Atomic Force Microscopy as a Tool to Study Transport Phenomena in Biological Systems
Abstract
:1. Introduction
2. Principles of AFM
2.1. Probe and Cantilever Design
2.2. Detection of Cantilever Deflection
2.3. Force Measurement
2.4. Scanning Modes
2.5. Functionalization of AFM Tips
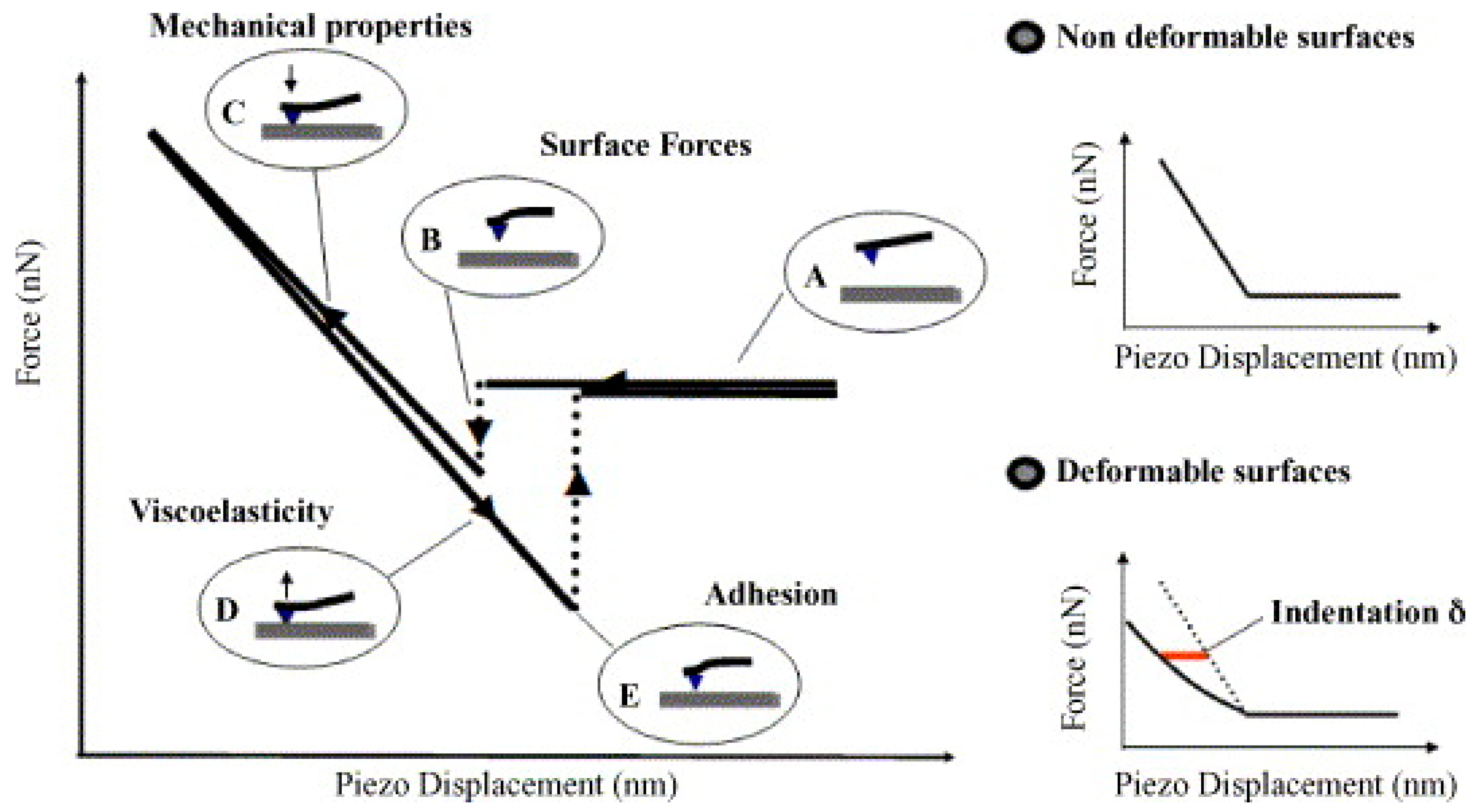
2.6. Investigating Mechanical Properties of Biological Materials
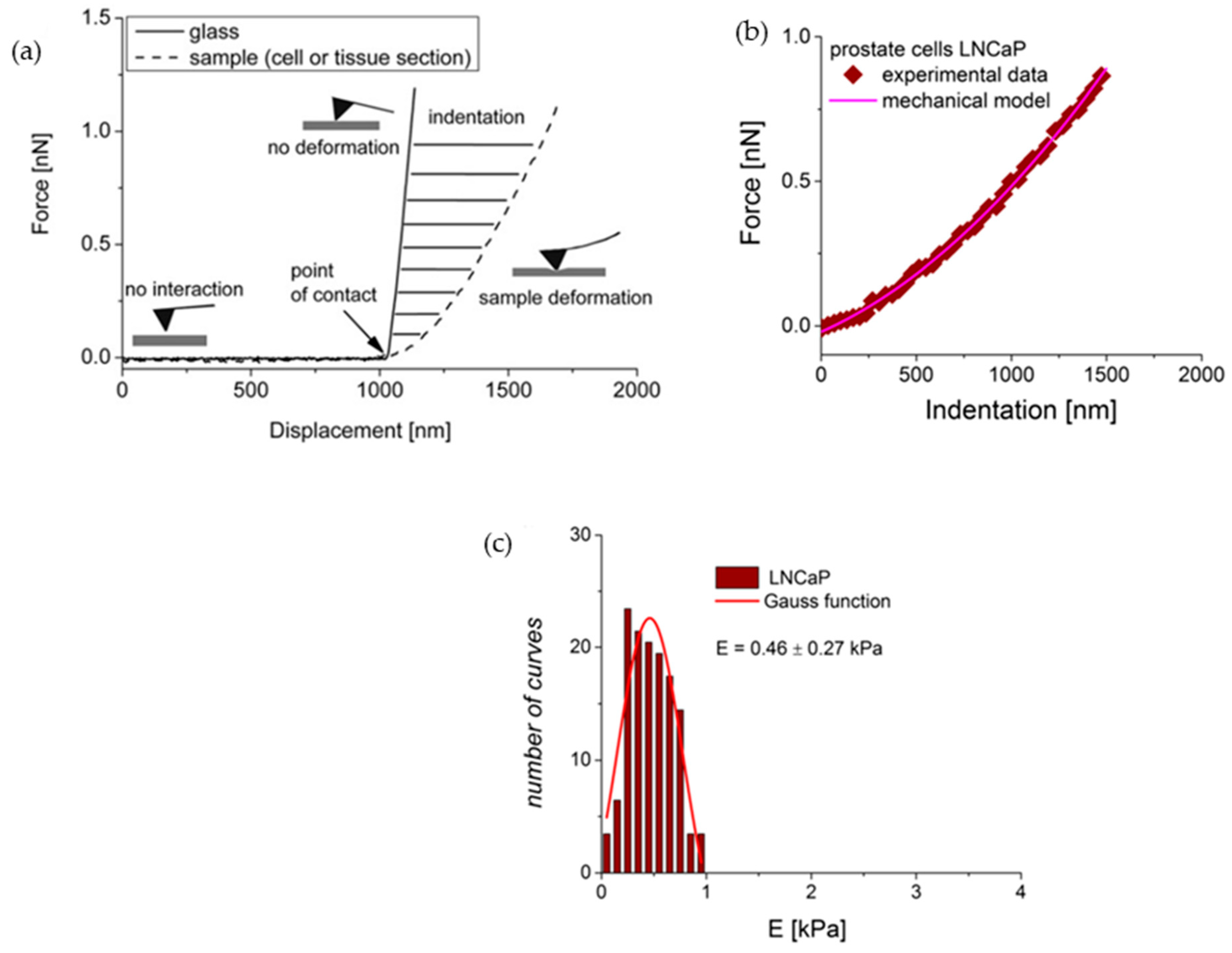
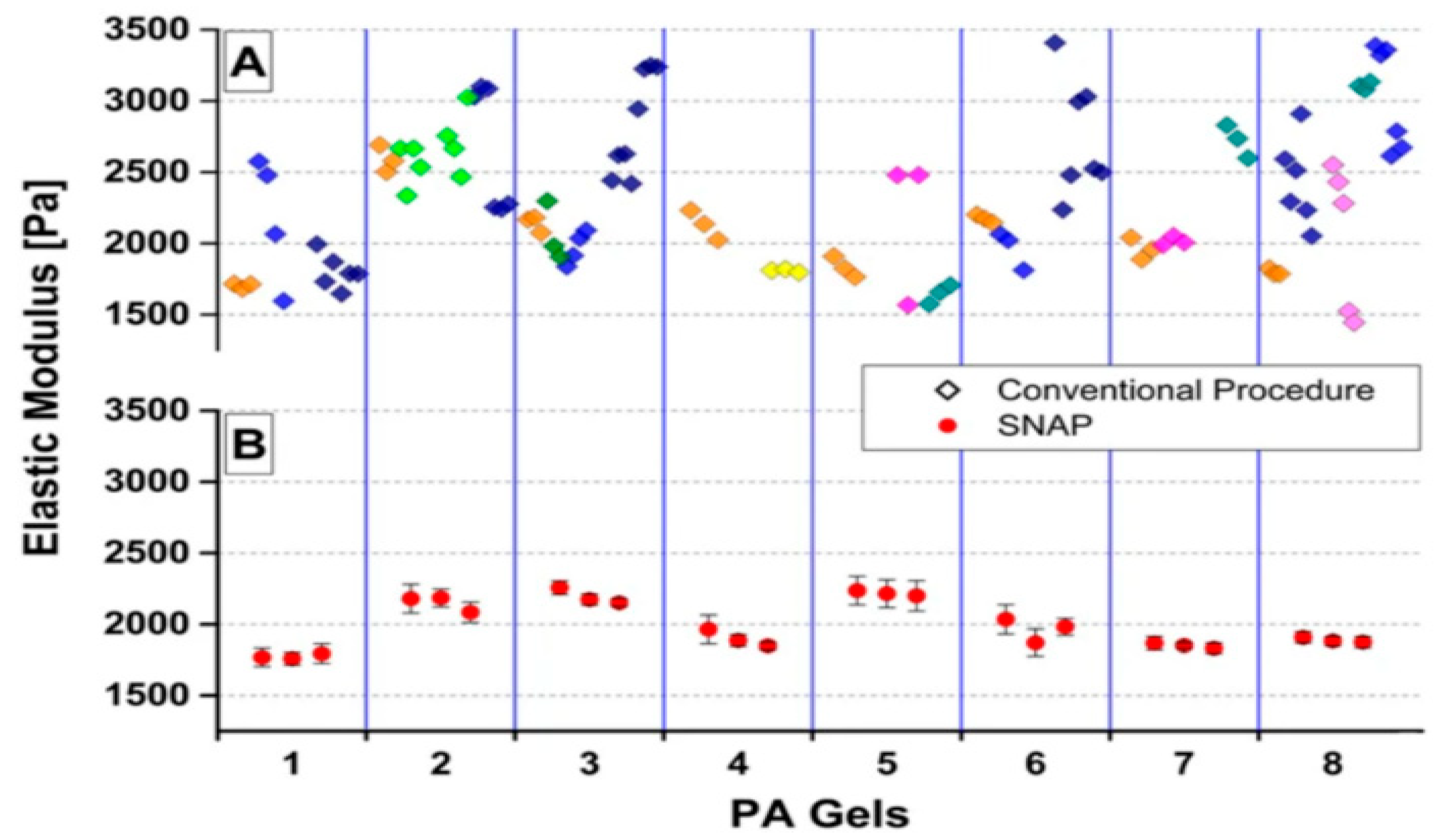
2.6.1. Elasticity and Young’s Modulus
2.6.2. Adhesion Force
2.7. Recent Advancement in AFM
3. Applications of AFM in the Study of Biomolecules
3.1. Protein
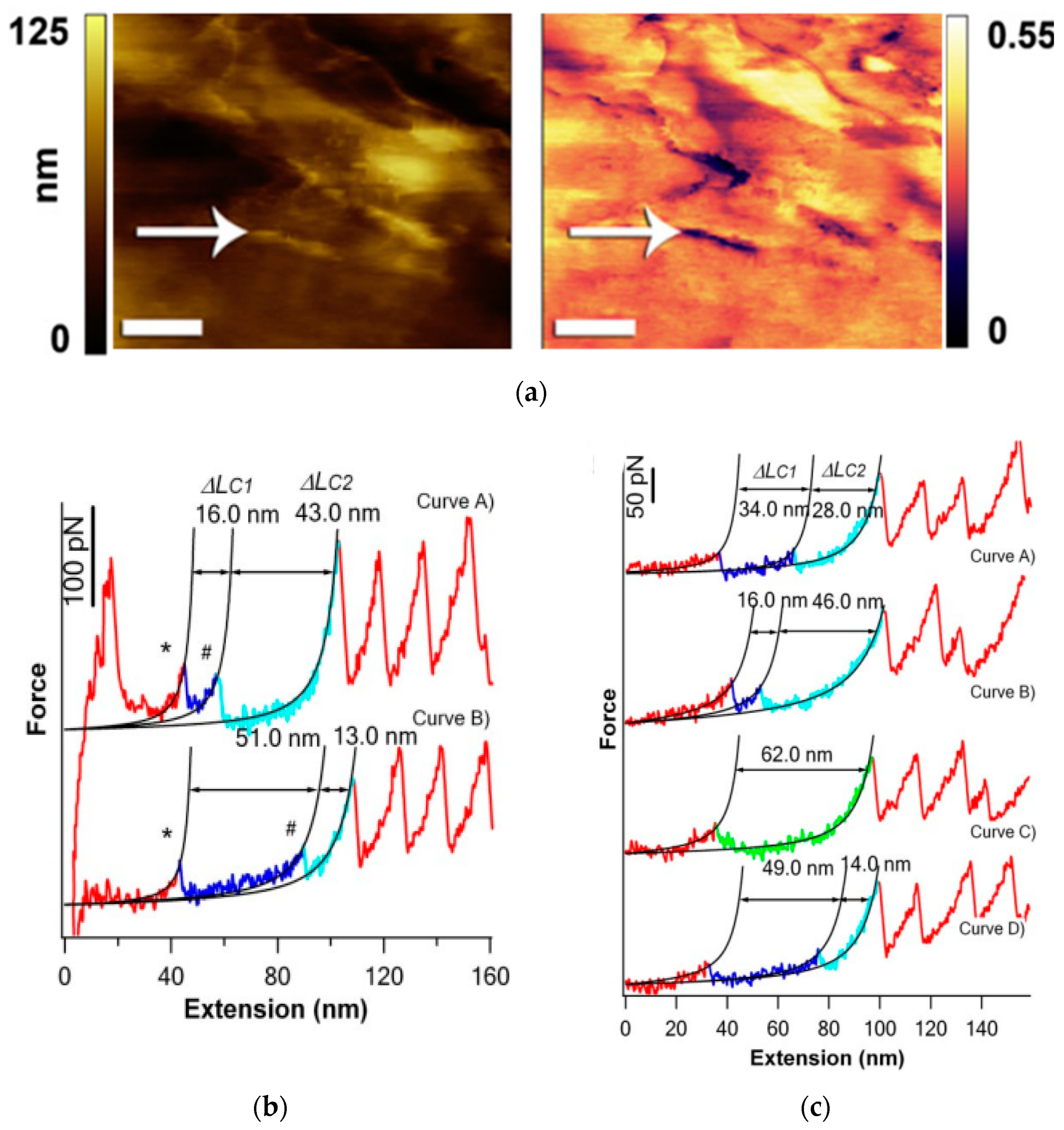
3.2. Nucleic Acids
3.3. Polysaccharides
3.4. Peptide
3.5. Enzymes
4. AFM for Studying Transport Phenomena in Biological Interactions
4.1. Unraveling the Mechanics of Antigen–Antibody Interactions
4.2. Aptamers: Versatile Tools for Molecular Probing
4.3. Understanding Interactions in Polymer Systems
4.4. Investigating Protein Interactions through AFM Manipulation
4.5. Carbohydrate Recognition in Biological Systems: Exploring Receptor Specificity
5. Unveiling the Mechanical Properties of Cells
6. Biomarkers
6.1. Cancer Biomarkers
6.2. Neurodegenerative Disease Biomarkers
6.3. Infectious Disease Biomarkers
6.4. Hematological Disorders
7. The Multifaceted Applications of AFM in Drug Therapy
7.1. Investigating Drug Delivery
7.2. Monitoring Treatment Efficiency
7.3. Investigating Binding Mechanisms
8. Conclusions and Perspective
Funding
Data Availability Statement
Conflicts of Interest
References
- Steffens, C.; Leite, F.L.; Bueno, C.C.; Manzoli, A.; De Paula Herrmann, P.S. Atomic force microscopy as a tool applied to nano/biosensors. Sensors 2012, 12, 8278–8300. [Google Scholar] [CrossRef] [PubMed]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic force microscope. Phys. Rev. Lett. 1986, 56, 930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zemła, J.; Danilkiewicz, J.; Orzechowska, B.; Pabijan, J.; Seweryn, S.; Lekka, M. Atomic force microscopy as a tool for assessing the cellular elasticity and adhesiveness to identify cancer cells and tissues. Semin. Cell Dev. Biol. 2018, 73, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Lyubchenko, Y.L.; Shlyakhtenko, L.S. AFM for analysis of structure and dynamics of DNA and protein–DNA complexes. Methods 2009, 47, 206–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schön, P. Imaging and force probing RNA by atomic force microscopy. Methods 2016, 103, 25–33. [Google Scholar] [CrossRef]
- Sajja, S.; Chandler, M.; Fedorov, D.; Kasprzak, W.K.; Lushnikov, A.; Viard, M.; Shah, A.; Dang, D.; Dahl, J.; Worku, B. Dynamic behavior of RNA nanoparticles analyzed by AFM on a mica/air interface. Langmuir 2018, 34, 15099–15108. [Google Scholar] [CrossRef]
- Connell, S.D.; Smith, D.A. The atomic force microscope as a tool for studying phase separation in lipid membranes. Mol. Membr. Biol. 2006, 23, 17–28. [Google Scholar] [CrossRef]
- Puech, P.-H.; Poole, K.; Knebel, D.; Muller, D.J. A new technical approach to quantify cell–cell adhesion forces by AFM. Ultramicroscopy 2006, 106, 637–644. [Google Scholar] [CrossRef]
- Allison, D.P.; Mortensen, N.P.; Sullivan, C.J.; Doktycz, M.J. Atomic force microscopy of biological samples. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 618–634. [Google Scholar] [CrossRef]
- Dufrêne, Y.F. Sticky microbes: Forces in microbial cell adhesion. Trends Microbiol. 2015, 23, 376–382. [Google Scholar] [CrossRef]
- Wagh, A.A.; Roan, E.; Chapman, K.E.; Desai, L.P.; Rendon, D.A.; Eckstein, E.C.; Waters, C.M. Localized elasticity measured in epithelial cells migrating at a wound edge using atomic force microscopy. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2008, 295, L54–L60. [Google Scholar] [CrossRef] [Green Version]
- Luo, Q.; Kuang, D.; Zhang, B.; Song, G. Cell stiffness determined by atomic force microscopy and its correlation with cell motility. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2016, 1860, 1953–1960. [Google Scholar] [CrossRef]
- Fang, T.; Alvelid, J.; Spratt, J.; Ambrosetti, E.; Testa, I.; Teixeira, A.I. Spatial regulation of T-cell signaling by programmed death-ligand 1 on wireframe DNA origami flat sheets. ACS Nano 2021, 15, 3441–3452. [Google Scholar] [CrossRef]
- Shi, X.; Zhang, X.; Xia, T.; Fang, X. Living cell study at the single-molecule and single-cell levels by atomic force microscopy. Nanomedicine 2012, 7, 1625–1637. [Google Scholar] [CrossRef]
- Zhang, Q.; Yang, J.; Tillieux, S.; Guo, Z.; Natividade, R.d.S.; Koehler, M.; Petitjean, S.; Cui, Z.; Alsteens, D. Stepwise Enzymatic-Dependent Mechanism of Ebola Virus Binding to Cell Surface Receptors Monitored by AFM. Nano Lett. 2022, 22, 1641–1648. [Google Scholar] [CrossRef]
- Neaves, K.J.; Cooper, L.P.; White, J.H.; Carnally, S.M.; Dryden, D.T.; Edwardson, J.M.; Henderson, R.M. Atomic force microscopy of the EcoKI Type I DNA restriction enzyme bound to DNA shows enzyme dimerization and DNA looping. Nucleic Acids Res. 2009, 37, 2053–2063. [Google Scholar] [CrossRef] [Green Version]
- Clark, C.G.; Sun, Z.; Meininger, G.A.; Potts, J.T. Atomic force microscopy to characterize binding properties of α7-containing nicotinic acetylcholine receptors on neurokinin-1 receptor-expressing medullary respiratory neurons. Exp. Physiol. 2013, 98, 415–424. [Google Scholar] [CrossRef] [Green Version]
- Bergler, F.; Fuentes, C.; Kadir, M.F.; Navarrete, C.; Supple, J.; Barrera, N.P.; Edwardson, J.M. Activation of P2X4 receptors induces an increase in the area of the extracellular region and a decrease in receptor mobility. FEBS Lett. 2020, 594, 4381–4389. [Google Scholar] [CrossRef]
- Murrough, J.W.; Huang, Y.; Hu, J.; Henry, S.; Williams, W.; Gallezot, J.-D.; Bailey, C.R.; Krystal, J.H.; Carson, R.E.; Neumeister, A. Reduced amygdala serotonin transporter binding in posttraumatic stress disorder. Biol. Psychiatry 2011, 70, 1033–1038. [Google Scholar] [CrossRef] [Green Version]
- Ruozi, B.; Tosi, G.; Leo, E.; Vandelli, M.A. Application of atomic force microscopy to characterize liposomes as drug and gene carriers. Talanta 2007, 73, 12–22. [Google Scholar] [CrossRef]
- Cardoso, F.L.; Brites, D.; Brito, M.A. Looking at the blood–brain barrier: Molecular anatomy and possible investigation approaches. Brain Res. Rev. 2010, 64, 328–363. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Peng, Z.; Seven, E.S.; Leblanc, R.M. Crossing the blood-brain barrier with nanoparticles. J. Control. Release 2018, 270, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Fotiadis, D. Atomic force microscopy for the study of membrane proteins. Curr. Opin. Biotechnol. 2012, 23, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Dhar-Chowdhury, P.; Malester, B.; Rajacic, P.; Coetzee, W. The regulation of ion channels and transporters by glycolytically derived ATP. Cell. Mol. Life Sci. 2007, 64, 3069–3083. [Google Scholar] [CrossRef] [PubMed]
- Alessandrini, A.; Facci, P. Phase transitions in supported lipid bilayers studied by AFM. Soft Matter 2014, 10, 7145–7164. [Google Scholar] [CrossRef]
- Viles, J.H. Imaging Amyloid-β Membrane Interactions; Ion-channel pores and Lipid-Bilayer Permeability in Alzheimer’s Disease. Angew. Chem. 2023, e202215785. [Google Scholar] [CrossRef]
- Yacoot, A.; Koenders, L. Aspects of scanning force microscope probes and their effects on dimensional measurement. J. Phys. D Appl. Phys. 2008, 41, 103001. [Google Scholar] [CrossRef]
- Meng, L.; Huang, T.; Wang, X.; Chen, S.; Yang, Z.; Ren, B. Gold-coated AFM tips for tip-enhanced Raman spectroscopy: Theoretical calculation and experimental demonstration. Opt. Express 2015, 23, 13804–13813. [Google Scholar] [CrossRef]
- Chan, K.A.; Kazarian, S.G. Tip-enhanced Raman mapping with top-illumination AFM. Nanotechnology 2011, 22, 175701. [Google Scholar] [CrossRef]
- Sadewasser, S.; Villanueva, G.; Plaza, J. Modified atomic force microscopy cantilever design to facilitate access of higher modes of oscillation. Rev. Sci. Instrum. 2006, 77, 073703. [Google Scholar] [CrossRef] [Green Version]
- Moore, S.I.; Ruppert, M.G.; Yong, Y.K. AFM cantilever design for multimode Q control: Arbitrary placement of higher order modes. IEEE/ASME Trans. Mechatron. 2020, 25, 1389–1397. [Google Scholar] [CrossRef]
- Joshi, M.; Rao, V.R.; Mukherji, S. A novel technique for microfabrication of ultra-thin affinity cantilevers for characterization with an AFM. J. Micromechanics Microengineering 2010, 20, 125007. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, Y.; Grierson, D.S.; Sridharan, K.; Shao, Y.; Jacobs, T.D.; Falk, M.L.; Carpick, R.W.; Turner, K.T. Tribochemical wear of diamond-like carbon-coated atomic force microscope tips. ACS Appl. Mater. Interfaces 2017, 9, 35341–35348. [Google Scholar] [CrossRef]
- Yu, F.; Liu, J.; Zhang, X.; Lin, A.-L.; Khan, N.; Pan, Y.; Gao, N.; Zou, Q.; Jeon, J. Design, fabrication, and characterization of polymer-based cantilever probes for atomic force microscopes. J. Vac. Sci. Technol. B Nanotechnol. Microelectron. Mater. Process. Meas. Phenom. 2016, 34, 06KI01. [Google Scholar] [CrossRef]
- Wilson, N.R.; Macpherson, J.V. Carbon nanotube tips for atomic force microscopy. Nat. Nanotechnol. 2009, 4, 483–491. [Google Scholar] [CrossRef]
- Meyer, G.; Amer, N.M. Novel optical approach to atomic force microscopy. Appl. Phys. Lett. 1988, 53, 1045–1047. [Google Scholar] [CrossRef]
- Tortonese, M.; Barrett, R.; Quate, C. Atomic resolution with an atomic force microscope using piezoresistive detection. Appl. Phys. Lett. 1993, 62, 834–836. [Google Scholar] [CrossRef]
- Villanueva, G.; Plaza, J.; Montserrat, J.; Perez-Murano, F.; Bausells, J. Crystalline silicon cantilevers for piezoresistive detection of biomolecular forces. Microelectron. Eng. 2008, 85, 1120–1123. [Google Scholar] [CrossRef]
- Shiba, Y.; Ono, T.; Minami, K.; Esashi, M. Capacitive AFM probe for high speed imaging. IEEJ Trans. Sens. Micromachines 1998, 118, 647–651. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, S.; Teran Arce, F.; Lal, R. Potential role of atomic force microscopy in systems biology. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011, 3, 702–716. [Google Scholar] [CrossRef] [Green Version]
- Gaboriaud, F.; Dufrêne, Y.F. Atomic force microscopy of microbial cells: Application to nanomechanical properties, surface forces and molecular recognition forces. Colloids Surf. B Biointerfaces 2007, 54, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Weymouth, A.J. Non-contact lateral force microscopy. J. Phys. Condens. Matter 2017, 29, 323001. [Google Scholar] [CrossRef] [PubMed]
- Ge, G.; Han, D.; Lin, D.; Chu, W.; Sun, Y.; Jiang, L.; Ma, W.; Wang, C. MAC mode atomic force microscopy studies of living samples, ranging from cells to fresh tissue. Ultramicroscopy 2007, 107, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Mohn, F.; Schuler, B.; Gross, L.; Meyer, G. Different tips for high-resolution atomic force microscopy and scanning tunneling microscopy of single molecules. Appl. Phys. Lett. 2013, 102, 073109. [Google Scholar] [CrossRef]
- Sahin, O.; Magonov, S.; Su, C.; Quate, C.F.; Solgaard, O. An atomic force microscope tip designed to measure time-varying nanomechanical forces. Nat. Nanotechnol. 2007, 2, 507–514. [Google Scholar] [CrossRef]
- Senapati, S.; Manna, S.; Lindsay, S.; Zhang, P. Application of catalyst-free click reactions in attaching affinity molecules to tips of atomic force microscopy for detection of protein biomarkers. Langmuir 2013, 29, 14622–14630. [Google Scholar] [CrossRef] [Green Version]
- Daza, R.; Colchero, L.; Corregidor, D.; Elices, M.; Guinea, G.V.; Rojo, F.J.; Pérez-Rigueiro, J. Functionalization of atomic force microscopy cantilevers and tips by activated vapour silanization. Appl. Surf. Sci. 2019, 484, 1141–1148. [Google Scholar] [CrossRef]
- Girish, C.; Binulal, N.; Anitha, V.; Nair, S.; Mony, U.; Prasanth, R. Atomic force microscopic study of folate receptors in live cells with functionalized tips. Appl. Phys. Lett. 2009, 95, 223703. [Google Scholar] [CrossRef]
- Lekka, M.; Gil, D.; Pogoda, K.; Dulińska-Litewka, J.; Jach, R.; Gostek, J.; Klymenko, O.; Prauzner-Bechcicki, S.; Stachura, Z.; Wiltowska-Zuber, J. Cancer cell detection in tissue sections using AFM. Arch. Biochem. Biophys. 2012, 518, 151–156. [Google Scholar] [CrossRef]
- Lekka, M. Discrimination between normal and cancerous cells using AFM. Bionanoscience 2016, 6, 65–80. [Google Scholar] [CrossRef] [Green Version]
- Schillers, H.; Rianna, C.; Schäpe, J.; Luque, T.; Doschke, H.; Wälte, M.; Uriarte, J.J.; Campillo, N.; Michanetzis, G.; Bobrowska, J. Standardized nanomechanical atomic force microscopy procedure (SNAP) for measuring soft and biological samples. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Xu, B. Transition model for ricin-aptamer interactions with multiple pathways and energy barriers. Phys. Rev. E 2014, 89, 022720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Guo, C.; Chen, G.; Park, B.; Xu, B. Following aptamer–ricin specific binding by single molecule recognition and force spectroscopy measurements. Chem. Commun. 2012, 48, 1644–1646. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.D.; Erickson, B.W.; Grossenbacher, J.; Brugger, J.; Nievergelt, A.; Fantner, G.E. Harnessing the damping properties of materials for high-speed atomic force microscopy. Nat. Nanotechnol. 2016, 11, 147–151. [Google Scholar] [CrossRef] [Green Version]
- Uchihashi, T.; Scheuring, S. Applications of high-speed atomic force microscopy to real-time visualization of dynamic biomolecular processes. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2018, 1862, 229–240. [Google Scholar] [CrossRef]
- Puppulin, L.; Kanayama, D.; Terasaka, N.; Sakai, K.; Kodera, N.; Umeda, K.; Sumino, A.; Marchesi, A.; Weilin, W.; Tanaka, H. Macrocyclic Peptide-Conjugated Tip for Fast and Selective Molecular Recognition Imaging by High-Speed Atomic Force Microscopy. ACS Appl. Mater. Interfaces 2021, 13, 54817–54829. [Google Scholar] [CrossRef]
- Yang, Q.; Ma, Q.; Herum, K.M.; Wang, C.; Patel, N.; Lee, J.; Wang, S.; Yen, T.M.; Wang, J.; Tang, H. Array atomic force microscopy for real-time multiparametric analysis. Proc. Natl. Acad. Sci. USA 2019, 116, 5872–5877. [Google Scholar] [CrossRef] [Green Version]
- Park, B.; Lee, S.; Kwon, J.; Kim, W.; Jung, S.; Kim, C. Dual-pulse photoactivated atomic force microscopy. Sci. Rep. 2021, 11, 17097. [Google Scholar] [CrossRef]
- Cheng, H.; Yu, J.; Wang, Z.; Ma, P.; Guo, C.; Wang, B.; Zhong, W.; Xu, B. Details of Single-Molecule Force Spectroscopy Data Decoded by a Network-Based Automatic Clustering Algorithm. J. Phys. Chem. B 2021, 125, 9660–9667. [Google Scholar] [CrossRef]
- Creasey, R.; Sharma, S.; Gibson, C.T.; Craig, J.E.; Ebner, A.; Becker, T.; Hinterdorfer, P.; Voelcker, N.H. Atomic force microscopy-based antibody recognition imaging of proteins in the pathological deposits in pseudoexfoliation syndrome. Ultramicroscopy 2011, 111, 1055–1061. [Google Scholar] [CrossRef]
- Best, R.B.; Li, B.; Steward, A.; Daggett, V.; Clarke, J. Can non-mechanical proteins withstand force? Stretching barnase by atomic force microscopy and molecular dynamics simulation. Biophys. J. 2001, 81, 2344–2356. [Google Scholar] [CrossRef] [Green Version]
- Kawakami, M.; Smith, D. A new atomic force microscope force ramp technique using digital force feedback control reveals mechanically weak protein unfolding events. Nanotechnology 2008, 19, 495704. [Google Scholar] [CrossRef]
- Peng, Q.; Li, H. Atomic force microscopy reveals parallel mechanical unfolding pathways of T4 lysozyme: Evidence for a kinetic partitioning mechanism. Proc. Natl. Acad. Sci. USA 2008, 105, 1885–1890. [Google Scholar] [CrossRef]
- Mahmood, I.A.; Moheimani, S.R.; Bhikkaji, B. A new scanning method for fast atomic force microscopy. IEEE Trans. Nanotechnol. 2009, 10, 203–216. [Google Scholar] [CrossRef]
- Hansma, H.G.; Bezanilla, M.; Zenhausern, F.; Adrian, M.; Sinsheimer, R.L. Atomic force microscopy of DNA in aqueous solutions. Nucleic Acids Res. 1993, 21, 505–512. [Google Scholar] [CrossRef] [Green Version]
- Zhu, R.; Howorka, S.; Pröll, J.; Kienberger, F.; Preiner, J.; Hesse, J.; Ebner, A.; Pastushenko, V.P.; Gruber, H.J.; Hinterdorfer, P. Nanomechanical recognition measurements of individual DNA molecules reveal epigenetic methylation patterns. Nat. Nanotechnol. 2010, 5, 788–791. [Google Scholar] [CrossRef] [Green Version]
- Shim, W.C.; Woo, S.; Park, J.W. Nanoscale Force-Mapping-Based Quantification of Low-Abundance Methylated DNA. Nano Lett. 2022, 22, 1324–1330. [Google Scholar] [CrossRef]
- Strobl, K.; Mateu, M.; de Pablo, P.J. Exploring nucleic acid condensation and release from individual parvovirus particles with different physicochemical cues. Virology 2023, 581, 1–7. [Google Scholar] [CrossRef]
- Abu-Lail, N.; Camesano, T. Polysaccharide properties probed with atomic force microscopy. J. Microsc. 2003, 212, 217–238. [Google Scholar] [CrossRef]
- Guo, C.; Fan, X.; Qiu, H.; Xiao, W.; Wang, L.; Xu, B. High-resolution probing heparan sulfate–antithrombin interaction on a single endothelial cell surface: Single-molecule AFM studies. Phys. Chem. Chem. Phys. 2015, 17, 13301–13306. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Xi, N.; Wang, Y.; Liu, L. Nanoscale multiparametric imaging of peptide-assembled nanofibrillar hydrogels by atomic force microscopy. IEEE Trans. Nanotechnol. 2019, 18, 315–328. [Google Scholar] [CrossRef]
- Gaspar, D.; Freire, J.M.; Pacheco, T.R.; Barata, J.T.; Castanho, M.A. Apoptotic human neutrophil peptide-1 anti-tumor activity revealed by cellular biomechanics. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2015, 1853, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhang, M.; Alexander Reese, R.; Zhang, H.; Xu, B. Real-time single molecular study of a pretreated cellulose hydrolysis mode and individual enzyme movement. Biotechnol. Biofuels 2016, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Wu, S.-C.; Zhou, W.; Xu, B. Imaging and measuring single-molecule interaction between a carbohydrate-binding module and natural plant cell wall cellulose. J. Phys. Chem. B 2012, 116, 9949–9956. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, B.; Xu, B. Measurements of single molecular affinity interactions between carbohydrate-binding modules and crystalline cellulose fibrils. Phys. Chem. Chem. Phys. 2013, 15, 6508–6515. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, B.; Xu, B. Mapping single molecular binding kinetics of carbohydrate-binding module with crystalline cellulose by atomic force microscopy recognition imaging. J. Phys. Chem. B 2014, 118, 6714–6720. [Google Scholar] [CrossRef]
- Kolodziejczyk, A.; Jakubowska, A.; Kucinska, M.; Wasiak, T.; Komorowski, P.; Makowski, K.; Walkowiak, B. Sensing of silver nanoparticles on/in endothelial cells using atomic force spectroscopy. J. Mol. Recognit. 2018, 31, e2723. [Google Scholar] [CrossRef]
- Dong, M.; Sahin, O. A nanomechanical interface to rapid single-molecule interactions. Nat. Commun. 2011, 2, 247. [Google Scholar] [CrossRef] [Green Version]
- Browning-Kelley, M.; Wadu-Mesthrige, K.; Hari, V.; Liu, G. Atomic force microscopic study of specific antigen/antibody binding. Langmuir 1997, 13, 343–350. [Google Scholar] [CrossRef]
- Hu, J.; Gao, M.; Wang, Z.; Chen, Y.; Song, Z.; Xu, H. Direct imaging of antigen–antibody binding by atomic force microscopy. Appl. Nanosci. 2021, 11, 293–300. [Google Scholar] [CrossRef]
- Hinterdorfer, P.; Baumgartner, W.; Gruber, H.J.; Schilcher, K.; Schindler, H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA 1996, 93, 3477–3481. [Google Scholar] [CrossRef] [PubMed]
- Berquand, A.; Xia, N.; Castner, D.G.; Clare, B.H.; Abbott, N.L.; Dupres, V.; Adriaensen, Y.; Dufrêne, Y.F. Antigen binding forces of single antilysozyme Fv fragments explored by atomic force microscopy. Langmuir 2005, 21, 5517–5523. [Google Scholar] [CrossRef] [PubMed]
- Kienberger, F.; Kada, G.; Mueller, H.; Hinterdorfer, P. Single molecule studies of antibody–antigen interaction strength versus intra-molecular antigen stability. J. Mol. Biol. 2005, 347, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zeng, G.; Chen, S.S.; Feng, Q.; Chen, Z.W. AFM force measurements of the gp120–sCD4 and gp120 or CD4 antigen–antibody interactions. Biochem. Biophys. Res. Commun. 2011, 407, 301–306. [Google Scholar] [CrossRef] [Green Version]
- Hianik, T. Aptamer-based biosensors. In Encyclopedia of Interfacial Chemistry; Wandelt, K., Ed.; Elsevier: Cambridge, MA, USA, 2018; Volume 7, pp. 11–19. [Google Scholar]
- Wang, C.; Yadavalli, V.K. Spatial recognition and mapping of proteins using DNA aptamers. Nanotechnology 2014, 25, 455101. [Google Scholar] [CrossRef]
- Junior, B.B.; Batistuti, M.R.; Pereira, A.S.; de Sousa Russo, E.M.; Mulato, M. Electrochemical aptasensor for NS1 detection: Towards a fast dengue biosensor. Talanta 2021, 233, 122527. [Google Scholar] [CrossRef]
- Leitner, M.; Poturnayova, A.; Lamprecht, C.; Weich, S.; Snejdarkova, M.; Karpisova, I.; Hianik, T.; Ebner, A. Characterization of the specific interaction between the DNA aptamer sgc8c and protein tyrosine kinase-7 receptors at the surface of T-cells by biosensing AFM. Anal. Bioanal. Chem. 2017, 409, 2767–2776. [Google Scholar] [CrossRef] [Green Version]
- Poturnayová, A.; Buríková, M.; Bízik, J.; Hianik, T. DNA aptamers in the detection of leukemia cells by the thickness shear mode acoustics method. ChemPhysChem 2019, 20, 545–554. [Google Scholar] [CrossRef] [Green Version]
- van Galen, M.; Kaniraj, J.P.; Albada, B.; Sprakel, J. Single-Molecule Force Spectroscopy of a Tetraaryl Succinonitrile Mechanophore. J. Phys. Chem. C 2022, 126, 1215–1221. [Google Scholar] [CrossRef]
- Qin, C.; Clarke, K.; Li, K. Interactive forces between lignin and cellulase as determined by atomic force microscopy. Biotechnol. Biofuels 2014, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Sahin, O. Imaging and three-dimensional reconstruction of chemical groups inside a protein complex using atomic force microscopy. Nat. Nanotechnol. 2015, 10, 264–269. [Google Scholar] [CrossRef] [Green Version]
- Viani, M.B.; Pietrasanta, L.I.; Thompson, J.B.; Chand, A.; Gebeshuber, I.C.; Kindt, J.H.; Richter, M.; Hansma, H.G.; Hansma, P.K. Probing protein–protein interactions in real time. Nat. Struct. Biol. 2000, 7, 644–647. [Google Scholar] [CrossRef]
- Zapotoczny, B.; Braet, F.; Wisse, E.; Lekka, M.; Szymonski, M. Biophysical nanocharacterization of liver sinusoidal endothelial cells through atomic force microscopy. Biophys. Rev. 2020, 12, 625–636. [Google Scholar] [CrossRef]
- Bonanni, B.; Kamruzzahan, A.; Bizzarri, A.; Rankl, C.; Gruber, H.; Hinterdorfer, P.; Cannistraro, S. Single molecule recognition between cytochrome C 551 and gold-immobilized azurin by force spectroscopy. Biophys. J. 2005, 89, 2783–2791. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, H.; Suzuki, Y.; Yokokawa, M.; Takeyasu, K.; Wyman, C. Protein–DNA interactions in high speed AFM: Single molecule diffusion analysis of human RAD54. Integr. Biol. 2011, 3, 1127–1134. [Google Scholar] [CrossRef] [Green Version]
- Touhami, A.; Hoffmann, B.; Vasella, A.; Denis, F.A.; Dufrêne, Y.F. Probing specific lectin-carbohydrate interactions using atomic force microscopy imaging and force measurements. Langmuir 2003, 19, 1745–1751. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, G.; Kumar, R.; Xu, B. Mapping out the structural changes of natural and pretreated plant cell wall surfaces by atomic force microscopy single molecular recognition imaging. Biotechnol. Biofuels 2013, 6, 147. [Google Scholar] [CrossRef] [Green Version]
- Faria, E.C.; Ma, N.; Gazi, E.; Gardner, P.; Brown, M.; Clarke, N.W.; Snook, R.D. Measurement of elastic properties of prostate cancer cells using AFM. Analyst 2008, 133, 1498–1500. [Google Scholar] [CrossRef]
- Li, M.; Liu, L.; Xi, N.; Wang, Y.; Dong, Z.; Xiao, X.; Zhang, W. Atomic force microscopy imaging and mechanical properties measurement of red blood cells and aggressive cancer cells. Sci. China Life Sci. 2012, 55, 968–973. [Google Scholar] [CrossRef] [Green Version]
- Ansardamavandi, A.; Tafazzoli-Shadpour, M.; Omidvar, R.; Nili, F. An AFM-based nanomechanical study of ovarian tissues with pathological conditions. Int. J. Nanomed. 2020, 4333–4350. [Google Scholar] [CrossRef]
- Giannetti, A.; Revilloud, J.; Verdier, C. Mechanical properties of 3D tumor spheroids measured by AFM. Comput. Methods Biomech. Biomed. Eng. 2020, 23, S125–S127. [Google Scholar] [CrossRef]
- Lekka, M.; Fornal, M.; Pyka-Fościak, G.; Lebed, K.; Wizner, B.; Grodzicki, T.; Styczeń, J. Erythrocyte stiffness probed using atomic force microscope. Biorheology 2005, 42, 307–317. [Google Scholar] [PubMed]
- Li, M.; Liu, L.; Xi, N.; Wang, Y.; Dong, Z.; Li, G.; Xiao, X.; Zhang, W. Measuring the physical properties of the lymphoma cells using atomic force microscopy. In Proceedings of the 2010 IEEE Nanotechnology Materials and Devices Conference, Monterey, CA, USA, 12–15 October 2010; pp. 310–314. [Google Scholar]
- Haase, K.; Pelling, A.E. Investigating cell mechanics with atomic force microscopy. J. R. Soc. Interface 2015, 12, 20140970. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Park, B.; Kwon, Y.; Xu, B. Determining the elastic properties of aptamer-ricin single molecule multiple pathway interactions. Appl. Phys. Lett. 2014, 104, 193702. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Liu, L.; Xiao, X.; Xi, N.; Wang, Y. Effects of methotrexate on the viscoelastic properties of single cells probed by atomic force microscopy. J. Biol. Phys. 2016, 42, 551–569. [Google Scholar] [CrossRef] [Green Version]
- Wei, F.; Yang, H.; Liu, L.; Li, G. A novel approach for extracting viscoelastic parameters of living cells through combination of inverse finite element simulation and Atomic Force Microscopy. Comput. Methods Biomech. Biomed. Eng. 2017, 20, 373–384. [Google Scholar] [CrossRef]
- Li, Q.; Lee, G.Y.; Ong, C.N.; Lim, C.T. AFM indentation study of breast cancer cells. Biochem. Biophys. Res. Commun. 2008, 374, 609–613. [Google Scholar] [CrossRef]
- Xu, W.; Mezencev, R.; Kim, B.; Wang, L.; McDonald, J.; Sulchek, T. Cell stiffness is a biomarker of the metastatic potential of ovarian cancer cells. PLoS One 2012, 7, e46609. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Xiao, X.; Liu, L.; Xi, N.; Wang, Y. Rapid recognition and functional analysis of membrane proteins on human cancer cells using atomic force microscopy. J. Immunol. Methods 2016, 436, 41–49. [Google Scholar] [CrossRef]
- Paul, D.; Roy, A.; Nandy, A.; Datta, B.; Borar, P.; Pal, S.K.; Senapati, D.; Rakshit, T. Identification of biomarker hyaluronan on colon cancer extracellular vesicles using correlative AFM and spectroscopy. J. Phys. Chem. Lett. 2020, 11, 5569–5576. [Google Scholar] [CrossRef]
- Park, K.; Lonsberry, G.E.; Gearing, M.; Levey, A.I.; Desai, J.P. Viscoelastic properties of human autopsy brain tissues as biomarkers for Alzheimer′s diseases. IEEE Trans. Biomed. Eng. 2018, 66, 1705–1713. [Google Scholar] [CrossRef]
- Nirmalraj, P.N.; Schneider, T.; Felbecker, A. Spatial organization of protein aggregates on red blood cells as physical biomarkers of Alzheimer’s disease pathology. Sci. Adv. 2021, 7, eabj2137. [Google Scholar] [CrossRef]
- Strijkova-Kenderova, V.; Todinova, S.; Andreeva, T.; Bogdanova, D.; Langari, A.; Danailova, A.; Krumova, S.; Zlatareva, E.; Kalaydzhiev, N.; Milanov, I. Morphometry and stiffness of red blood cells—Signatures of neurodegenerative diseases and aging. Int. J. Mol. Sci. 2022, 23, 227. [Google Scholar] [CrossRef]
- Patil, S.; Martinez, N.F.; Lozano, J.R.; Garcia, R. Force microscopy imaging of individual protein molecules with sub-pico Newton force sensitivity. J. Mol. Recognit. Interdiscip. J. 2007, 20, 516–523. [Google Scholar] [CrossRef]
- Gilbert, Y.; Deghorain, M.; Wang, L.; Xu, B.; Pollheimer, P.D.; Gruber, H.J.; Errington, J.; Hallet, B.; Haulot, X.; Verbelen, C. Single-molecule force spectroscopy and imaging of the vancomycin/D-Ala-D-Ala interaction. Nano Lett. 2007, 7, 796–801. [Google Scholar] [CrossRef]
- Rankl, C.; Kienberger, F.; Wildling, L.; Wruss, J.; Gruber, H.J.; Blaas, D.; Hinterdorfer, P. Multiple receptors involved in human rhinovirus attachment to live cells. Proc. Natl. Acad. Sci. USA 2008, 105, 17778–17783. [Google Scholar] [CrossRef]
- Fantner, G.E.; Barbero, R.J.; Gray, D.S.; Belcher, A.M. Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat. Nanotechnol. 2010, 5, 280–285. [Google Scholar] [CrossRef] [Green Version]
- Faustino, A.F.; Carvalho, F.A.; Martins, I.C.; Castanho, M.A.; Mohana-Borges, R.; Almeida, F.C.; Da Poian, A.T.; Santos, N.C. Dengue virus capsid protein interacts specifically with very low-density lipoproteins. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 247–255. [Google Scholar] [CrossRef]
- Herman-Bausier, P.; Dufrêne, Y.F. Atomic force microscopy reveals a dual collagen-binding activity for the staphylococcal surface protein SdrF. Mol. Microbiol. 2016, 99, 611–621. [Google Scholar] [CrossRef]
- Newton, R.; Delguste, M.; Koehler, M.; Dumitru, A.C.; Laskowski, P.R.; Müller, D.J.; Alsteens, D. Combining confocal and atomic force microscopy to quantify single-virus binding to mammalian cell surfaces. Nat. Protoc. 2017, 12, 2275–2292. [Google Scholar] [CrossRef]
- Kiss, B.; Kis, Z.; Pályi, B.; Kellermayer, M.S. Topography, spike dynamics, and nanomechanics of individual native SARS-CoV-2 virions. Nano Lett. 2021, 21, 2675–2680. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Jin, H.; Lu, Y.; Wang, Q.; Pan, Y.; Cai, J.; Wang, H. Detection of erythrocytes in patient with elliptocytosis complicating ITP using atomic force microscopy. Micron 2011, 42, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.A.; Connell, S.; Miltenberger-Miltenyi, G.; Pereira, S.V.; Tavares, A.; Ariëns, R.A.; Santos, N.C. Atomic force microscopy-based molecular recognition of a fibrinogen receptor on human erythrocytes. ACS Nano 2010, 4, 4609–4620. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, J. Detection of erythrocytes in patients with Waldenström macroglobulinemia using atomic force microscopy. Acta Biochim. Biophys. Sin. 2014, 46, 420–425. [Google Scholar] [CrossRef]
- Li, M.; Liu, L.; Xiao, X.; Xi, N.; Wang, Y. Viscoelastic properties measurement of human lymphocytes by atomic force microscopy based on magnetic beads cell isolation. IEEE Trans. NanoBiosci. 2016, 15, 398–411. [Google Scholar] [CrossRef]
- Jazvinšćak Jembrek, M.; Vlainić, J.; Čadež, V.; Šegota, S. Atomic force microscopy reveals new biophysical markers for monitoring subcellular changes in oxidative injury: Neuroprotective effects of quercetin at the nanoscale. PLoS ONE 2018, 13, e0200119. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, M.; Li, X.; Li, M.; Xing, X.; Liu, L. Nanomechanical signatures of extracellular vesicles from hematologic cancer patients unraveled by atomic force microscopy for liquid biopsy. Nano Lett. 2023, 23, 1591–1599. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, L.; Pan, J. Probing the binding of the flavonoid diosmetin to human serum albumin by multispectroscopic techniques. J. Agric. Food Chem. 2012, 60, 2721–2729. [Google Scholar] [CrossRef]
- Domingues, M.M.; Silva, P.M.; Franquelim, H.G.; Carvalho, F.A.; Castanho, M.A.; Santos, N.C. Antimicrobial protein rBPI21-induced surface changes on Gram-negative and Gram-positive bacteria. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 543–551. [Google Scholar] [CrossRef]
- Rajendran, A.; Endo, M.; Hidaka, K.; Teulade-Fichou, M.-P.; Mergny, J.-L.; Sugiyama, H. Small molecule binding to a G-hairpin and a G-triplex: A new insight into anticancer drug design targeting G-rich regions. Chem. Commun. 2015, 51, 9181–9184. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Xiao, X.; Liu, L.; Xi, N.; Wang, Y. Nanoscale quantifying the effects of targeted drug on chemotherapy in lymphoma treatment using atomic force microscopy. IEEE Trans. Biomed. Eng. 2015, 63, 2187–2199. [Google Scholar] [CrossRef]
- Ren, J.; Huang, H.; Liu, Y.; Zheng, X.; Zou, Q. An atomic force microscope study revealed two mechanisms in the effect of anticancer drugs on rate-dependent Young’s modulus of human prostate cancer cells. PLoS ONE 2015, 10, e0126107. [Google Scholar] [CrossRef]
- Alhalhooly, L.; Mamnoon, B.; Kim, J.; Mallik, S.; Choi, Y. Dynamic cellular biomechanics in responses to chemotherapeutic drug in hypoxia probed by atomic force spectroscopy. Oncotarget 2021, 12, 1165. [Google Scholar] [CrossRef]
- Taranta, M.; Bizzarri, A.R.; Cannistraro, S. Probing the interaction between p53 and the bacterial protein azurin by single molecule force spectroscopy. J. Mol. Recognit. Interdiscip. J. 2008, 21, 63–70. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, G.; Song, C.; Li, Y.; Qiao, H.; Zhu, P.; Hinterdorfer, P.; Zhang, B.; Tang, J. Single molecular recognition force spectroscopy study of a luteinizing hormone-releasing hormone analogue as a carcinoma target drug. J. Phys. Chem. B 2012, 116, 13331–13337. [Google Scholar] [CrossRef]
- Xiao, H.; Zhang, J.; Xu, Z.; Feng, Y.; Zhang, M.; Liu, J.; Chen, R.; Shen, J.; Wu, J.; Lu, Z. Metformin is a novel suppressor for transforming growth factor (TGF)-β1. Sci. Rep. 2016, 6, 28597. [Google Scholar] [CrossRef]
- Li, M.; Liu, L.; Xiao, X.; Xi, N.; Wang, Y. The dynamic interactions between chemotherapy drugs and plasmid DNA investigated by atomic force microscopy. Sci. China Mater. 2017, 3, 269–278. [Google Scholar] [CrossRef] [Green Version]
- Yun, X.; Tang, M.; Yang, Z.; Wilksch, J.J.; Xiu, P.; Gao, H.; Zhang, F.; Wang, H. Interrogation of drug effects on HeLa cells by exploiting new AFM mechanical biomarkers. RSC Adv. 2017, 7, 43764–43771. [Google Scholar] [CrossRef] [Green Version]
- Song, G.; Tian, F.; Liu, H.; Li, G.; Zheng, P. Pioglitazone inhibits metal cluster transfer of mitoNEET by stabilizing the labile Fe–N bond revealed at single-bond level. J. Phys. Chem. Lett. 2021, 12, 3860–3867. [Google Scholar] [CrossRef]












Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kandapal, S.; Xu, B. Atomic Force Microscopy as a Tool to Study Transport Phenomena in Biological Systems. Processes 2023, 11, 2430. https://doi.org/10.3390/pr11082430
Kandapal S, Xu B. Atomic Force Microscopy as a Tool to Study Transport Phenomena in Biological Systems. Processes. 2023; 11(8):2430. https://doi.org/10.3390/pr11082430
Chicago/Turabian StyleKandapal, Sneha, and Bingqian Xu. 2023. "Atomic Force Microscopy as a Tool to Study Transport Phenomena in Biological Systems" Processes 11, no. 8: 2430. https://doi.org/10.3390/pr11082430
APA StyleKandapal, S., & Xu, B. (2023). Atomic Force Microscopy as a Tool to Study Transport Phenomena in Biological Systems. Processes, 11(8), 2430. https://doi.org/10.3390/pr11082430






