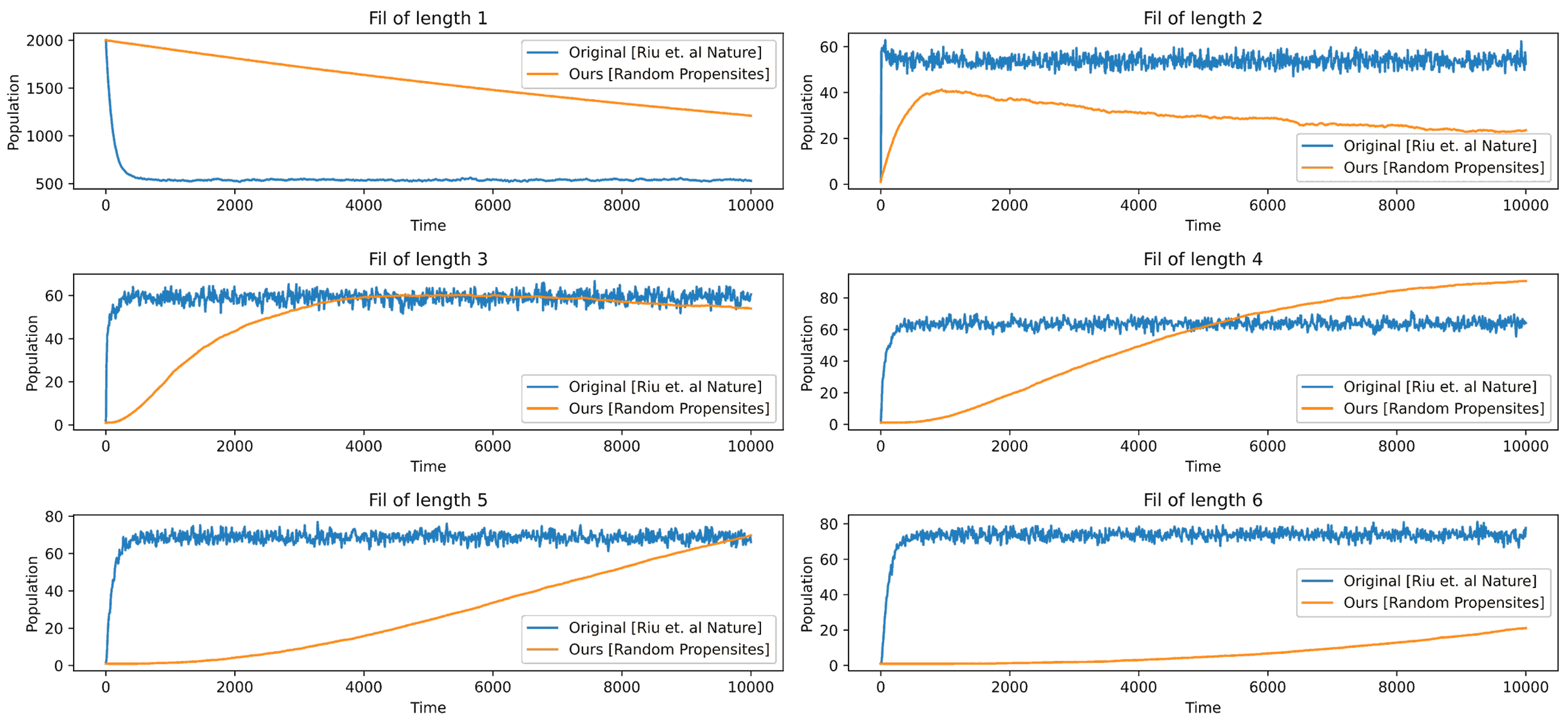
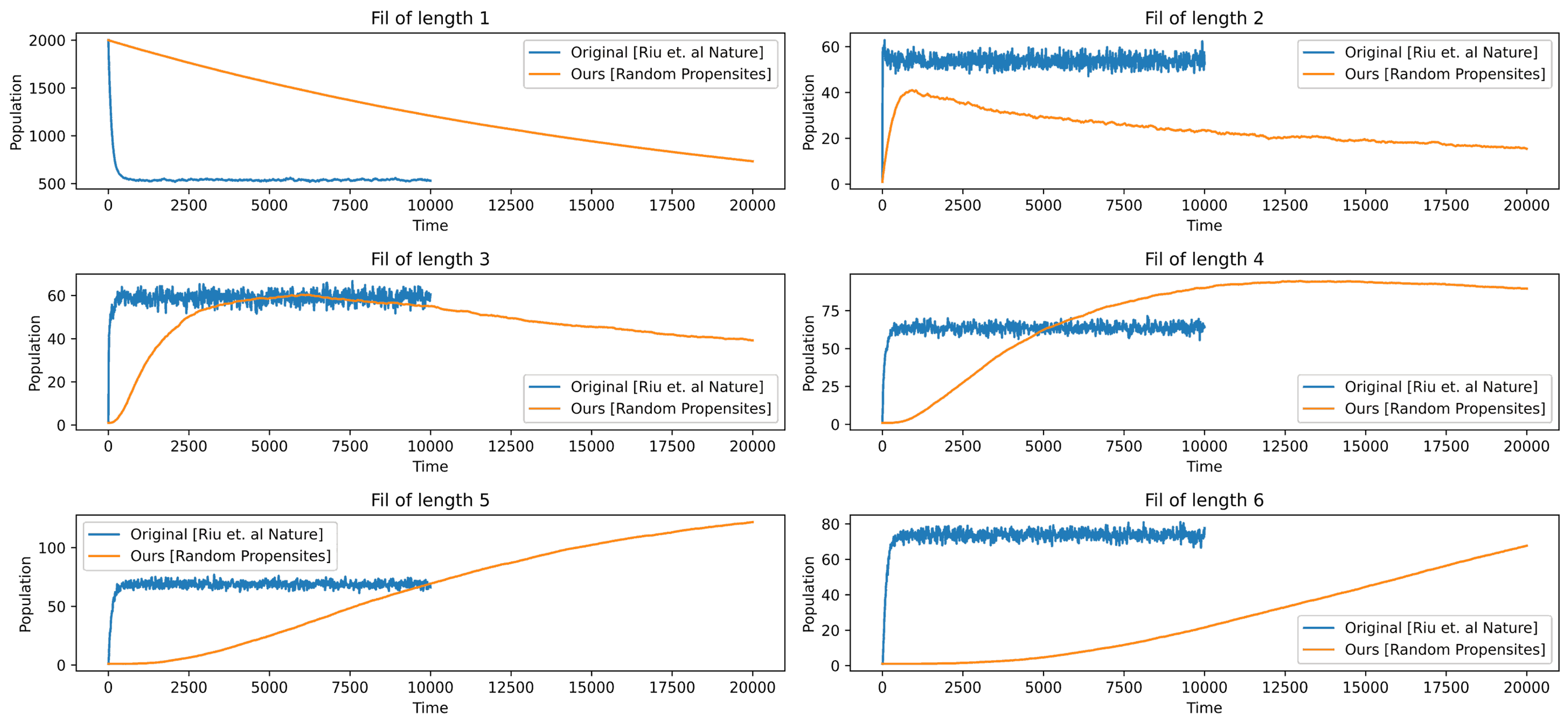
In
Figure 12, we use random probabilities instead of the ones given in [
13]. We can notice a sharp decline in the free protein, whereas ours has a gradual decline. Overall, the results are comparable, but one thing that we can instantly realize is that for our configuration to converge, we need to run the simulation for a longer time. In
Figure 13, we run the simulation for longer time, and we can see that
, and
first increase and then decrease. The highest convergence is in the filament of length six. The above results are obtained when we use their stoichiometry but using random propensities for the events instead of their fixed propensities. To produce these results, we use the starting probability or the probability of the occurrence of the first event elongation to be
, then each subsequent event on the kinetics will have exactly half the probability than the one it preceded.
When we use our stoichiometry, which consists of 54 different possible reactions along with different propensities, although the filaments are not distributed comparably, their trajectories look similar. The result of this is given in the extra materials, the corresponding figures are
Figures S7 and S8 of Supplementary Material.
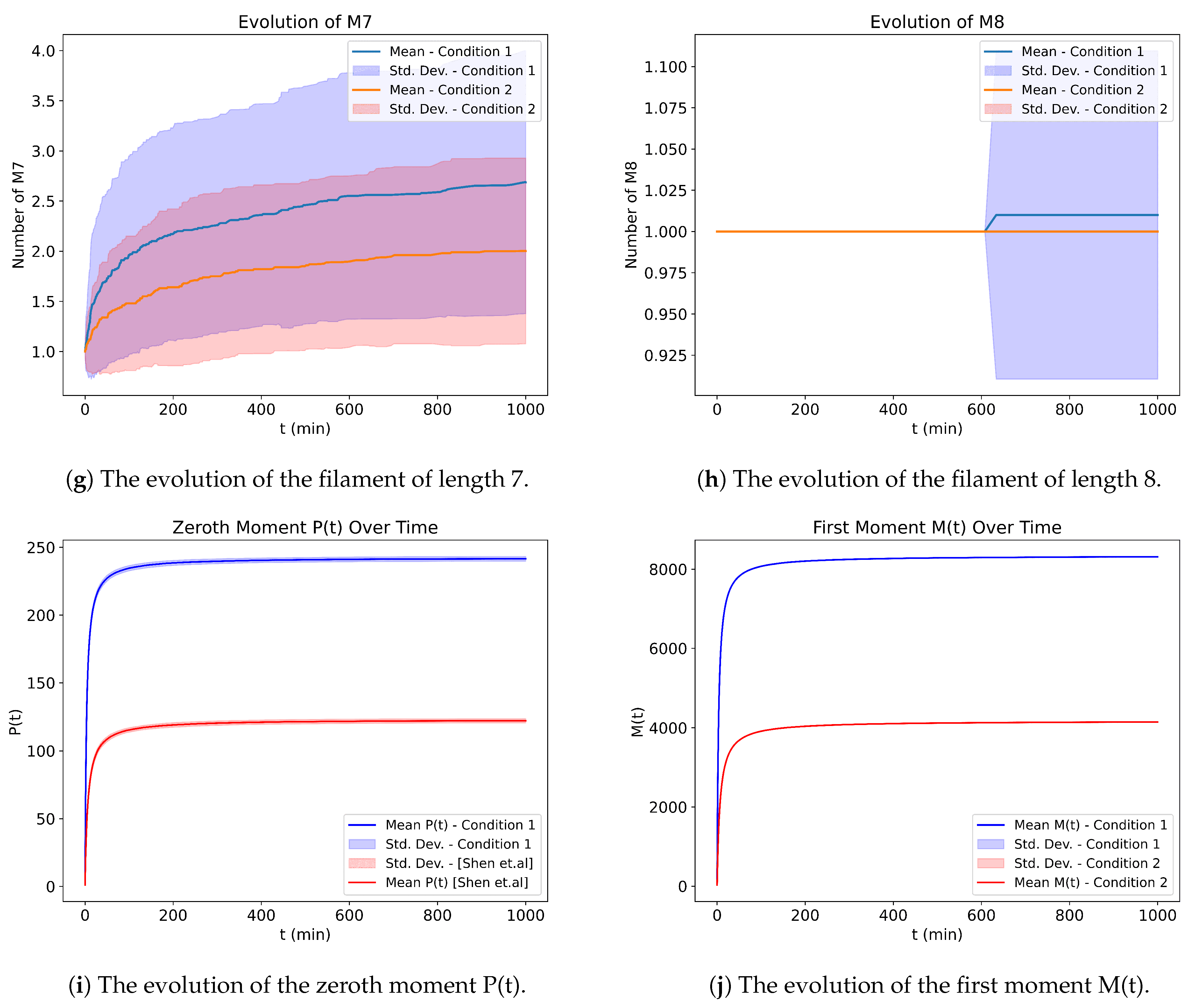
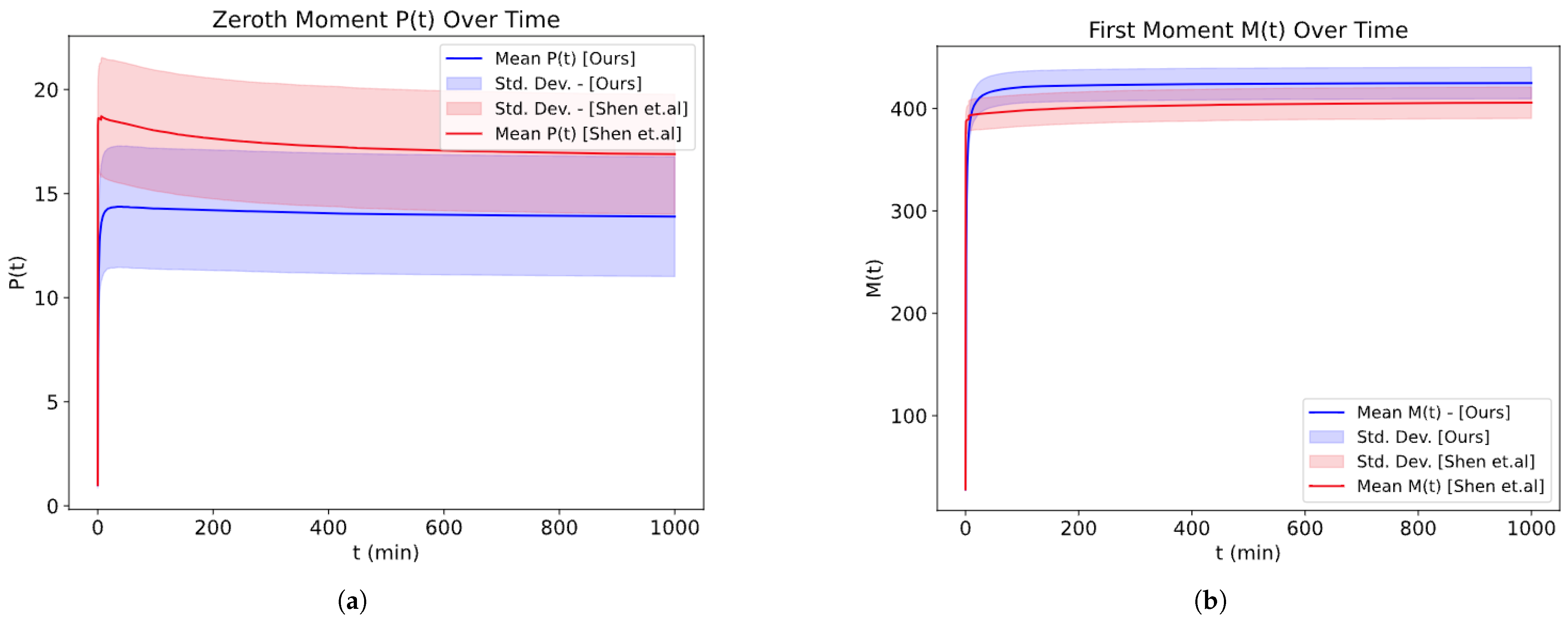
We further compare our work with [
6], and as can be seen in
Figure 14, we obtain a comparable result.
In
Figure 14, there are two separate simulations: one uses the parameters of [
6], whereas the other uses ours. Since [
6] uses different units for time and the protein mass, we cannot compare them exactly, so we changed the parameters a little bit to make them comparable. Even though the probability used is not exactly the same, the output looks comparable in terms of convergence. However, it should be noted that we used our own stoichiometry matrix, as the author of [
6] did not provide the stoichiometry used for their stochastic simulation.
6. Conclusions
Our study underscores the crucial role of stochastic modeling in capturing the nuances of biological processes, particularly in scenarios where deterministic models fall short. Deterministic approaches, while effective in representing the average dynamics of macroscopic experiments, often fail to encapsulate the inherent stochastic fluctuations that are especially pronounced in microscopic cellular processes. To bridge this gap, we employed stochastic modeling to simulate the dynamics of amyloid-beta aggregation in Alzheimer’s disease, a process fraught with randomness and complexity. The findings from our stochastic models yielded significant insights. We observed that the evolution of amyloid-beta filaments of varying lengths is intricately tied to the specific reaction rates employed in the model. This link underscores the sensitivity of the aggregation process to these rates, paving the way for a deeper understanding of the mechanisms driving amyloid-beta aggregation.
A particularly striking observation is the tendency of the system to converge towards filaments of a specific length, in our case, length six. This convergence suggests a dominance of certain reaction events over others, an aspect that could be pivotal in understanding the progression of Alzheimer’s disease at a molecular level. Moreover, the influence of reaction probabilities on the system’s final convergence and stability was evident, reinforcing the importance of these parameters in the aggregation process. Additionally, our exploration of random reaction propensities revealed marked differences in convergence patterns, highlighting the impact of stochastic variability on the system’s behavior. This aspect of our study points to the potential of stochastic models to reveal subtleties that deterministic models might overlook.
In conclusion, our comprehensive stochastic simulations not only enhance our current understanding of amyloid-beta aggregation but also set the stage for future research. They open new avenues for employing sophisticated stochastic modeling techniques to unravel the intricate dynamics of amyloid-beta aggregation. This comprehensive simulation could be instrumental in understanding and developing more effective strategies for combating diseases, such as Alzheimer’s, which are directly or indirectly related to the protein aggregation process by providing a more detailed understanding of the underlying molecular processes.